Abstract
A two-hybrid screen was used to identify Saccharomyces cerevisiae genes encoding proteins that interact with MSH2. One gene was found to encode a homologue of Schizosaccharomyces pombe EXO1, a double-stranded DNA-specific 5′–3′ exonuclease. S. cerevisiae EXO1 interacted with both S. cerevisiae and human MSH2 in two-hybrid and coimmunoprecipitation experiments. exo1 mutants showed a mutator phenotype, and epistasis analysis was consistent with EXO1 functioning in the MSH2-dependent mismatch repair pathway. exo1 mutations were lethal in combination with rad27 mutations, and overexpression of EXO1 suppressed both the temperature sensitive and mutator phenotypes of rad27 mutants.
Keywords: cancer, hereditary nonpolyposis colorectal carcinoma, mutagenesis, mutS
Genetic and biochemical studies have indicated eukaryotes contain a mismatch repair (MMR) pathway related to the bacterial MutHLS pathway (reviewed in refs. 1 and 2). However, recent evidence suggests that eukaryotic MMR is more complex. In Saccharomyces cerevisiae there are two MMR pathways that require MSH2, a MutS homologue that recognizes mispaired bases (3, 4). One is a single base substitution mispair pathway that requires a complex of MSH2 and the MutS homologue MSH6 (also called GTBP or p160 in humans) (1–3). There is also an insertion/deletion mispair pathway that requires either a complex of MSH2 and MSH6 or a complex of MSH2 and MSH3, a third MutS homologue (1–3). Additionally, four S. cerevisiae MutL homologues have been identified, PMS1 (PMS2 in humans) and MLH1–MLH3; PMS1 and MLH1 function in MMR and have been shown to form a heterodimer (1, 2).
In vitro studies in Escherichia coli have shown that the excision step of MMR can occur either 5′ to 3′ or 3′ to 5′ of the initiating nick and requires the combination of a helicase (UvrD) and one of three single-stranded DNA exonucleases (Exo I, Exo VII or RecJ) (reviewed in ref. 2). In eukaryotic MMR, proteins involved in excising the mispair have not been identified, although some candidates have been suggested. These include S. cerevisiae RAD27 (RTH1, YKL510), a 5′–3′ exonuclease and flap endonuclease (5), and Schizosaccharomyces pombe EXO1 and its Drosophila homologue Tosca, which are members of the same family of endo- and exonucleases as RAD27 (6, 7).
The importance of determining the mechanism of MMR is underscored by its association with hereditary nonpolyposis colorectal carcinoma (HNPCC) (reviewed in refs. 2 and 8). HNPCC is associated primarily with germ-line mutations in two human MMR genes, MSH2 and MLH1, whereas mutations in other MMR genes are rare (ref. 9; reviewed in refs. 2 and 8). Somatic mutations in MMR genes have been found in some sporadic tumors, suggesting some sporadic cancers could be due to acquired mutations in MMR genes (reviewed in ref. 8, and see ref. 10). However, not all of HNPCC or sporadic cancers with mutator phenotypes can be accounted for by known MMR genes (9, 10). Consequently, there has been interest in identifying additional MMR genes. Here we describe the use of a two-hybrid screen to identify proteins that interact with MSH2 and function in MMR.
MATERIALS AND METHODS
Strains.
Four series of isogenic strains were constructed by disrupting genes of interest using standard techniques. The first series of strains were in the MGD background (RKY2575-MATa, ade2, ura3-52, leu2-3, 112, trp1-289, his3Δ1, lys2-Bgl, hom3-10) with the following differences: RKY2537-MATα; RKY2587 and RKY2663-exo1Δ690::HIS3; RKY2558-msh2::hisG; RKY2588msh2::hisG, exo1Δ690::HIS3. The second series was in the S288c background (RKY2321 MATα, ura3-52, his3Δ200, leu2Δ1) and was RKY2662-exo1Δ690::HIS3. The third series was in the S288c background (RKY2672 MATa, ura3-52, his3Δ200, trp1Δ63, leu2Δ1, ade2Δ1, ade8, lys2-Bgl, hom3-10) and were RKY3044exo1Δ690::HIS3 and RKY2608-rad27::URA3. The fourth series was in the S288C background (RKY2664 MATa, ura3-52, his3Δ200, trp1Δ63 or RKY2666 MATα, ura3-52, his3Δ200, trp1Δ63) and were RKY3057-rad27::URA3 and RKY3056exo1Δ690::HIS3, respectively. The msh2 and rad27 mutations have been described (4, 11).
General Genetic Techniques.
