Abstract
Members of the POU-homeodomain gene family encode transcriptional regulatory molecules that play important roles in terminal differentiation of many organ systems. Sperm-1 (Sprm-1) is a POU domain factor that is exclusively expressed in the differentiating male germ cell. We show here that the Sprm-1 protein is expressed in the haploid spermatid and that 129/Sv Sprm-1(−/−) mice are subfertile when compared with wild-type or heterozygous littermates yet exhibit normal testicular morphology and produce normal numbers of mobile spermatozoa. Our data suggest that the Sprm-1 protein plays a discrete regulatory function in the haploid spermatid, which is required for the optimal function, but not the terminal differentiation, of the male germ cell.
It is estimated that 15% of all married couples in the United States experience fertility problems, of which half of the cases are thought to result from infertility in the male (1). The diagnosis in 40–60% of cases of male infertility is idiopathic infertility, which, as the name implies, has an enigmatic etiology. Many of these males exhibit oligozoospermia, which is characterized by a reduction of sperm concentration in the ejaculate from greater than 50 million per ml to less than 10–20 million sperm per ml. But many still exhibit relatively normal concentrations of mobile spermatozoa.
Spermatogenesis is a complex cellular differentiation process whereby a single diploid cell gives rise to thousands of highly specialized haploid spermatozoa (2, 3). The stem cells of the testis arise by invasion of the embryonic genital ridges by migrating primordial germ cells. These stem cells, known as type A spermatogonia, proliferate either to self-renew or to generate type B spermatogonia. Puberty is initiated by signals from the hypothalamic-pituitary axis, which direct the progression of type B spermatogonia to primary spermatocytes. The final cellular division in this process is the meiotic reduction of secondary spermatocytes that generates haploid spermatids. These spermatids exist as a syncytial bundle that undergoes dramatic morphological and genetic changes to produce functional, mobile spermatozoa in a process known as spermiogenesis. During this entire differentiation process germ cells maintain intimate contact with the supporting Sertoli cells, which, in response to hormonal stimuli from the pituitary and Leydig cells, provide the appropriate environment for germ cell maturation.
It was thought previously that a surge of RNA synthesis immediately before the meiotic reduction, along with RNA storage and translational regulation, provided sufficient resources during spermiogenesis to obviate the need for de novo haploid gene transcription (4). However, advances in cloning strategies have led to the identification of many genes that are not transcribed until after the meiotic reduction (5). Indeed, it has been shown recently that many haploid-expressed genes are regulated by the cyclic AMP-responsive element modulator protein CREM (6, 7).
Results of the analyses of mice carrying both targeted mutations and natural mutations of POU domain factors offer strong evidence that this subclass of homeodomain genes has evolved to serve functions in terminal cellular differentiation in many regions of the developing neuroendocrine system. Recent studies using gene disruption techniques have illustrated that, in addition to the described roles of Pit-1 in mouse and human pituitary development and Unc-86 in Caenorhabditis elegans neural development, many other POU domain factors play critical roles in cellular differentiation. Targeted mutations in Brn-2, Brn-3.1, Brn-3.2, and Tst-1/Oct-6/SCIP result in a blockage of cellular differentiation in the hypothalamus, the hair cells of the inner ear, the retinal ganglion cells, and in myelinating Schwann cells, respectively (8–13).
We have previously identified an Oct-3/4-like POU factor known as Sprm-1, which is specifically expressed in the developing male germ cell immediately before the first meiotic division (14). We show here that although Sprm-1 transcripts are detected before the meiotic reduction, the Sprm-1 protein is seen predominantly in the post-meiotic spermatid. Because of this unique and specific expression pattern of Sprm-1, along with the classically defined importance of other POU domain proteins in cellular differentiation, it is a good candidate for the study of transcriptional regulation in the haploid germ cell. To investigate the role of Sprm-1 in the development of the male germ cell we have generated mice containing a targeted disruption of the Sprm-1 gene. We show that mutant mice exhibit normal testicular morphology and produce normal numbers of sperm yet display subnormal fertility, indicating that spermatozoa produced in the absence of functional Sprm-1 are functionally compromised.
MATERIALS AND METHODS
Immunohistochemical Analysis.
Rabbit polyclonal antibodies were raised against the NH2 terminus (amino acids 1–116), the COOH terminus (amino acids 263–335), and to the holo-Sprm-1 (amino acids 1–335) proteins. Animals were immunized with glutathione S-transferase fusion proteins that were purified over glutathione agarose (Sigma) or by preparative SDS/PAGE. Rabbits were injected with the proteins three times at regular intervals (4 weeks between each injection). Animals were boosted with cleaved protein that was passed over a glutathione agarose column to remove contaminating anti-glutathione S-transferase antibodies. Testes-1 protein was detected using a polyclonal antibody raised to bacculovirus-expressed holo-Tst-1 protein as previously described (15).
