Abstract
IL-13 and IL-4 are central T helper 2 (Th2) cytokines in the immune system and potent activators of inflammatory responses and fibrosis during Th2 inflammation. Recent studies using Il13ra1−/− mice have demonstrated a critical role for IL-13 receptor (IL-13R) α1 in allergen-induced airway responses. However, these observations require further attention especially because IL-4 can induce similar lung pathology to IL-13, independent of IL-13, and is still present in the allergic lung. Thus, we hypothesized that IL-13Rα1 regulates IL-4-induced responses in the lung. To dissect the role of IL-13Rα1 and the type I and II IL-4Rs in experimental asthma, we examined lung pathology induced by allergen, IL-4, and IL-13 challenge in Il13ra1−/− mice. We report that IL-13Rα1 is essential for baseline IgE production, but Th2 and IgE responses to T cell-dependent antigens are IL-13Rα1-independent. Furthermore, we demonstrate that increased airway resistance, mucus, TGF-β, and eotaxin(s) production, but not cellular infiltration, are critically dependent on IL-13Rα1. Surprisingly, our results identify a CCR3- and IL-13Rα1-independent pathway for lung eosinophilia. Global expression profiling of lungs from mice stimulated with allergen or IL-4 demonstrated that marker genes of alternatively activated macrophages are differentially regulated by the type I and type II IL-4R. Taken together, our data provide a comprehensive mechanistic analysis of the critical role by which IL-13Rα1 mediates allergic lung pathology and highlight unforeseen roles for the type II IL-4R.
Keywords: inflammation, cytokines, eosinophils, chemokines, mucus
Interleukin 13 is a central immune regulator of many hallmark type 2 disease characteristics, including IgE synthesis, mucus hypersecretion, airway hyperreactivity, and fibrosis (1). IL-13 shares overlapping biological functions with IL-4 (1, 2), and both signal via a complex network of receptors. IL-4 mediates its effects through either the type I IL-4 receptor (IL-4R) (i.e., IL-4Rα and the common γ chain) or the type II IL-4R (i.e., IL-4Rα and IL-13Rα1). In contrast, IL-13 is hypothesized to execute its IL-4Rα-dependent effects solely through the type II IL-4R but may use a signaling complex that does not require IL-4Rα (3). In addition, IL-13Rα2, an IL-13 decoy receptor (4), has been recently reported to also mediate IL-13 signaling and induce TGF-β production (5, 6). Thus, the assumption that IL-13Rα1 is the main signaling receptor for IL-13 needs definitive proof.
Although IL-4 and IL-13 initiate similar intracellular signaling cascades, IL-13 is capable of exerting specific and IL-4-independent signals (4, 7). In addition, IL-4 can induce lung pathology even in the absence of IL-13, and treatment with an IL-13 antagonist does not inhibit the effects of IL-4 (8). Yet it is currently unknown whether these IL-13-independent effects of IL-4 are mediated via the type I or type II IL-4R (9).
A valuable way to distinguish the role of these two receptors is by genetic deletion of the IL-13Rα1 chain, because such genetically engineered mice would harbor a functional deletion of the type II IL-4R but have an intact type I IL-4R.
A recent study using Il13ra1−/− mice has shown that these mice are protected from Schistosoma mansoni egg antigen-induced airway hyperreactivity and mucus hypersecretion (10). However, these results require further clarification especially because IL-4 is still up-regulated in the lungs of these mice and could potentially induce airway hyperreactivity and mucus production (8).
Results
Il13ra1−/− Mice Display Enhanced Circulating Soluble IL-13Rα2 and IL-13.
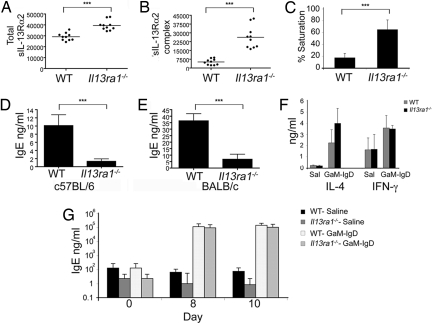
Soluble IL-13Rα2 (sIL-13Rα2) has been proposed to counterregulate IL-13 activities (4, 11–13) via an autoregulatory pathway initiated by IL-13Rα1 (5). Contrary to the current paradigm, which would have predicted decreased levels of sIL-13Rα2 (5, 14), Il13ra1−/− [supporting information (SI) Fig. S1] mice had elevated circulating total sIL-13Rα2, increased sIL-13Rα2:IL-13 complexes (indicating increased IL-13 levels), and markedly increased saturation of IL-13Rα2 with IL-13 (Fig. 1 A–C).