Yeast extract/peptone/dextrose (YPD) media, sporulation media, synthetic drop-out (SD) media, 5-fluoroorotic acid, and canavanine media were as described (11). Mutation rates were calculated using the method of the median and at least five independent colonies as described (11).
exo1Δ690::HIS3 deletion strains were constructed using an adaptation of a published method (12). A disruption construct was generated by amplifying the HIS3 gene present in pPS729 (from P. Silver, Dana–Farber Cancer Institute) by PCR using primers (HIS3 sequences are in lowercase) 22244 (5′-AAAGGAGCTCGAAAAAACTGAAAGGCGTAGAAAGGAATGGGTATCCAAGGTggcctcctctagtactc) and 21964 (5′-CCTCCGATATGAAACGTGCAGTACTTAACTTTTATTTACCTTTATAAACAAATTGGGgcgcgcctcgttcagaatg) and Taq DNA polymerase. The resulting PCR product was used to transform RKY2321 (a S288c strain containing the hisΔ200 allele) resulting in strain RKY2662 in which the DNA sequence encoding amino acids 6–695 of the EXO1 open reading frame (ORF) was replaced with the HIS3 gene. This disruption was introduced into other strains by transformation with a DNA fragment containing the HIS3 gene bordered by several hundred base pairs of EXO1 sequence generated by PCR using primers 22388 (5′-CCGGCCCGAGAAGGAGAAGTA) and 22237 (5′-TGCGGAGAATAAAAGGTTGTGACG) and genomic RKY2662 DNA as template. The correct integration of the exo1Δ690::HIS3 mutation was verified by PCR analysis.
DNA Analysis.
All DNA sequencing was performed using an Applied Biosystems model 373A DNA sequencer (13). Sequence comparisons, alignments, and phylogenetic trees were generated using DNAstar (Madison, WI) software. Percent identity between homologs was determined using the fasta algorithm available over the Internet (http://genome.eerie.fr/bin/align-guess.cgi) and database searches were performed using the blast algorithm (http://www.ncbi.nlm.nih.gov/BLAST/).
Plasmids.
The yMSH2 (y = S. cerevisiae) bait (pRDK371) was constructed by cloning the entire yMSH2 ORF into pEG202. A BamHI/KpnI fragment containing the yMSH2 gene was excised from pEN11 (4) and inserted into pGEM7Zf(+) (Promega). This construct was digested with XhoI to release a fragment containing the entire yMSH2 ORF that was inserted into the SalI site of pEG202 in the orientation giving an in-frame fusion of the yMSH2 ORF to LexA. The hMSH2 (h = human) bait (pRDK372) plasmid was constructed by digesting pMSH22 (14) with BamHI and NcoI to obtain a fragment containing the entire hMSH2 ORF. This fragment was inserted into pEG202 digested with BamHI and NcoI, resulting in an in-frame fusion of LexA to hMSH2. The yMSH2 and hMSH2–LexA fusion constructs were tested for expression of full-length fusion proteins by Western blot analysis using an anti-LexA antibody (15) supplied by P. Silver.
Full-length EXO1 coding sequence was introduced into the “prey” plasmid using a modified version of pJG4-5, pRDK483, which contains AvrII and BssHII sites introduced between the EcoRI and XhoI sites of pJG4-5 (to be described elsewhere). The EXO1 ORF was amplified using Klentaq/Pfu polymerases, pRKD480 (see “Cloning of EXO1” below) as template, and primers 22418 (5′-GCTTATCTCGAGTTATTTACCTTTATAAACAAATTGGGAAAGCAAGGA) and 25430 (5′-CAGGGGCCTAGGCATGGGTATCCAAGGTCTTCTTCC) which contain XhoI and AvrII sites, respectively. The resulting plasmid was named pRDK502.
The EXO1 ORF was subcloned into pET-28b vector (Novagen) as described for construction of pRDK483 (see above) using PCR primers 22370 (5′-GAAGGAGATATACCATGGGTATCCAAGGTCTTCTTCCTCAGTTA) and 22418 (5′-GCTTATCTCGAGTTATTTACCTTTATAAACAAATTGGGAAAGCAAGGA) containing NcoI and XhoI sites, respectively, yielding pRDK379.
Two-Hybrid Techniques.
A two-hybrid screen for yMSH2 interactors was performed essentially as described (16, 17). The LexA–yMSH2 bait plasmid pRDK371 (a HIS3 2μ plasmid) was cotransformed into strain EGY48 along with pSH18–34 (a URA3 2μ plasmid), and the resulting strain was transformed with an acid activation-tagged S. cerevisiae genomic library [constructed by P. Watt (John Radcliffe Hospital, Oxford, U.K.) and obtained from P. Silver] in the 2μ TRP1 plasmid pJG4-5. A total of 500,000–1,000,000 independent transformants were screened for galactose-dependent growth and galactose-dependent expression of LacZ activity. DNA was prepared from positive strains, and the insert was amplified by PCR using primers 19260 (5′-GATACCAGCCTCTTGCTGAGTGGAGATGCCTCC) and 19261 (5′-CGTAAATTTGGCAAGGTAGACAAGCCGACAACC) and partially sequenced. Plasmids of interest were rescued and transformed back into the parent strain (EGY48 + pSH18–34 + pRDK371) for retesting and into strain RFY206 for interaction mating tests (18) for target specificity. The resulting RFY206 strains were mated to an array of EGY48 strains expressing different baits: LexA–Cdc2, LexA–ftz, LexA–bcd2–160, LexA–Max, LexA–hairy, LexA–yMSH2, LexA–hMSH2, LexA alone, and a pEG202 construct in which LexA was removed (17–19). β-Galactosidase assays were performed as described (20); two to three independent cultures were used per experiment, and four independent experiments were performed.