Immunohistochemical detection was performed on slide-mounted formalin fixed frozen for Sprm-1, or Bouin’s fixed paraffin embedded sections for the Tst-1 staining. Prepared sections were blocked with 10% goat serum in PBS + Tween 20 (0.1%) (PBT) for 30 min at room temperature and subsequently probed with diluted primary antibody (1:500 for Sprm-1 and 1:1,000 for Tst-1) in PBT + 2% BSA at 4°C overnight in a humidified chamber. Sections were washed and probed with an anti-rabbit horseradish peroxidase-coupled secondary antibody. Peroxidase activity was detected with 3′,3′-diaminobenzidine tetrahydrochloride dihydrate (DAB).
Construction of Sprm-1 Targeting Vector and Generation of Germ-Line Transmitting Mice.
The entire rat Sprm-1 cDNA was random-primed with [α-32P]dCTP and used to probe 106 plaques from a J1 129/Sv mouse genomic library (Stratagene l Fix-II vector). The 5′ flanking region comprising an 8-kb NotI–NcoI fragment and a 3-kb PstI 3′ flanking fragment were subcloned into the corresponding cloning sites of the neomycin-containing vector pBM2.0, where the expression of the neomycin gene is driven by the phosphoglycerate kinase promoter. This created a deletion of the entire coding region of Sprm-1. The flanked phosphoglycerate kinase-neomycin cassette was subcloned into a herpes simplex virus thymidine kinase plasmid, resulting in the positioning of thymidine kinase genes at both ends of the homologous Sprm-1-flanking DNA regions. The J1 embryonic stem cell line (kindly provided by En Li, Whitehead Institute for Biomedical Research, Cambridge, MA), was cultured on mitomycin C-treated neomycin-resistant mouse embryonic fibroblasts and grown in the presence of DMEM high-glucose media containing 15% fetal calf serum (HyClone) and supplemented with 1,000 units/ml of exogenously added leukemia inhibitory factor (ESGRO, GIBCO/BRL). Targeting vector DNA was linearized (25 μg) and electroporated into 2 × 107 embryonic stem cells in 0.8 ml of electroporation buffer at 250 V and 500 μF using a genepulser (Bio-Rad). Cells were grown for 7–9 days in 250 mg/ml of neomycin and 2 mM gancyclovir (Syntex), and 155 double drug-selected clones were grown for 3 additional days. All clones were frozen and their genomic DNA isolated for Southern blot analysis.
Homologous recombinant cell lines were identified by Southern blot analysis using the 3′ external probe (B), an 800-bp HindIII fragment, which hybridizes to an 8-kb wild-type allele and to a 6-kb recombinant allele after a StuI/EcoRV genomic DNA double-digestion. Recombination on the 5′ side of the gene was verified using a 1.1-kb SmaI fragment (probe A) that hybridizes to a 14-kb wild allele and/or to a 11-kb recombinant allele after a SmaI/SphI double-digestion.
Two embryonic stem cell lines, which were positive for homologous recombination at the Sprm-1 locus, were microinjected into e3.5 C57BL/6 blastocysts, which were transferred to pseudopregnant females. Chimeric male mice were mated to C57BL/6 females, and germ-line transmission was recognized by agouti coat color in the progeny. Mice generated from the recombinant cell line were genotyped by PCR using internal primers for the neomycin gene and to the Sprm-1 gene in the same reaction.
Morphology.
Tissue samples were fixed in either Bouin’s fixative or in 4% glutaraldehyde. For light microscopy analysis Bouin’s fixed samples were embedded in paraffin wax and sectioned. Five-micrometer sections were mounted onto polyfrost(+) slides and dehydrated. Tissues were subsequently deparaffinized and stained with either hematoxylin and eosin for nuclear and cytoplasmic analysis or with bisbenzamide (Hoechst stain) for specific nuclear visualization. Slides were analyzed on a Zeiss Axioskop-50 microscope using either Nomarski or phase-contrast optics and were photographed with Kodak Ectachrome 160T (tungsten) film. In addition to our analysis of Sprm-1(−/−) testis we have conducted an extensive morphological analysis of other tissues, including brain, skin, muscle, salivary glands, lymph nodes, thymus, thyroid, heart, lung, stomach, bone, spinal cord, pancreas, intestine, kidney, adrenal, and ovaries. All tissues were found to have normal morphology. Additionally, blood samples from 20 mice, including six homozygous mutants, were analyzed for white cell content and chemistry and found to be normal. For electron microscopy, glutaraldehyde-fixed tissues were embedded in plastic and processed into thin < 1-μm sections. Sections were mounted onto grids and imaged on an electron microscope.
Analysis of Fertility.