Fig. 1.
Assessment of baseline serum IL-13Rα2 and IgE. (A and B) Assessment of total sIL-13Rα2 (free and bound to IL-13) and IL-13/sIL-13Rα2 complexes. The y axes in are shown in pg/ml where each dot represents a different mouse (n = 10 mice per group). ***, P < 0.001. (C) A graphic representation of the saturation status of sIL-13Rα2. (D and E) Total serum Ig concentrations were determined. ***, P < 0.01. IL-4, IFN-γ (F, day 5) and serum IgE levels (G) were monitored after goat anti-mouse IgD antiserum (GαM-IgD) injection. n = 2 (six mice per experimental group).
IL-13Rα1 Is Critical for Maintenance of Homeostatic IgE Independent of Changes in IL-4.
Il13ra1−/− mice had barely detectable IgE (Fig. 1 D and E) and displayed a minor increase in IgG2a levels but no changes in IgA, IgM, IgG1, IgG3, or IgG2b levels (Fig. S2 A–F).
Given the crucial role of IL-4 in IgE production (15–17), we examined IL-4 levels and signaling components in Il13ra1−/− mice. Serum IL-4 and IFNγ levels, IL-4Rα expression, and STAT6 phosphorylation in response to IL-4 were comparable between WT and Il13ra1−/− splenocytes (Fig. S3 A–E). In response to CD3/CD28 stimulation, Il13ra1−/− splenocytes produced normal amounts of IL-4 whereas IFNγ production was decreased (Fig. S3 F–H).
IL-13Rα1 Is Dispensable for Polarized T helper 2 (Th2) Responses in Vivo.
Subsequently, we examined the ability of Il13ra1−/− mice to manifest an acquired Th2 response. After treatment with goat anti-mouse IgD (GαM-IgD), a potent Th2 polarizing agent (18), both Il13ra1−/− and WT mice had comparable serum IL-4 and IFN-γ levels (Fig. 1F) and, accordingly, similar serum IgE levels (Fig. 1G).
Assessment of IL-13Rα1-Mediated Responses in a Model of IL-13-Induced Airway Inflammation.
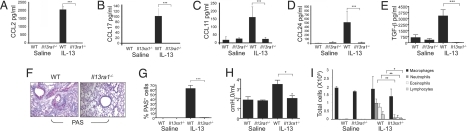
To directly define the role of IL-13Rα1 in IL-13-induced lung responses, IL-13 was administered intratracheally to Il13ra1−/− mice and lung inflammation was assessed. IL-13 strongly induced chemokine expression (i.e., CCL2, CCL17, CCL11, and CCL24) in WT mice but not Il13ra1−/− mice (Fig. 2 A–D).
Fig. 2.
Assessment of IL-13Rα1-mediated responses in a model of IL-13-induced airway inflammation. Forty-eight hours after the final IL-13 challenge, the mice were assessed for BALF chemokine (A–D) and active TGF-β (E) levels, mucus production (F and G), airway resistance (H), and BALF cellular infiltration (I). Data are representative of three experiments (six to eight mice per experimental group). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
No induction of TGF-β was observed in IL-13-challenged Il13ra1−/− mice, whereas WT mice displayed significantly elevated TGF-β levels in response to IL-13 challenge (Fig. 2E).
Strikingly, no mucus induction was observed in Il13ra1−/− mice whereas WT mice displayed many PAS+ cells (Fig. 2 F and G). Furthermore, Il13ra1−/− mice were completely protected from the ability of IL-13 to induce airway resistance (Fig. 2H).
Assessment of IL-13's effects on inflammatory cell recruitment showed a marked reduction in cellular infiltration in Il13ra1−/− mice (Fig. 2I and data not shown).
To further test the specific role of IL-13Rα1 in the pulmonary effects of IL-13, we conducted similar experiments in Il13ra2-deficient mice. In contrast to the essential role of IL-13Rα1 in mediating IL-13-induced lung responses, Il13ra2−/− mice displayed a phenotype identical to that of IL-13-challenged WT mice (Fig. S4). No change was observed in chemokines, TGF-β levels (Fig. S4 A–D), mucus production (Fig. S4 E and F), airway resistance (Fig. S4G), or IL-13-mediated cellular infiltration (Fig. S4H).