Cloning of EXO1.
Oligonucleotide primers 21955 (5′-TCCGAAGATTCTGACCTCCTC) and 21956 (3′-TCTTCCGAATAGGGCAAAATAC) were used to amplify a 369-bp fragment in the most 5′ region of the DHS1 sequence obtained from GenBank. A 32P-labeled hybridization probe was prepared from this fragment and was used to screen a YEp213 library of cloned S. cerevisiae chromosomal DNA fragments (from B. Futcher, Cold Spring Harbor Laboratory) using standard procedures. The resulting clone used for further analysis which contained the entire EXO1 ORF was named pRDK480.
Preparation of Yeast Extracts.
Extracts were prepared from EGY48 strains containing “bait” and “prey” plasmids essentially as described (21) using 50 ml cultures grown to an A600 of 1 to 1.5 in SD Ura−His−Trp− glucose medium then induced for 4 h in SD Ura−His−Trp− galactose medium.
Immunoprecipitation Experiments.
Hemagglutinin (HA)-tagged preys were immunoprecipitated essentially as described (21). Briefly, 500-μl reactions containing 150 mM NaCl were precleared by incubation with protein G-Sepharose; these protein G-Sepharose beads were saved for later analysis and are referred to as “preclear.” The precleared extracts were immunoprecipitated using 12CA5 antibody and protein G-Sepharose, then analyzed by Western blotting and visualized using anti-LexA antibody and an ECL kit (Amersham). The membranes were then stripped and visualized using 12CA5 antibody.
Overexpression of EXO1 Protein and Exonuclease Assays.
Both pRDK379 and pET-28b were transformed into NovaBlue (DE3) cells (Novagen). Cultures (500 ml) of both transformants were grown at 37°C to OD = 0.4 followed by induction with 1 mM isopropyl β-d-thiogalactoside for 4 h. The cells were harvested and lysed as described (22). A 60% ammonium sulfate fraction was prepared and resuspended to a conductivity equivalent of 0.1 M NaCl in 19 ml of buffer A (20 mM Tris, pH 7.5/0.1 mM EDTA/1 mM DTT/0.5 mM phenylmethylsulfonyl fluoride). A 15 cm × 1 cm PBE94 (Pharmacia) column was equilibrated with buffer A/0.1 M NaCl. After loading, the column was washed with 40 ml of buffer A/0.1 M NaCl and eluted with a 200 ml linear gradient from 0.1–0.5 M NaCl in buffer A at 0.17 ml/min. Protein concentrations were determined using the Bio-Rad protein assay. Exonuclease assays were similar to previously described assays (23, 24). Mixtures (200 μl) contained 20 mM Tris (pH 7.5), 5 mM MgCl2, 1 mM DTT, 500 ng of [3H]-T7 DNA (SA = 3.1 × 107 cpm/pmol) substrate, and 5 μl of each fraction. Following a 10-min incubation at 37°C, the formation of acid soluble nucleotides was measured (24).
RESULTS
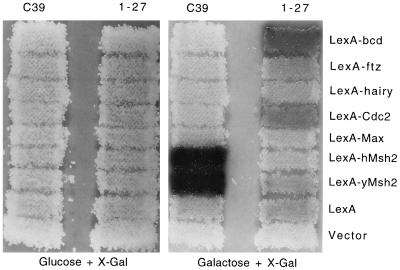
A two-hybrid screen was performed in S. cerevisiae using LexA fused to the entire ORF of yMSH2 as bait and an activation-tagged library of S. cerevisiae genomic DNA as prey. Sixty-five Leu+ clones expressing LacZ were partially sequenced and a subset were examined for target specificity using an interaction mating test (18). One clone, C39, encoded a fusion protein that interacted specifically with both yMSH2 and hMSH2 (Fig. 1). To further quantify this interaction, β-galactosidase assays were performed; the original positive strain expressing C39 in combination with yMSH2 displayed 75 Miller units of β-galactosidase activity, a 125-fold increase over the yMSH2 plus prey vector control (data not shown).
Figure 1.
Interaction mating test showing target specificity of yMSH2 interactor C39. Also shown is the apparently nonspecific weak yMSH2 interactor 1-27, which expresses an activation-tagged C-terminal region of MOT1 beginning at amino acid 1579. The interaction mating test was performed as described (18). Briefly, haploid MATa strains that contain a lacZ reporter plasmid (URA3 vector) and different bait plasmids (HIS3 vector) were streaked in horizontal rows on a Ura−His− plate, and MATα strains that contain galactose-induced yMSH2 interactor plasmids (TRP1 vector) C39 and 1-27 were streaked in vertical columns on a Trp− plate. Both plates were replica plated onto a single yeast extract/peptone/dextrose (YPD) plate to allow mating, replica plated to a Ura−His−Trp− glucose plate to select diploids, then replica plated to Ura−His−Trp− 5-bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) glucose and galactose indicator plates and incubated for 2 days at 30°C (shown). The plasmids contained in each strain tested are indicated at the top of each column and at the right of each row.