Reproductive capacity of 129/Sv Sprm-1(−/−) males was assayed by cohabitation with Sprm-1(+/+), (+/−), or (−/−) females. Briefly, sets of 10 males of each genotype were mated with various females for 2 months. During this time the mating pairs were observed for sexual activity. After 2 months, the females were removed to holding cages and isolated for 21 days to allow them to birth any pups they were carrying. After this waiting period the females were placed with males again except that the females that previously were with Sprm-1 mutants now were mated to wild-type or heterozygous males and vice versa. The mating period for the entire experiment was 6 months. For the analysis of the results we counted the total number of litters sired during the entire mating period, regardless of the size or viability of the litter, and averaged the number of litters in a 2-month period.
In Vitro Fertilization.
CB6F1 females were super-ovulated by injection with 5 units of pregnant mare serum followed, after 48 hr, by 5 units of human chorionic gonadotropin. Eggs were isolated from the oviducts and cumulus cells removed by treatment with 15 mg/ml hyaluronidase in M2 (pH 7.4) medium. Sperm was isolated from the vas deferens and caudal epididymis of 129/Sv Sprm-1(−/−), (+/−), or (+/+) males and capacitated in Brinster’s medium (GIBCO/BRL) for 30 min to 1 hr in an incubator at 37°C and 5% CO2. Sperm from each animal was quantitized by counting three separate samples in a hemocytometer and averaging the values. Serial dilutions were made to equalize the sperm concentrations between the animals. The final concentrations of spermatozoa were 5 × 106, 5 × 105, and 1 × 105 sperm/ml in Brinster’s medium. Twenty microliters of each sperm preparation was added to a 20-μl microdroplet containing 10–15 eggs in M16 medium (pH 7.4). Eggs were exposed to sperm for 3 hr, after which they were washed twice and transferred to fresh M16 media where they were cultured at 37°C and 5% CO2 for 3 days under equilibrated paraffin oil. Embryos were scored after 3 days as either fertilized, unfertilized, or parthenogenetic based on their morphology.
Northern Blot Analysis.
Poly(A)+ RNA from 129/Sv Sprm-1(+/−) or Sprm-1(−/−) testes (2–4 μg) were denatured and fractionated on 0.9% agarose gels, and transferred to bio-trans(+) nylon membranes (ICN) in 20× SSC, as described previously. The membranes were baked and hybridized with 2 × 106 cpm of random primer-labeled probe per ml of hybridization solution (5× standard saline phosphate/EDTA, 5× Denhardt’s solution, 50% formamide, 100 μg/ml sonicated salmon sperm DNA, and 0.1% SDS). All probes used were generated by reverse transcriptase-PCR on wild-type testis poly(A)+ RNA using specific oligonucleotides. Amplified products were separated on 1.5% agarose gels, and bands of the appropriate size were purified and 32P random primer labeled. Hybridized blots were washed down to 0.1× SSC, 0.1% SDS, and 45°C, and exposed to autoradiographic film overnight.
Zoo Blot.
DNA from human, hamster, mouse, and Drosophila were digested to completion with HindIII, EcoRI, or BamHI restriction endonucleases, separated by agarose gel electrophoresis, then transferred to a nylon membrane. The blot was hybridized with a 32P-labeled probe to the entire rat Sprm-1 cDNA under low-stringency conditions, and subsequently washed at 0.2× SSC, 0.1% SDS, and 42°C, and exposed overnight to autoradiographic film.
RESULTS
Protein Expression of POU Domain Factors in the Testes.
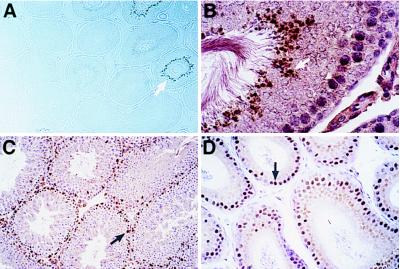
Our previous in situ hybridization analysis of Sprm-1 revealed that the transcript is expressed in a stage-specific manner during spermatogenesis and that this expression is predominantly in the secondary spermatocytes immediately before meiosis I (14). Other POU domain genes have been previously described in the testes, including Testes-1 (Tst-1/Oct-6/SCIP) (16–18). Tst-1 is also expressed in other tissues, including several regions of the brain and glia, and has been implicated in myelination (8, 9, 16). Unfortunately, our mice carrying mutations in the Tst-1 gene did not survive long enough for us to evaluate the potential role of this factor in spermatogenesis (8). Because both Sprm-1 and Tst-1 are expressed in the testes and are likely to function as transcriptional activators, we analyzed the protein expression patterns of each to determine the possibility of a functionally redundant relationship between these two factors. Immunohistochemical analysis using specific rabbit polyclonal antibodies raised to bacterially expressed and purified amino terminus of rat Sprm-1 showed that the Sprm-1 protein is expressed in a stage-specific manner within the seminiferous tubules as we had expected from our previous in situ hybridization analysis (Fig. 1 A and B). Specifically, the protein was expressed in haploid spermatids after the meiotic reduction (Fig. 1B), which is consistent with our previous report that the mRNA expression begins immediately before the meiotic reduction (14). However, because of the long interval between the initiation of meiosis and final reductive division, we were surprised that the protein did not show a wider expression pattern beginning earlier in germ cell development. These data are consistent with a delay in the translation of the Sprm-1 mRNA until after the meiotic reduction: a type of translational control not uncommon to genes that are expressed in this time frame (19). Analysis of Tst-1 protein expression in the testes revealed expression in the Sertoli cells closer to the basal membrane of the seminiferous tubules, whereas no expression was detected in any of the germ cells (Fig. 1C). Additionally, Tst-1 expression was detected in the cells lining the epididymis (Fig. 1D). These cells are responsible for secreting factors that create the proper environment for maturation of the spermatozoa. Thus, although both Sprm-1 and Tst-1 are expressed in the testes, the protein expression patterns suggest that they may serve independent functions in the process of male germ cell maturation.