Assessment of IL-13Rα1-Mediated Responses in Allergen-Induced Airway Inflammation.
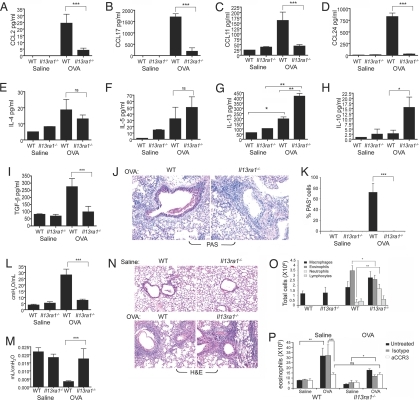
Next, we examined the contribution of IL-13Rα1 to lung pathology in allergen (OVA)-induced experimental asthma (19). Il13ra1−/− mice displayed a complete (i.e., ≈99%) reduction in CCL2, CCL11, and CCL24 and an 82% reduction in CCL17 (Fig. 3 A–D). Assessment of the major Th2 cytokines in the bronchoalveolar lavage fluid (BALF) of OVA-challenged mice indicated that Il13ra1−/− mice displayed levels of IL-4 and IL-5 similar to those of WT mice. Nevertheless, Il13ra1−/− mice had increased BALF IL-10 and IL-13 levels (Fig. 3 E–H) but did not display any TGF-β induction (Fig. 3I). Although Il13ra1−/− mice showed slightly (but statistically significantly) lower levels of IgE induction, they were still capable of inducing a prominent IgE response (Fig. S5).
Fig. 3.
Assessment of IL-13Rα1-mediated responses in allergen-induced airway inflammation. Twenty-four hours after the final allergen challenge, the mice were examined for BALF chemokine (A–D) and cytokine (E–H) production, active TGF-β production (I), mucus production (J and K), airway resistance (L), lung compliance (M), and BALF and lung cellular infiltration (N and O). Data are representative of one of three experiments (6–17 mice per experimental group). ns, not significant. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Chemotaxis was assessed with eosinophils in response to BALF from allergen-challenged WT or Il13ra−/− mice (P). “Isotype” indicates isotype-matched control, and “aCCR3” indicates anti-CCR3. Data are representative of three experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Remarkably, allergen-challenged Il13ra1−/− mice revealed complete abrogation of goblet cell hyperplasia and mucus production (Fig. 3 J and K). Furthermore, physiological measurements of airway resistance and compliance revealed that Il13ra1−/− mice were totally protected from allergen-induced airway resistance (Fig. 3L) and decreased lung compliance (Fig. 3M).
Despite the fact that Il13ra1−/− mice displayed near ablation of eosinophil-specific chemokines, only a minor decrease in BALF eosinophilia was observed, whereas neutrophil counts were increased (Fig. 3 N and O). Using an in vitro chemotaxis assay, BALF of allergen-challenged Il13ra1−/− mice displayed chemotactic activity toward eosinophils, albeit lower than BALF obtained from allergen-challenged WT mice (Fig. 3P). The chemotactic ability of allergen-challenged WT BALF was partially dependent on CCR3, because anti-CCR3 was able to significantly reduce eosinophil chemotaxis. In contrast, eosinophil chemotactic activity in the BALF of Il13ra1−/− mice was completely CCR3-independent. Furthermore, after CCR3 neutralization, allergen-challenged WT BALF displayed chemotactic activity similar to that of BALF obtained from allergen-challenged Il13ra1−/− mice.
Assessment of IL-13Rα1-Mediated Responses in a Murine Model of IL-4-Induced Airway Inflammation.
Because IL-4 was still induced in the BALF of Il13ra1−/− mice and theoretically able to mediate the same cardinal features of disease (8, 9), we hypothesized that IL-4 may also be mediating its affects in the lung via the type II IL-4R. Notably, prior studies concerning IL-4's action in the lung have not distinguished between type I and type II IL-4 receptors. Thus, we examined IL-4 responses in Il13ra1−/− mice by direct administration of a long-acting formulation of IL-4 (20).