Sequence analysis showed that C39 contained a region of the S. cerevisiae DHS1 gene (25). We cloned the DHS1 gene, and sequence analysis (GenBank accession no. U86134; ORF name YORO33c) showed that the published DHS1 sequence contained an error (a 1-bp deletion) and that the full-length DHS1 ORF encodes a predicted protein of 80,171 molecular weight that is homologous to S. cerevisiae DIN7, S. pombe EXO1, and D. melanogaster Tosca (Fig. 2A; refs. 6, 7, and 26) and also shares homology with other members of the XPG/RAD2 family via the two conserved regions designated the N (N-terminal) and I (internal) regions (Fig. 2B). DIN7 is remarkably homologous to full-length DHS1 in the N and I region (67% identity); however, it is a considerably shorter protein (Fig. 2B), and it is slightly less homologous to S. pombe EXO1 than to full-length DHS1 (45% and 43% identity with S. pombe EXO1 in the N and I regions, respectively; also see Fig. 2B). A phylogenetic tree indicates that full-length DHS1 protein and DIN7 are members of the subgroup of the XPG/RAD2 including S. pombe EXO1 and D. melanogaster Tosca (Fig. 2C; refs. 6 and 7). As full-length DHS1 protein is slightly more homologous to S. pombe EXO1 than to DIN7, and for reasons discussed below, we suggest that the DHS1 gene is more appropriately named EXO1.
Figure 2.
Homology among the XPG/RAD2/EXO1 family of endo- and exonucleases. (A) Alignment of the N-terminal conserved regions of S. pombe EXO1, S. cerevisiae EXO1 and DIN7, and D. melanogaster Tosca. (B) Illustration of the sequence relatedness of the XPG/RAD2/EXO1 family of endo- and exonucleases adapted from figure 1 of Szankasi and Smith (7). The highly conserved N and I regions are represented by boxes filled with forward and backward diagonal lines, respectively. (C) Phylogenetic tree of the XPG/RAD2/EXO1 family of endo- and exonucleases generated from alignment of the first 240 amino acids of these proteins. YEN1, a related but divergent protein sequence found during database searches, is also included for clarity.
The C39 clone did not contain the full length EXO1 gene, but instead contained an insert encoding the carboxyl half of EXO1 protein beginning at amino acid 368. Thus the MSH2-interacting region is outside the N and I regions (Fig. 2B) thought to be required for exonuclease activity (6, 27). To confirm that EXO1 interacts with yMSH2, we constructed a prey vector that expressed full-length EXO1 protein and tested its interaction with LexA–yMSH2. The results showed full length EXO1 and LexA–yMSH2 did interact by both two-hybrid criteria (data not shown) and by coimmunoprecipitation experiments (see below).
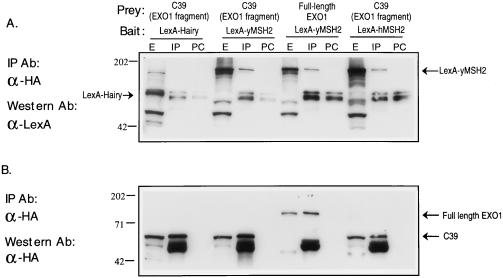
To independently verify the interaction between EXO1 and MSH2, extracts prepared from two-hybrid strains expressing LexA–yMSH2 or LexA–hMSH2 and various HA-tagged preys were analyzed by immunoprecipitation. The results showed that both LexA–yMSH2 and LexA–hMSH2 coimmunoprecipitated with the EXO1 fragment expressed by C39, and LexA–yMSH2 also coimmunoprecipitated with full-length EXO1 (Fig. 3). LexA–Hairy did not coimmunoprecipitate with the EXO1 fragment expressed by C39 (Fig. 3), consistent with the two-hybrid results shown in Fig. 1. In a separate experiment, LexA–yMSH2 did not coimmunoprecipitate with HA-tagged NPL3 while it did with the EXO1 fragment expressed by C39 (data not shown), indicating that LexA–yMSH2 is not nonspecifically “sticky.”
Figure 3.
Immunoprecipitation of proteins expressed in two-hybrid strains. (A) Cell extracts (200 μg) were prepared from the two-hybrid strains expressing LexA-tagged baits and HA-tagged preys. The extracts were precleared with protein-G Sepharose, immunoprecipitated with monoclonal 12CA5 anti-HA antibody and protein-G Sepharose, fractionated by SDS/PAGE, and analyzed by Western blotting with rabbit anti-LexA antiserum. The two bands seen in every IP and PC lane are proteins nonspecifically precipitated by protein-G Sepharose. (B) To ensure appropriate expression and immunoprecipitation of HA-tagged preys, the immunoblot was stripped and reprobed with 12CA5 antibody. E, extract; IP, immunoprecipitation; PC, proteins eluted from protein G-Sepharose used to preclear.