Figure 1.
Immunohistochemical analysis of Sprm-1 and Testes-1. (A) Low magnification (×100) of testis section probed with rabbit anti-Sprm-1 antibodies showing distribution in approximately 10% of the seminiferous tubules. (B) High magnification (×400) of a similar section that has been counter-stained with hematoxylin and eosin to show nuclear and cytoplasmic morphology. Arrow shows Sprm-1- positive spermatid. (C) High magnification (×200) light-field view of mouse testis labeled with anti-Tst-1 antibodies. Brown signal indicating Tst-1 immunoreactivity is seen in Sertoli cells at the basal edge of the tubules (arrow). (D) High magnification (×400) of mouse epididymis probed with anti-Tst-1 antibody and counter-stained with hematoxylin and eosin. Tst-1 immunoreactivity is seen in the cells lining the epididymal tubules (arrow).
Testicular Morphology of Sprm-1(−/−) Animals.
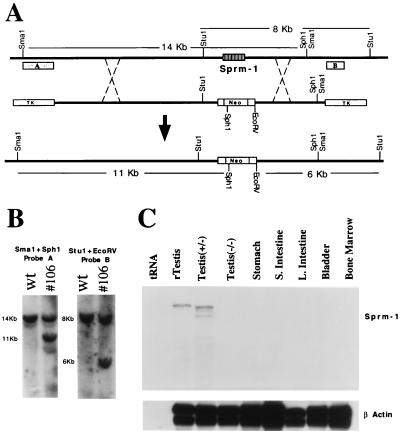
Because of the uniqueness of the Sprm-1 expression pattern and the important roles of other POU domain proteins in cellular differentiation, Sprm-1 is an excellent candidate for a haploid gene regulator. We tested this possibility by generating a targeting vector that was used to replace the Sprm-1 locus with a neomycin gene in embryonic stem cells (Fig. 2). Two targeted cell lines were injected into blastocysts to generate mice capable of transmitting the disrupted Sprm-1 locus through the germ line. Two founder males were used to start colonies in both the C57BL/6 and the 129/Sv genetic backgrounds. Disruption of the Sprm-1 gene was subsequently verified by RNase protection assay (Fig. 2C) and by Western blot analysis (data not shown).
Figure 2.
Targeted disruption of mouse Sprm-1 gene. (A) Schematic representation of the mouse Sprm-1 allele (Top), the targeting vector (Middle), and the targeted recombinant allele (Bottom). The single Sprm-1 exon is shown as a striped box. External probes A and B, shown under the Sprm-1 allele, were used to analyze for homologous recombination on the 5′ and 3′ ends of the allele respectively. Probe A hybridizes to a 14-kb SmaI/SphI double-digested fragment on the wild-type allele. The neomycin cassette introduces an additional SphI site, reducing the recognized fragment to 11 kb. Probe B hybridizes to an 8-kb StuI/EcoRV double-digested fragment on the wild-type allele, whereas a polylinker introduced into the 3′ end of the neomycin cassette added an EcoRV site that reduces the recognized fragment size to 6 kb. (B) Southern blots show the restriction fragment length polymorphism analysis of one positive cell line (No. 106), which was injected into e3.5 mouse blastocysts to generate chimeric animals. (C) RNase protection assay showing lack of Sprm-1 mRNA expression in mutant animals. A rat anti-cRNA probe to the Sprm-1 POU domain was used to probe the indicated tissue total RNAs. All heterologous tissue RNA analyzed is from wild-type animals. Smaller bands in the mouse testes RNA is due to a few nucleotide mismatches between mouse and rat sequences. A β-actin probe was included in the reaction mixture to verify the integrity of the RNA.