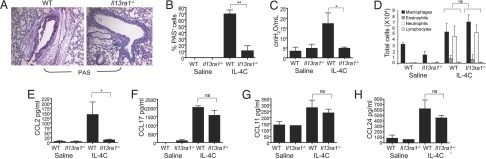
IL-4-challenged Il13ra1−/− mice displayed markedly decreased mucus production (an ≈80% reduction in PAS+ cells) (Fig. 4 A and B), and IL-4 was unable to increase airway resistance in the absence of Il13ra1 (Fig. 4C).
Fig. 4.
Assessment of IL-13Rα1-mediated responses in a model of IL-4-induced airway inflammation. Twenty-four hours after the final IL-4C challenge the mice were assessed for mucus production (A and B), airway resistance (C), BALF cellular infiltration (D), and chemokine production (E–H). The data, representative of one of two experiments, are presented as mean ± SD (nine mice per experimental group). *, P < 0.05; ***, P < 0.001.
Il13ra1 deficiency did not alter inflammatory cell recruitment to the BALF and lung by IL-4; Il13ra1−/− mice displayed preserved leukocyte recruitment similar to that of IL-4-treated WT mice (Fig. 4D and data not shown). Further assessment of BALF chemokine levels revealed that CCL2 induction depended on IL-13Rα1 (Fig. 4E). Nevertheless, consistent with our findings demonstrating preserved leukocyte recruitment, in the absence of Il13ra1, IL-4 induced CCL17, CCL11, and CCL24 (Fig. 4 F–H) at comparative levels to WT mice. These effects were not due to IL-4-dependent IL-13 induction because IL-13 expression was not detected in IL-4-treated mice (data not shown).
Identification of OVA-Induced IL-13Rα1-Dependent Genes.
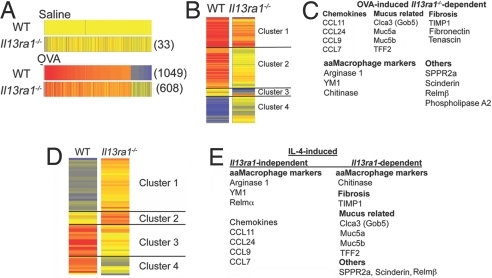
To gain mechanistic insight into the action of IL-13Rα1 in asthma pathogenesis, we identified IL-13Rα1-dependent and -independent pathways using global microarray analysis. Il13ra1−/− mice displayed alteration of 33 transcripts at baseline (Fig. 5A). Among these, mucin-associated gene Clca3 (Gob5), Ear11 (eosinophil ribonuclease A11), and Chi3l4 (chitinase 3-like 4) were markedly down-regulated (34.7-, 5.6-, and 3.47-fold, respectively). This indicates a central role for the type II IL-4R in baseline lung homeostasis.
Fig. 5.
Identification of IL-13Rα1-dependent genes. DNA microarray analysis of allergen-challenged lungs from Il13ra1−/− or WT mice identifies 33 altered genes (shown in parentheses) at baseline in the Il13ra1−/− mice, 1,049 altered genes in the allergen-challenged WT mice, and 608 altered genes in the allergen-challenged Il13ra1−/− mice (in comparison with saline-treated mice) (A). Comparison of microarray data obtained from the lungs of allergen-challenged mice reveals a subset of 205 altered genes that are depicted in four clusters as shown (B). Several allergen-induced IL-13Rα1-dependent genes are shown (C). Comparison of microarray data obtained from IL-4-challenged mice reveals a subset of 63 altered genes that are dysregulated (D). Several IL-4-induced IL-13Rα1-dependent genes are depicted (E).
After allergen challenge, the expression of 1,049 genes was changed ≥2-fold in WT mice (compared with saline-treated WT mice). In contrast, 608 transcripts were changed in allergen-challenged Il13ra1−/− mice (compared with saline-treated Il13ra1−/− mice) (Fig. 5A).
Comparison of allergen-challenged WT with Il13ra1−/− mice identified a set of 205 IL-13Rα1-dependent genes (e.g., dysregulated ≥2-fold between allergen-challenged WT and Il13ra1−/− mice) (Fig. 5B). These genes were segregated into the following four clusters: cluster 1, up-regulated genes (in Il13ra1−/− mice but to a lesser extent, i.e., less responsive); cluster 2, unaltered genes (in Il13ra1−/− mice, i.e., nonresponsive); cluster 3, down-regulated genes (only in Il13ra1−/− mice); and cluster 4, down-regulated genes in WT mice but unaltered in Il13ra1−/− mice (Fig. 5B).
