Abstract
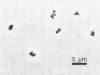
A moderately halophilic methanogenic bacterium was enriched with trimethylamine and isolated from the sediment of a solar salt pond (total dissolved solids of pond water, 250 g/liter; pH 7.5). The isolate (strain SF1, DSM 3243) was an irregular coccus which stained gram negative, with a diameter of 1 μm and a thin monolayered cell wall. The organism grew singly, in pairs, and in irregular clumps. Colonies were tannish yellow, circular, with entire edges, and about 1 mm in diameter within 1 week. Only methylamines or methanol was used for growth and methanogenesis. Most rapid growth (doubling time, 10.2 h) occurred at a temperature of 37°C and a pH of 7.4. The optimum NaCl concentration was 2.1 M. Yeast extract or rumen fluid was required. The isolate was lysed by sodium dodecyl sulfate (0.1 g/liter) and was sensitive to chloramphenicol. The G+C content of the DNA was 41 (±1) mol%.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baresi L., Mah R. A., Ward D. M., Kaplan I. R. Methanogenesis from acetate: enrichment studies. Appl Environ Microbiol. 1978 Jul;36(1):186–197. doi: 10.1128/aem.36.1.186-197.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway de Macario E., Macario A. J., Wolin M. J. Specific antisera and immunological procedures for characterization of methanogenic bacteria. J Bacteriol. 1982 Jan;149(1):320–328. doi: 10.1128/jb.149.1.320-328.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson T. J., Mah R. A. Isolation and characterization of an h(2)-oxidizing thermophilic methanogen. Appl Environ Microbiol. 1983 Jan;45(1):265–274. doi: 10.1128/aem.45.1.265-274.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig K., Gottschalk G. Methanogenesis from Choline by a Coculture of Desulfovibrio sp. and Methanosarcina barkeri. Appl Environ Microbiol. 1983 Jan;45(1):161–168. doi: 10.1128/aem.45.1.161-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- KELLENBERGER E., RYTER A., SECHAUD J. Electron microscope study of DNA-containing plasms. II. Vegetative and mature phage DNA as compared with normal bacterial nucleoids in different physiological states. J Biophys Biochem Cytol. 1958 Nov 25;4(6):671–678. doi: 10.1083/jcb.4.6.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M., Klug M. J., Lovley D. R. Metabolism of acetate, methanol, and methylated amines in intertidal sediments of lowes cove, maine. Appl Environ Microbiol. 1983 Jun;45(6):1848–1853. doi: 10.1128/aem.45.6.1848-1853.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King G. M. Metabolism of trimethylamine, choline, and glycine betaine by sulfate-reducing and methanogenic bacteria in marine sediments. Appl Environ Microbiol. 1984 Oct;48(4):719–725. doi: 10.1128/aem.48.4.719-725.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumann E., Hippe H., Gottschalk G. Betaine: New Oxidant in the Stickland Reaction and Methanogenesis from Betaine and l-Alanine by a Clostridium sporogenes-Methanosarcina barkeri Coculture. Appl Environ Microbiol. 1983 Feb;45(2):474–483. doi: 10.1128/aem.45.2.474-483.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oremland R. S., Polcin S. Methanogenesis and sulfate reduction: competitive and noncompetitive substrates in estuarine sediments. Appl Environ Microbiol. 1982 Dec;44(6):1270–1276. doi: 10.1128/aem.44.6.1270-1276.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J. F., Boone D. R. Analytical determination of the buoyant density of DNA in acrylamide gels after preparative CsCl gradient centrifugation. FEBS Lett. 1973 Dec 1;37(2):321–324. doi: 10.1016/0014-5793(73)80487-9. [DOI] [PubMed] [Google Scholar]
- SCHILDKRAUT C. L., MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its buoyant density in CsCl. J Mol Biol. 1962 Jun;4:430–443. doi: 10.1016/s0022-2836(62)80100-4. [DOI] [PubMed] [Google Scholar]
- Sowers K. R., Ferry J. G. Isolation and Characterization of a Methylotrophic Marine Methanogen, Methanococcoides methylutens gen. nov., sp. nov. Appl Environ Microbiol. 1983 Feb;45(2):684–690. doi: 10.1128/aem.45.2.684-690.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TARR H. L. Microbiological deterioration of fish post mortem, its detection and control. Bacteriol Rev. 1954 Mar;18(1):1–15. doi: 10.1128/br.18.1.1-15.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Magrum L. J., Fox G. E. Archaebacteria. J Mol Evol. 1978 Aug 2;11(3):245–251. doi: 10.1007/BF01734485. [DOI] [PubMed] [Google Scholar]
- Yancey P. H., Clark M. E., Hand S. C., Bowlus R. D., Somero G. N. Living with water stress: evolution of osmolyte systems. Science. 1982 Sep 24;217(4566):1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]