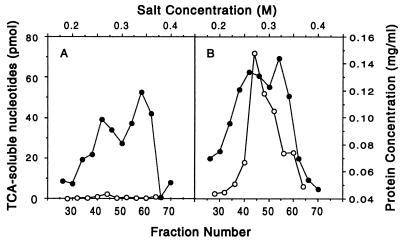
To confirm that S. cerevisiae EXO1 is an exonuclease, it was overproduced in E. coli under control of a T7 promoter. Induction by isopropyl β-d-thiogalactoside resulted in the expression of a 80-kDa polypeptide, the predicted size of EXO1. Fractionation of extracts prepared from induced cells by chromatography on a PBE94 column showed that when the EXO1 gene was present there was an ≈50-fold induction of exonuclease activity compared with the vector control (Fig. 4). This exonuclease had a 3- to 5-fold preference for double-stranded vs. single-stranded DNA templates (data not shown). Further purification and characterization of EXO1 will be published elsewhere.
Figure 4.
Expression of EXO1 in E. coli results in induction of an exonuclease activity. Extracts from E. coli expressing either pET28b vector alone (A) or RDK480 (B) were chromatographed on a PBE94 column. Fractions were analyzed for protein (•), exonuclease activity (○), and conductivity (NaCl, top scale). Activity is pmol acid-soluble nucleotides released per 5 μl of each fraction.
S. pombe EXO1 has been proposed to play a role in recombination and to possibly function in either short patch or long patch MMR (7). To test the hypothesis that S. cerevisiae EXO1 functions in MMR, haploid exo1 mutants were tested for defects in DNA repair. exo1 mutants did not display any growth defects, nor were they sensitive to UV-irradiation or methylmethanesulfonate (data not shown). In plate assays exo1 mutants showed increased accumulation of canavanine resistant (Canr) mutations and reversion of hom3–10 mutations relative to wild-type strains, although this increase was lower than that seen in msh2 mutants (data not shown). Strains that contained both exo1 mutations and mutations in msh2, mlh1, or pms1 showed the same increase in spontaneous mutation frequency as the msh2, mlh1, or pms1 single mutants (data not shown). These results suggest that msh2, mlh1, and pms1 mutations are all epistatic to the exo1 mutation with regard to spontaneous mutation frequency, and that EXO1 functions in the same pathway of DNA repair as these genes.
In fluctuation tests (Table 1), exo1 mutants showed moderate increases in the rate of accumulation of Canr mutations (8-fold) and the rate of reversion of hom3-10 mutations (6-fold) relative to the wild-type strain. These effects contrast with the larger increases in the rate of accumulation of Canr mutations (25-fold) and the rate of reversion of hom3-10 mutations (850-fold) seen in the msh2 mutants (Table 1). The exo1,msh2 double mutant had mutation rates similar to the msh2 single mutant (Table 1), consistent with MSH2 being epistatic to EXO1.
Table 1.
Mutation rates in exo1 mutants
| Strain | Mutation rate
|
|
|---|---|---|
| Canr | Hom+ | |
| Wild type | ||
| RKY2575 | 8.4 × 10−8 | 7.7 × 10−9 |
| RKY2575 | 2.2 × 10−7 | 1.2 × 10−8 |
| RKY2537 | 1.3 × 10−7 (1) | 8.2 × 10−9 (1) |
| exo1 | ||
| RKY2587 | 8.1 × 10−7 | 4.0 × 10−8 |
| RKY2587 | 1.3 × 10−6 | 2.4 × 10−8 |
| RKY2663 | 1.2 × 10−6 (7.6) | 1.1 × 10−7 (6.2) |
| msh2 | ||
| RKY2558 | 3.6 × 10−6 (25) | 7.9 × 10−6 (850) |
| exo1, msh2 | ||
| RKY2588 | 3.9 × 10−6 (27) | 7.2 × 10−6 (774) |
The rate of accumulation of Canr mutants and Hom+ revertants per cell division is given. For wild-type and exo1 strains, data are shown from three experiments employing two isolates of the same genotype. For the msh2 and exo1, msh2 strains, data are from one experiment. The numbers in parentheses represent the fold induction relative to the wild-type value; for wild type and exo1, this number is the average of all three experiments.
It has been suggested that RAD27 (RTH1, YKL510), a flap endonuclease and 5′ to 3′ exonuclease, may function in MMR (5, 28). To determine whether EXO1 and RAD27 might play similar roles in the cell, we attempted to construct exo1 rad27 double mutants by crossing an exo1::HIS3 mutant to a rad27::URA3 mutant (data not shown). A high proportion of dead spores or microcolonies were observed (12.5% and 17.3%, respectively, of 26 tetrads analyzed) compared with the exo1 × wild-type cross (0% inviable spores) or the rad27 × wild-type cross (5.5% inviable spores). The percentage of dead spores plus microcolonies (30%) is very close to the expected percentage of dead spores (25%) assuming the combination of exo1 and rad27 mutations is lethal. After extended incubation it was possible to genotype the microcolonies formed in the exo1 × rad27 cross; these represented all the His+Ura+ (exo1 rad27) clones recovered. These data show the combination of exo1 and rad27 mutations is effectively lethal at 30°C.