Among the less responsive genes, various CC chemokines, mucin-associated genes, and alternatively activated macrophage (aaMΦ) marker genes such as Arg-1 (arginase 1) and Chi3l3 (YM1) were identified. Interestingly, the expression of other aaMΦ markers, including Retnla (Relm-α) and MglI (macrophage galactose-type calcium-type lectin 1/CD301a), were independent of IL-13Rα1 (Table 1 and Table S1) (21, 22). Furthermore, numerous other genes including Sppr2a, Corin, and Il13ra2 (Fig. 5C and Tables S2–S5) were unresponsive to induction by allergen challenge in Il13ra1−/− mice.
Table 1.
Comparison of allergen- and IL-4-induced aaMΦ markers in WT and Il13ra1−/− mice
| Description | Gene symbol | OVA | IL-4 |
|---|---|---|---|
| Scinderin | Scin | 16.13 | 16.27 |
| Resistin-like β | Retnlb | 8.88 | 8.84 |
| Chitinase, acidic | Chia | 6.705 | 6.77 |
| Similar to gel-forming mucin | Muc5ac | 6.896 | 7.02 |
| Small proline-rich protein 2A | Sppr2a | 5.09 | 5.78 |
| Intelectin a | Itlna | 5.225 | 5.48 |
| Calpain 9 | Capn9 | 3.775 | 3.8 |
| Solute carrier member 1 | Slc5a1 | 5.856 | 5.98 |
| Tissue inhibitor of metalloproteinase 1 | Timp1 | 3.65 | NC |
| Dentin matrix protein 1 | Dmp1 | 5.205 | NI |
| Corin | Corin | 7.165 | NI |
Comparison of allergen (OVA)-induced and IL-4 induced gene expression between WT mice relative to their expression in Il13ra1−/− mice. Values are expressed by increased fold change. NC, not changed; NI, not induced.
In addition, we identified 367 genes that were IL-13Rα1-independent (i.e., similarly regulated in both OVA-challenged Il13ra1−/− and WT mice). These included C-X-C chemokines such as CXCL9 and CXCL10, which are up-regulated ≈13- and 8.5-fold, respectively, in both WT and Il13ra1−/− mice (Table S1). Interestingly, 15-lipooxygenase (15-LO), a recently described IL-13-induced gene and a key enzyme in arachidonic acid metabolism, was also IL-13Rα1-independent (23).
Identification of IL-4-Induced IL-13Rα1-Dependent Genes.
Next, we aimed to identify the relative contribution of the different IL-4R chains in IL-4-induced lung responses. By means of a global microarray approach, we compared the genetic signature of IL-4-treated WT mice with IL-4-treated Il13ra1−/− mice (Fig. 5D). This comparison identified a set of 63 genes that were induced by IL-4 and dependent on IL-13Rα1 (e.g., dysregulated ≥2-fold between WT and Il13ra1−/− mice) (Fig. 5D). Interestingly, most genes associated with aaMΦ, such as Chi3l3, Arg-1, and Retnla, were independent of Il13ra1 expression (Table 1). In contrast, IL-4-induced Chia (chitinase) expression was dependent on IL-13Rα1. Consistent with our findings, several mucus-associated genes such as Muc5ac, Clca3 (Gob5), and Tff2 were dependent on Il13ra1 (i.e., found on clusters 3 or 4) whereas CC chemokine induction (but not Ccl2) was largely independent of Il13ra1 (Fig. 5E). Furthermore, other genes implicated in asthma pathogenesis such as Scin (scinderin), Itlna (intelectin), and Sppr2a were also dependent on IL-13Rα1 (Fig. 5E and Tables S6–S9).
Comparison of Allergen- and IL-4-Induced IL-13Rα1-Dependent Genes.
Because both allergen- and IL-4-induced airway resistance and mucus production were dependent on the type II IL-4R, we identified IL-13Rα1-dependent genes that were similarly regulated after IL-4 and OVA (Table S10). These genes include Chia, Scin, Retnlb (Relm-β), Itlna, and Capn9 (Calpain 9). Although IL-13Rα1 commonly regulated several allergen- and IL-4-induced genes, our analysis revealed several pathways that were differentially regulated. Furthermore, by examining aaMΦ signature genes (10, 21), we identified a subset of genes that were dependent on IL-13Rα1 after allergen challenge (i.e., Arg-1 and Chia) but not after IL-4-challenge (i.e., Arg-1, MglI, and Retnla) (Table 1).