Spermatogenesis is a very tightly organized differentiation process that progresses in waves along the long axis of the seminiferous tubules. Within a section of seminiferous tubule the differentiation process is synchronized to produce very specific cellular associations permitting specific classification of the seminiferous epithelium into discrete stages (2), and a change in duration of any event during the spermatogenic cellular progression would result in a corresponding change in the number of tubules at the affected stage. Staining with hematoxylin and eosin revealed that mutant testes had normal cellularity and that all stages of the cycle of the seminiferous epithelium were present and were indistinguishable in character or number from normal testes (Fig. 3 A and B). Because we showed that Sprm-1 protein is present in the post-meiotic spermatid it is possible that it could regulate the process of nuclear condensation, which involves a transition in expression of DNA packing proteins from histones to transition proteins and finally to protamines. However, a nuclear analysis using bisbenzamide nucleolar stain illustrates that the dramatic nuclear compaction occurring after the meiotic reduction is grossly unaffected (Fig. 3 C and D). Other post-meiotic events include the organization of the acrosome, a structure at the tip of the sperm head that is involved in the fertilization process, and of the sperm tail. To test the potential role for Sprm-1 in the process of organizing these structures we used electron microscopy to analyze the subcellular details of testicular spermatids and spermatozoa. Subcellular analysis revealed normal acrosome development (Fig. 3 E and F) as well as normal organization of the midpiece and the filaments within the flagella of the Sprm-1(−/−) germ cells.
Figure 3.
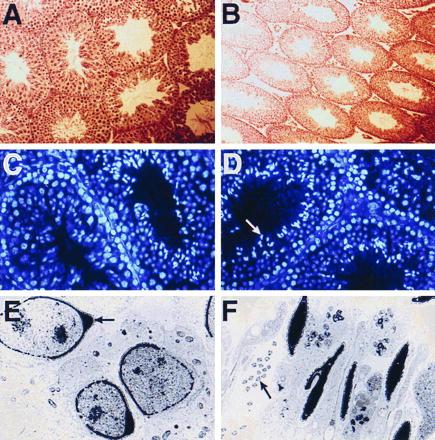
Morphology of Sprm-1(−/−) testis. (A and B) Sprm-1 mutant testis display normal morphology. Five-micrometer sections of Bouin’s fixed 129/Sv wild-type (A) or mutant (B) testis counter-stained with hematoxylin and eosin show that Sprm-1 mutants have normal morphology and contain all stages of the cycle of the seminiferous epithelium. Mutant testis section is shown at a lower magnification to show a greater number and variety of tubules. (C and D) Sprm-1 mutant spermatids undergo normal post-meiotic nuclear condensation. Biz-benzamide nucleolar stain was used to compare the process of nuclear compaction in wild-type (C) and mutant (D) testis. Compacted nuclei are visualized as bright staining sickle-shaped structures in the lumen of the tubules (arrow, D). (E and F) Sprm-1-deficient testes have normal ultrastructural morphology. (E) Electron microscopy analysis of a tubule from a 129/Sv Sprm-1 mutant showing normal acrosome formation (arrow, E), and sperm tail morphology (arrow, F).
Decreased Fertility in the Sprm-1(−/−) Males.
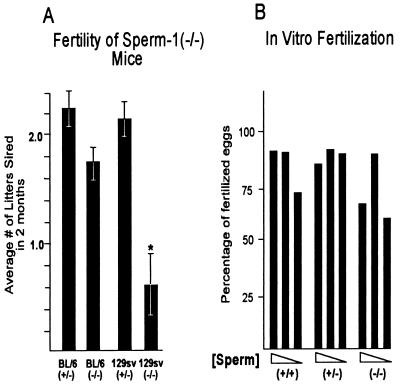
The normal appearance of the differentiating germ cells indicates that, unlike other tissue-specific POU domain factors, Sprm-1 is not required for normal morphological differentiation. To evaluate the functionality of the differentiated spermatozoa in vivo we analyzed the reproductive capacity of Sprm-1(−/−) males using described methods (20) by cohabitation with females of various genotypes, using wild-type and heterozygous males for comparison. Animals were allowed to mate for 2 months after which the females were removed. Females that had been mated to the knockout animals were subsequently switched to a wild-type mate and those previously with wild-type or heterozygous males were mated with Sprm-1(−/−) males after a 22-day delay, to allow any pregnant mothers to deliver their pups. The results of this controlled mating experiment showed clearly that the mutant animals exhibited a significantly reduced fertility compared with their heterozygous or wild-type counterparts (Fig. 4A). Additionally, a majority of the litters that were sired by the Sprm-1(−/−) mice were subsequently cannibalized by the parents, probably due to the small litter sizes seen in many litters sired by mutant fathers, thereby reducing the effective fertility even further than is illustrated. In fact, the effective reproduction of Sprm-1(−/−) males is so low that we have been consistently unable to sustain a 129/Sv colony using only mutant males and females. Interestingly, this phenotype is specific to the 129/Sv background as the Sprm-1 mutants backcrossed into the C57BL/6 background have relatively normal fertility. During the mating experiments, mutant males exhibited normal aggressiveness and sexual activity suggesting that the observed defect is not the result of an indirect hormonal or general systemic problem with the mice.
Figure 4.