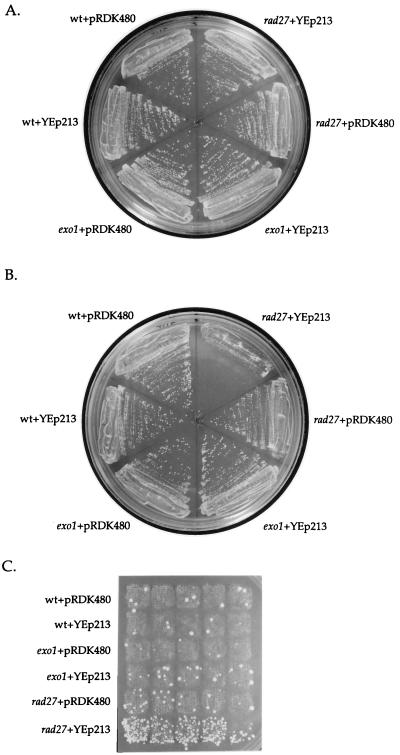
The synthetic lethality between exo1 and rad27 suggested that overexpressing EXO1 might suppress the phenotypes of rad27 mutants (29). Expression of EXO1 on a 2μ vector (pRDK480) suppressed the temperature sensitive phenotype of rad27 mutants at 37°C (Fig. 5 A and B). We then tested whether pRDK480 would suppress the mutator phenotype observed in rad27 mutants (28). Patch assays (Fig. 5C) showed that pRDK480 suppressed the accumulation of Canr mutations in both exo1 and rad27 mutants. This result was further quantified by fluctuation analysis (Table 2). Experiments using a GAL-induced EXO1 overexpressor gave essentially the same results (data not shown), indicating these results are not due to overexpression of another ORF on the EXO1 genomic clone.
Figure 5.
Overexpression of EXO1 suppresses both the temperature sensitive and mutator phenotypes of rad27(rth1) mutants. Isogenic wild-type (wt) (RKY2672), exo1 (RKY3044), or rad27 (RKY2608) strains were transformed with either YEp213 (a 2μ LEU2-marked plasmid) or pRDK480 (YEp213 with a yeast genomic insert containing the EXO1 gene) and the resulting strains streaked out on SD Leu− plates and grown for 48 h at either 30°C (A) or 37°C (B), or they were patched onto a SD Leu− plate, grown at 30°C for 2 days, then replica plated to a SD Leu− canavanine plate and grown at 30°C for 3 days to detect the presence of Canr mutations, which appear as papillae (C).
Table 2.
Overexpression of EXO1 decreases the rad27 mutation rate
| Strain | Rate
|
|
|---|---|---|
| Canr | Lys+ | |
| Wild type + YEp213 | 3.5 × 10−7 (1) | 1.1 × 10−8 (1) |
| Wild type + pRDK480 | 5.1 × 10−7 (1.5) | 1.4 × 10−8 (1.3) |
| exo1 + YEp213 | 1.7 × 10−6 (4.9) | 2.3 × 10−8 (2.1) |
| exo1 + pRDK480 | 6.1 × 10−7 (1.7) | 1.3 × 10−8 (1.2) |
| rad27 + YEp213 | 1.5 × 10−5 (43) | 3.0 × 10−7 (27) |
| rad27 + pRDK480 | 1.5 × 10−6 (4.3) | 3.7 × 10−8 (3.4) |
Mutation rates per cell division are given. SD Leu− medium was used to ensure plasmid maintenance. The isogenic strains were wild type (RKY2672), exo1 (RKY3044), and rad27 (RKY2608) transformed with either YEp213 (a 2μ LEU-marked plasmid) or pRDK480 (YEp213 with a yeast genomic insert containing the EXO1 gene). The numbers in parentheses represent the fold induction relative to wild type + YEp213.
DISCUSSION
The interaction of S. cerevisiae EXO1 with MSH2 (Figs. 1 and 3), the mutator phenotype of exo1 mutants and the epistasis relationship between EXO1 and MSH2, PMS1 and MLH1 (Table 1 and data not shown) suggest that EXO1 functions as a 5′–3′ exonuclease in the MSH2 pathway of MMR. In support of the view that EXO1 encodes such a nuclease are the observations that overproduction of EXO1 protein results in overproduction of a exonuclease (Fig. 4), that this exonuclease copurifies with the EXO1 protein (data not shown), and that exo1 mutants are missing a previously described 5′ to 3′ double-stranded DNA-specific exonuclease (27).