Discussion
The pathological effects of IL-4 and IL-13 in Th2 immunity have been a focus of intense research in the last decade (1, 7, 17, 19). Even so, the receptor–ligand interactions responsible for the central roles of IL-4 and IL-13 remain to be elucidated. To fully dissect the molecular mechanisms that are regulated by IL-13Rα1 in the lung in response to allergen challenge and the relative contribution of this receptor to IL-13- and IL-4-induced pathology, we examined diverse Th2 responses in Il13ra1−/− mice. We report that IL-13Rα1 regulates baseline IgE (independent of changes in IL-4). However, IgE responses to T cell-dependent antigens are IL-13Rα1-independent. Integrating the data obtained from the in vivo models with global microarray analysis of allergen- and IL-4-challenged lungs enabled us to conclude the following: (i) IL-13Rα1 is the chief receptor for IL-13 in the lung; (ii) airway resistance, mucus production, and profibrogenic mediator induction are nearly totally dependent on IL-13Rα1, which serves as a signaling molecule for both IL-4 and IL-13; (iii) IL-13 and IL-13Rα1 dependence of the CC chemokine response (especially eotaxin generation) predominantly reflects greater production of IL-13 than IL-4; (iv) IL-4 efficiently utilizes the type I IL-4R to induce inflammatory cell recruitment, even though IL-4 is present at lower levels than IL-13; and (v) aaMΦ induction (defined by the expression of their classic gene products) depends on both the type I and type II IL-4Rs (see Table 2). In addition, we demonstrate that key pathogenic molecules associated with asthma severity, such as chitinase (24), are entirely dependent on IL-13Rα1.
Table 2.
Summary of the differential regulation of various pathological changes in the lung and their dependency on the type I or type II IL-4Rs
| Stimuli | AR | Mucus | PF | CC chemokines | Eosinophilia | aaMac genes |
||
|---|---|---|---|---|---|---|---|---|
| Arg | Retnla | Chia | ||||||
| IL-13 | Type II | Type II | Type II | Type II | Type II | Type II | Type II | Type II |
| OVA | Type II | Type II | Type II | Type II | Type I | Type II | Type I | Type II |
| IL-4 | Type II | Type II | ? | Type I | Type I | Type I | Type I | Type II |
AR, airway resistance; PF, profibrogenic mediators; aaMac, alternatively activated macrophages.
Our data demonstrate that baseline IgE expression depends on IL-13Rα1. Nevertheless, Il13ra1−/− mice can still mount a normal Th2 cytokine and IgE response. Baseline natural, but not antigen-specific, IgE has been recently attributed to a unique population of ε-germ-line transcript-positive B2 cells (25). We propose that IL-13Rα1 may control natural IgE production by this subpopulation of B cells, which may express the IL-13Rα1 and type II IL-4R along with or instead of γc and the type I IL-4R.
Our studies also evaluated the relative roles of IL-13Rα1 and IL-13Rα2 in induction of TGF-β. IL-13 mediated TGF-β induction, and TGF-β production in liver fibrosis after S. mansoni infection has been proposed to be independent of IL-13Rα1 (10). Yet findings demonstrate that IL-13- and allergen-induced TGF-β production is completely dependent on IL-13Rα1. The finding that IL-13Rα1 is the key regulator of TGF-β production has therapeutic implications related to allergen-driven fibrotic reactions.
Importantly, mucus production and increased airway resistance were nearly completely dependent on IL-13Rα1. Similarly, whereas the chemokine response (except for CCL2) elicited by IL-4 was mostly IL-13Rα1-independent, all of the examined CC chemokines in the allergen-stimulated lung were IL-13Rα1-dependent. This dependency suggests that the CC chemokine response is regulated mostly by IL-13 and not IL-4, presumably because both the type I and type II IL-4Rs can induce CC chemokine production and more IL-13 is produced than IL-4. Despite this, and consistent with observations made with stat6−/− and Il13ra1−/− mice (10, 26), BALF eosinophilia was only modestly affected in allergen-challenged Il13ra1−/− mice.