Fertility and sperm count analysis in Sprm-1(−/−) mice. (A) Reduced fertility of Sprm-1(−/−) males. Bar graph shows the results of cohabitation experiments where sets of 10–14 males of each genotype were allowed to mate with females of proven fertility. Because no mutants in the 129/Sv background ever sired litters within 1 month of mating, the results are expressed as the average number of litters sired in 2 months. The values shown are the mean ± 1 SEM. (∗, P < 0.01.) (B) Sprm-1 mutant sperm can bind the zona pellucida and affect fertilization in vitro. The graph shows the results from one representative in vitro fertilization assay. Bars represent the percentage of eggs fertilized in the presence of approximately 10,000, 1,000, or 200 sperm per egg (in a consistent volume of 40 μl) where the sperm were isolated from wild-type, heterozygous, or mutant animals.
To investigate the subnormal fertility observed in the 129/Sv Sprm-1(−/−) males we conducted a sperm count analysis where mature sperm were isolated from the vas deferens and caudal epididymis of 2-month-old males and counted. Comparison of wild-type with heterozygote and mutant sperm counts, all from 129/Sv background, revealed that there was not a significant reduction in the number or motility of spermatozoa produced by the Sprm-1 deficient males (data not shown). Also, histological analysis of the epididymis revealed similar quantities of mature spermatozoa between mutant and wild-type litter mates (data not shown).
The relatively normal sperm counts in the mutant animals led us to believe that the sperm produced could be less capable of fertilizing eggs due to defects in binding and penetration of the zona pellucida, thereby explaining the results of our cohabitation experiment. To test this we isolated viable swimming sperm from 129/Sv wild-type, heterozygous, and from mutant mice and analyzed their ability of affect fertilization in vitro. Serial dilutions of the sperm were made from 10,000 down to only about 200 sperm per egg. Each dilution of each genotype was added to 10–15 eggs. After 3 days each fertilization chamber was scanned, and the embryos were scored as unfertilized, parthenogenetic, or fertilized. Surprisingly, the differences observed in the ability of the spermatozoa from each animal to bind and penetrate the zona pellucida was not significant enough to account for the reduction in reproductive capacity observed in the cohabitation experiments (Fig. 4B).
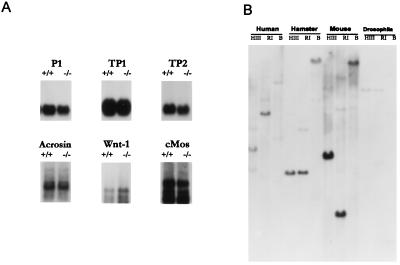
The reduction in fertility of the Sprm-1 mutant animals indicates that this factor plays an important physiological role after the meiotic reduction. Because Sprm-1 protein is expressed after the meiotic division, it is an excellent candidate for a regulator of haploid gene transcription. To investigate the potential molecular mechanism behind this fertility loss we probed mRNA blots of 129/Sv wild-type versus Sprm-1(−/−) testes for a series of genes that have been described as being activated in the haploid germ cell. These haploid expressed genes can be divided into three functional categories: (i) basic histone-like proteins that are involved in normal compaction and condensation of the sperm nucleus that include the protamine P1 and the transition proteins TP1 and TP2; (ii) protooncogenes whose role in the spermiogenic process is not clear, including cMos, and Wnt1; and (iii) genes involved in the actual fertilization process, including acrosin that acts to affect penetration of the egg. Despite the physiological affect of the Sprm-1 mutation in the 129/Sv background none of the markers tested were altered significantly in the knockout animals as compared with heterozygous litter mates (Fig. 5A). These results suggest that there are as-yet-unidentified genes that are regulated by Sprm-1 and have subtle, but important, roles in the function of the male germ cell.
Figure 5.
Haploid markers in Sprm-1(−/−) testes and zoo blot analysis. (A) Expression of candidate target genes in Sprm-1(−/−) mice. Two micrograms of testes poly(A)+ RNA from either wild-type or Sprm-1(−/−) animals was fractionated on an agarose gel, transferred, and hybridized with a probe for the indicated gene markers. Markers are protamine 1 (P1), transition protein 1 and 2 (TP1, TP2), acrosin, Wnt1, and cMos. Additional bands that hybridized to the cMos and Wnt-1 probes result from alternate splicing variants of these genes in the testes. (B) Zoo blot analysis. DNA from the indicated species was digested with HindIII (HIII), EcoRI (RI), or BamHI (B) and subsequently fractionated on an agarose gel, transferred, and probed with the entire rat Sprm-1 coding region under low-stringency conditions.
Sprm-1 in Higher Organisms.