Biochemical analysis of MMR in E. coli has shown that the excision reaction involves redundant 5′ to 3′ and 3′ to 5′ exonucleases (reviewed in ref. 2). The observation of bidirectional excision in eukaryotic in vitro MMR systems suggests a similar redundancy of exonucleases (2). S. cerevisiae exo1 mutants show a moderate mutator phenotype that is significantly lower than that caused by mutations in MSH2, MLH1, and PMS1 (Table 1; refs. 1 and 3). Additionally, the mutations that arise in exo1 mutants are single base deletion and substitution mutations similar to those seen in msh2 mutants (ref. 3 and data not shown). These results are consistent with eukaryotic MMR employing redundant exonucleases. Eukaryotic 3′–5′ exonucleases involved in MMR remain to be identified. Mutations in Ustilago maydis REC1, which encodes a 3′–5′ exonuclease, cause a mutator phenotype (30), raising the possibility that an S. cerevisiae counterpart of REC1 could function in MMR. One such candidate is the S. cerevisiae RAD17 gene, which encodes a REC1 homolog (31). The remarkable homology between S. cerevisiae EXO1 and DIN7 (Fig. 2) makes DIN7 another candidate for a exonuclease which functions in MMR. However, preliminary experiments have indicated that neither rad17 nor din7 mutants display a mutator phenotype and that the combination of either exo1 and din7 mutations or exo1 and rad17 mutations does not appear to have synergistic effects on spontaneous mutation frequency (D.X.T. and R.D.K., unpublished data).
The RAD27(RTH1) gene encodes the S. cerevisiae homologue of FEN-1, an endo/exonuclease required for processing Okazaki fragments and proposed to be involved in processing branched DNA structures formed by various DNA repair pathways (reviewed in ref. 28). It has been suggested that RAD27 functions in the MSH2 pathway of MMR (5, 28); however, recent data has indicated that the major role of RAD27 is in DNA replication fidelity and that it has little, if any, role in MMR (11). The rad27 mutator phenotype instead appears to result from the failure to process Okazaki fragments, resulting primarily in duplication mutations (11).
The observation that rad27 and exo1 mutations are lethal in combination with each other can be interpreted in two ways. One is that RAD27 and EXO1 encode redundant functions in an essential pathway like the processing of Okazaki fragments. Alternately, mutations in one gene could cause DNA damage repair of which requires the action of the other gene product (32). Overexpression of EXO1 suppresses both the temperature sensitive and the spontaneous mutator phenotype of rad27 mutants (Fig. 5 and Table 2) indicating that EXO1 can substitute for RAD27’s function in processing Okazaki fragments. Alternately, overexpression of EXO1 could increase the ability of cells to repair the kinds of lesions generated in rad27 mutants. Although EXO1 is required for viability in the absence of RAD27, EXO1 does not appear to normally play a major role in the processing of Okazaki fragments; unlike rad27 mutants, exo1 mutants do not show growth defects, methylmethanesulfonate sensitivity, or the accumulation of duplication mutations (ref. 11 and unpublished data).
The observation that EXO1 interacts with S. cerevisiae MSH2 and appears to act in MSH2-dependent MMR has important implications for the mechanism of eukaryotic MMR. First, this interaction suggests that MMR involves the formation of a complex containing components required for many aspects of MMR. MSH2, which functions in mismatch recognition, is a reasonable candidate for an important protein in the organization of such a complex (4). Second, the use of a double-stranded DNA-specific exonuclease in the excision step in eukaryotic MMR is different from what is observed in E. coli, where excision requires the combined action of a helicase and a single-stranded DNA-specific exonuclease (2).
The D. melanogaster EXO1 homologue Tosca is expressed specifically in the female germ line during meiosis and in early embryogenesis, suggesting a role for Tosca in recombination and MMR (6). Additionally, S. cerevisiae exo1 mutants have a defect in an in vitro recombination assay and in in vivo recombination between direct repeats (27), a phenotype observed in msh2 and msh3 mutants (33). Given that MMR and recombination are highly interrelated processes (reviewed in ref. 34) and that MSH2 has recently been shown to bind to Holliday junctions (35), it seems reasonable to propose that EXO1 plays roles in both MMR and recombination.
Acknowledgments
We thank P. Silver, R. Brent, and R. Finley for reagents and advice; B. Futcher for the yeast genomic DNA library; D. Carroll for information about D. melanogaster EXO1 homologue; P. Noirot for [3H]T7 DNA; and G. Marsischky for helpful discussions. This work was supported by National Institutes of Health Grants GM50006 to R.D.K. and Core Grants CA06516 and AI28691 to the Dana–Farber Cancer Institute.
ABBREVIATIONS
- SD medium
synthetic drop-out medium
- MMR
mismatch repair
- HA
hemagglutinin
- y
Saccharomyces cerevisiae
- h
human
- Canr
canavanine resistant
References
- 1.Crouse G F. In: DNA Damage and Repair. Nickoloff J A, Hoekstra M F, editors. Vol. 1. Clifton, NJ: Humana; 1997. , in press. [Google Scholar]
- 2.Kolodner R. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 3.Marsischky G T, Filosi N, Kane M F, Kolodner R. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 4.Alani E, Chi N-W, Kolodner R D. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 5.Johnson R E, Gopala K K, Prakash L, Prakash S. Science. 1995;269:238–240. doi: 10.1126/science.7618086. [DOI] [PubMed] [Google Scholar]
- 6.Digilio F A, Pannuti A, Lucchesi J C, Furia M, Polito L C. Dev Biol. 1996;178:90–100. doi: 10.1006/dbio.1996.0200. [DOI] [PubMed] [Google Scholar]
- 7.Szankasi P, Smith G R. Science. 1995;267:1166–1169. doi: 10.1126/science.7855597. [DOI] [PubMed] [Google Scholar]
- 8.Kolodner R D. Trends Biochem. 1995;20:397–402. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Parsons R, Papadopoulos N, Nicolaides N C, Lynch H T, Watson P, Jass J R, Dunlop M, Wyllie A, Peltomaki P, de la Chapelle A, Hamilton S R, Vogelstein B, Kinzler K W. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 10.Moslein G, Tester D J, Lindor N M, Honchel R, Cunningham J M, French A J, Halling K C, Schawb M, Goretzki P, Thibodeau S N. Hum Mol Genet. 1996;5:1245–1252. doi: 10.1093/hmg/5.9.1245. [DOI] [PubMed] [Google Scholar]
- 11.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 12.Baudin A, Ozier-Kalogeropoulos, Denouel A, Lacroute F, Cullin C. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolodner R D, Hall N R, Lipford J R, Kane M F, Morrison P, Finan P J, Burn J, Chapman P, Earabino C, Merchant E, Bishop D T. Cancer Res. 1995;55:242–248. [PubMed] [Google Scholar]
- 14.Fishel R A, Lescoe M K, Rao M R S, Copland N, Jenkins N, Garber J, Kane M, Kolodner R. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 15.Golemis E A, Brent R. Mol Cell Biol. 1992;12:3006–3014. doi: 10.1128/mcb.12.7.3006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gyuris J, Golemis E, Chertkov H, Brent R. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 17.Zervos A S, Gyuris J, Brent R. Cell. 1993;72:223–232. doi: 10.1016/0092-8674(93)90662-a. [DOI] [PubMed] [Google Scholar]
- 18.Finley R L, Brent R. Proc Natl Acad Sci USA. 1994;91:12980–12984. doi: 10.1073/pnas.91.26.12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paroush Z, Finley R L, Kidd T, Wainwright S M, Ingham P W, Brent R, Ish-Horowicz D. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 20.Xiao W, Sampson L. Nucleic Acids Res. 1992;20:3599–3606. doi: 10.1093/nar/20.14.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elion E A, Satterberg B, Kranz J E. Mol Biol Cell. 1993;4:495–510. doi: 10.1091/mbc.4.5.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall S D, Kane M F, Kolodner R D. J Bacteriol. 1993;175:277–287. doi: 10.1128/jb.175.1.277-287.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson A W, Kolodner R D. J Biol Chem. 1991;266:14046–14054. [PubMed] [Google Scholar]
- 24.Szankasi P, Smith G R. J Biol Chem. 1992;267:3014–3023. [PubMed] [Google Scholar]
- 25.Lee S L, Shimizu J, Yoda K, Yamasaki M. Biosci Biotechnol Biochem. 1994;58:391–395. doi: 10.1271/bbb.58.391. [DOI] [PubMed] [Google Scholar]
- 26.Mieczkowski P, Fikus M, Ciesla Z. Mol Gen Genet. 1997;253:655–665. doi: 10.1007/s004380050369. [DOI] [PubMed] [Google Scholar]
- 27.Fiorentini P, Huang K N, Tishkoff D X, Kolodner R D, Symington L S. Mol Cell Biol. 1997;17:2764–2773. doi: 10.1128/mcb.17.5.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lieber M R. BioEssays. 1996;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 29.Reagan M S, Pittenger C, Siede W, Friedberg E C. J Bacteriol. 1995;177:364–371. doi: 10.1128/jb.177.2.364-371.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Onel K, Thelen M P, Ferguson D O, Bennett R L, Holloman W K. Mol Cell Biol. 1995;15:5329–5338. doi: 10.1128/mcb.15.10.5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siede W, Nusspaumer G, Portillo V, Rodriguez R, Friedberg E C. Nucleic Acids Res. 1996;24:1669–1675. doi: 10.1093/nar/24.9.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morrison A, Johnson A L, Johnston L H, Sugino A. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saparbaev M, Prakash L, Prakash S. Genetics. 1996;142:727–736. doi: 10.1093/genetics/142.3.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stahl F. Cell. 1996;87:965–968. doi: 10.1016/s0092-8674(00)81791-2. [DOI] [PubMed] [Google Scholar]
- 35.Alani E, Lee S, Kane M F, Griffith J G, Kolodner R. J Mol Biol. 1997;265:289–301. doi: 10.1006/jmbi.1996.0743. [DOI] [PubMed] [Google Scholar]