The results of our study indicate that Sprm-1 is required for maximal fertility. Because of the obvious importance of male fertility for species propagation we predict that Sprm-1 activity should be important for all mammals. To investigate the conservation of Sprm-1 between species, a Southern blot containing DNA from human, hamster, mouse, and Drosophila was probed with rat Sprm-1 cDNA (Fig. 5B). Bands observed in the mouse DNA lanes show a simple banding pattern, indicating that this probe is specifically recognizing Sprm-1 sequences and is only weakly crosshybridizing with other Oct-3/4-related genes. The increasing complexity of the weaker bands in the human DNA is consistent with a previous study that showed a greater complexity of Oct-3/4-like genes in the human genome (21). A probe for the human Oct-3/4-like gene, OTF3C, hybridized only to the weaker Sprm-1 hybridizing bands. The relatively strong signals and simple banding pattern in all lanes from human to Drosophila is consistent with the argument that Sprm-1 shows evolutionary conservation.
DISCUSSION
In this manuscript we show that although the Sprm-1 transcripts are detected before the first meiotic division of the male germ cell, the protein product is only detected in haploid spermatids after the meiotic reduction. This is consistent with a delay in translation of the Sprm-1 message, similar to that seen for other genes expressed in this window (5–7). This expression pattern indicates that Sprm-1 may function in the unique process of haploid gene regulation. We used homologous recombination to disrupt the Sprm-1 locus and show that Sprm-1 mutant mice exhibit subnormal fertility in cohabitation experiments. Surprisingly, this fertility defect is specific for the 129/Sv genetic background, indicating that the higher fecundity of the C57BL/6 mice compensates for the Sprm-1 defect in the laboratory environment. We further show that although the mutant mice show a physiological defect they produce relatively normal numbers of sperm and exhibit normal testicular morphology. This indicates that the physiological defect is very late in the process of spermatogenic differentiation and is not due to a blockage in germ cell development, but rather is due to the production of a cell that is less capable of performing its function than its wild-type counterpart. We further show that the sperm produced by mutant mice are capable of binding to the zona pellucida of eggs and affecting fertilization in vitro. This is not entirely surprising because we do observe rare litters sired by 129/Sv Sprm-1 mutants, indicating that the mutant sperm must be capable of effecting fertilization even in vivo, albeit at a lower frequency than wild-type sperm. It should be mentioned that because mammalian spermatogenesis occurs in a syncytium crosstalk between haploid nuclei could rescue Sprm-1(−) spermatids in heterozygous 129/Sv matings.
There have recently been several gene knockout studies that have observed male sterility due to azoospermia, notably the retinoid X receptor β (RXRβ) (22) and CREM (6, 7). Genetic disruption of either RXRβ or CREM results in male sterility due to a failure of release of the spermatozoa from the germinal epithelium (22) or from a blockage in cellular progression in the spermatid (6, 7), respectively. The expression of RXRβ in the Sertoli cells suggest that this effect is a result of a disruption in the environment of germ cell maturation and is not an autonomous effect within the germ cell itself. CREM mutant animals lack virtually all post-meiotic markers, suggesting that CREMτ is a primary factor responsible for haploid gene activation. However, the total lack of late spermatids in these mice along with the increased germ cell apoptosis and the dynamic nature of spermiogenesis makes it difficult to determine whether the absent haploid gene markers are a result of a lack of direct activation by CREMτ or of a lack of progression past a developmental checkpoint normally required for their expression.
Our data clearly show that Sprm-1 plays a much more subtle role in the spermiogenic process than either RXRβ or CREMτ. In fact, the lack of fertility defects in the C57BL/6 Sprm-1(−/−) males argues that mouse strains of higher fecundity are capable of overcoming the defect in the laboratory environment. Because of these results, we propose that Sprm-1 regulates a haploid gene pathway that is required for optimum fertility and that disruption of this pathway will compromise, but not destroy, male fertility. It has been previously demonstrated that rodents exhibit male hyperfertility presumably to allow for continued reproduction even during nonideal conditions. Male rats, for example, will inject 10- to 100-fold excess sperm than is required to affect fertility, whereas humans are thought to inject only between 2- and 10-fold excess. Others have demonstrated that mouse strains of lower fecundity are more susceptible to reproductive toxicities from ethylene glycol monomethyl ether than those of higher fertility (20). These studies argue that partial defects in fertility that may be relevant to human infertility are better studied in mouse strains that have a lower natural fecundity. This observation may, in fact, explain other cases where mice exhibited apparently normal fertility even with seemingly devastating genetic alterations such as a disruption of the acrosin gene or a reduction of the number of sperm receptors (mZP3) on the egg (23, 24).
The existence of many infertile human males that exhibit normal testicular morphology and relatively normal concentrations of mobile spermatozoa raises the possibility that the reduction in fertility of the Sprm-1(−/−) mice, without abnormal morphology or a decrease of sperm concentration, would make them potentially useful models for studying the subtle events in mammalian reproduction, which may be effected in a subset of the infertile human population.
Acknowledgments
We are very grateful to C. Wayne Bardin for his assistance with the evaluation of the morphology of the mutant testes. We are also grateful to B. Andersen, M. Ayers, M. Schonemann, and R. McEvilly for critical advice, reagents, and technical assistance. R.V.P. is supported by a grant from the National Institute of Mental Health. M.G.R. is an Investigator with the Howard Hughes Medical Institute.
ABBREVIATION
- CREM
cyclic AMP-responsive element modulator
References
- 1.Lipshultz L I, Howards S S. Infertility in the Male. 2nd Ed. St. Louis: Mosby; 1991. [Google Scholar]
- 2.Leblond C P, Clermont Y. Ann NY Acad Sci. 1952;55:548–573. doi: 10.1111/j.1749-6632.1952.tb26576.x. [DOI] [PubMed] [Google Scholar]
- 3.Parvinen M. Endocr Rev. 1982;3:404–417. doi: 10.1210/edrv-3-4-404. [DOI] [PubMed] [Google Scholar]
- 4.Erickson R P. Trends Genet. 1990;6:264–269. doi: 10.1016/0168-9525(90)90209-o. [DOI] [PubMed] [Google Scholar]
- 5.Hecht N B. J Reprod Fertil. 1990;88:679–693. doi: 10.1530/jrf.0.0880679. [DOI] [PubMed] [Google Scholar]
- 6.Blendy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Nature (London) 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 7.Nantel F, Monaco L, Foulkes N S, Masquilier D, LeMeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Nature (London) 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 8.Bermingham J R, Jr, Scherer S S, O’Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 9.Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. Science. 1996;273:507–510. doi: 10.1126/science.273.5274.507. [DOI] [PubMed] [Google Scholar]
- 10.Schonemann M D, Ryan A K, McEvilly R J, O’Connell S M, Arias C A, Kalla K A, Li P, Sawchenko P E, Rosenfeld M G. Genes Dev. 1995;9:3122–3135. doi: 10.1101/gad.9.24.3122. [DOI] [PubMed] [Google Scholar]
- 11.Nakai S, Kawano H, Yudate T, Nishi M, Kuno J, Nagata A, Jishage K, Hamada H, Fujii H, Kawamura K, Shiba K, Noda T. Genes Dev. 1995;9:3109–3121. doi: 10.1101/gad.9.24.3109. [DOI] [PubMed] [Google Scholar]
- 12.Gan L, Xiang M, Zhou L, Wagner D S, Klein W H, Nathans J. Proc Natl Acad Sci USA. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erkman L, McEvilly R J, Luo L, Ryan A K, Hooshmand F, O’Connell S M, Keithley E M, Rapaport D H, Ryan A F, Rosenfeld M G. Nature (London) 1996;381:603–606. doi: 10.1038/381603a0. [DOI] [PubMed] [Google Scholar]
- 14.Andersen B, Pearse R V D, Schlegel P N, Cichon Z, Schonemann M D, Bardin C W, Rosenfeld M G. Proc Natl Acad Sci USA. 1993;90:11084–11088. doi: 10.1073/pnas.90.23.11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renner K, Leger H, Rosenfeld M G. Proc Natl Acad Sci USA. 1994;91:6433–6437. doi: 10.1073/pnas.91.14.6433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinstein D E, Burrola P G, Lemke G. Mol Cell Neurosci. 1995;6:212–229. doi: 10.1006/mcne.1995.1018. [DOI] [PubMed] [Google Scholar]
- 17.He X, Gerrero R, Simmons D M, Park R E, Lin C J, Swanson L W, Rosenfeld M G. Mol Cell Biol. 1991;11:1739–1744. doi: 10.1128/mcb.11.3.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoykova A S, Sterrer S, Erselius J R, Hatzopoulos A K, Gruss P. Neuron. 1992;8:541–558. doi: 10.1016/0896-6273(92)90282-i. [DOI] [PubMed] [Google Scholar]
- 19.Schafer M, Nayernia K, Engel W, Schafer U. Dev Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- 20.Chapin R E, Morrissey R E, Gulati D K, Hope E, Barnes L H, Russell S A, Kennedy S R. Fundam Appl Toxicol. 1993;21:8–14. doi: 10.1006/faat.1993.1065. [DOI] [PubMed] [Google Scholar]
- 21.Takeda J, Seino S, Bell G I. Nucleic Acids Res. 1992;20:4613–4620. doi: 10.1093/nar/20.17.4613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kastner P, Mark M, Leid M, Gansmuller A, Chin W, Grondona J M, Decimo D, Krezel W, Dierich A, Chambon P. Genes Dev. 1996;10:80–92. doi: 10.1101/gad.10.1.80. [DOI] [PubMed] [Google Scholar]
- 23.Liu C, Litscher E S, Wassarman P M. Mol Biol Cell. 1995;6:577–585. doi: 10.1091/mbc.6.5.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baba T, Azuma S, Kashiwabara S, Toyoda Y. J Biol Chem. 1994;269:31845–31849. [PubMed] [Google Scholar]