Because therapeutic targeting of IL-4Rα substantially decreases eosinophilia in response to allergen challenge (27), an IL-13Rα1-independent pathway for eosinophil recruitment that is efficiently induced by IL-4 through the type I IL-4R must exist. Our in vitro chemotaxis studies support the existence of a CC chemokine-independent pathway for eosinophil recruitment under these conditions. Arachidonic acid metabolites produced in response to IL-4 such as induction of 15-LO may be responsible for CC chemokine-independent pulmonary eosinophilia. Although 15-LO can be induced by IL-13 (23), its induction is independent of IL-13Rα1 after IL-4 or allergen challenge. This suggests 15-LO production by a unique cell type compared with mucus- and eotaxin-producing cells and that these 15-LO-producing cells have more type I than type II IL-4R.
Incorporating global transcript expression analysis of the lungs of allergen- and IL-4-challenged Il13ra1−/− mice provided an opportunity to dissect the contribution of IL-13Rα1 and the type II IL-4R to the asthma phenotype. Remarkably, the expression of Chia, a marker of aaMΦ development and a marker and causative molecule for asthma severity (10, 22, 24), was entirely dependent on IL-13Rα1. This result is of particular interest because Arg-1, a hallmark aaMΦ gene (21, 22), was independent of IL-13Rα1 after IL-4 administration but dependent on IL-13Rα1 after allergen challenge whereas other genes (i.e., Retnla and MglI) were entirely independent of IL-13Rα1. Thus, IL-4 may require both the type I and type II IL-4Rs to induce full development of aaMΦ in the lung. Furthermore, these data demonstrate that the precise phenotype of aaMΦ in the allergic lung depends on the stoichiometric relationship between IL-4 and IL-13. This could explain the different results regarding lung expression of arginase 1 in our study (in which IL-13 levels are greater than IL-4 levels) and that by Ramalingam et al. (10) (in which IL-4 levels were greater than IL-13 levels). Alternatively, it is possible that subsets of aaMΦ exist that produce either arginase 1 or chitinase or that the latter molecule is produced by other cells in the lung such as epithelial cells.
In addition, we identified a subset of genes that were induced by allergen challenge and IL-4 and were commonly regulated by IL-13Rα1 such as Scin (Scnderin), Capn9 (Calpain 9), and solute carrier family member 1 (Slc5a1). These newly identified pathways may be important for regulating airway resistance and mucus production in the asthmatic lung.
In summary, our results establish that the critical role for IL-13Rα1 in asthma pathogenesis is mediated by its interactions with both IL-4 and IL-13. Furthermore, we dissociate mechanisms that stimulate cellular infiltration from those that induce airway resistance and goblet cell hyperplasia and emphasize IL-13Rα1 blockade as a potent target for the treatment of increased airway resistance, mucus production, and fibrosis in asthma. As such, these data highlight IL-13Rα1 as a dominant target for disrupting IL-13-, IL-4-, and allergen-mediated effects in the lung.
Materials and Methods
Measurement of the IL-13/Soluble IL-13Rα2 Complex.
Total sIL-13Rα2 and serum levels of IL-13/sIL-13R were measured (28).
Serum IL-4 and IFN-γ Level Determination.
Determination of serum IL-4 and IFN-γ levels were assessed by the in vivo cytokine capture assay (29).
Th2 Polarization.
Goat anti-mouse IgD was injected i.p. (18), and serum IL-4, IFN-γ, and IgE levels were assessed (29).
Cytokine-Induced Airway Inflammation.
Three doses (10 μg per mouse) of IL-13 were administered intratracheally every other day for 4 days. A long-acting form of IL-4 produced by mixing recombinant mouse IL-4 (PeproTech) with a neutralizing mAb (BVD4-1D11) at a 2:1 molar ratio (IL-4C) was administered every other day for 7 days.
Allergen-Induced Airway Inflammation.
Experimental asthma was induced as described (19). Twenty-four hours after the final challenge, the mice were anesthetized and the trachea was cannulated for airway resistance measurements. Subsequently, bronchoalveolar lavage was performed, and the lungs were excised for histological measurements.
Ig and Mediator Assessment.
Serum Igs and BALF cytokines were measured with kits purchased from the following sources: IgA, IgM, IgG1, IgG2a, IgG2b, and IgG3 from Southern Biotech (lower detection limits: 7.8, 15.6, 31.2, 7.8, 7.8, and 15.6 pg/ml, respectively); IgE from BD Bioscience (lower detection limit: 15 pg/ml); and CCL11, CCL24, CCL2, CCL17, IL-4, IL-13, IL-5, IL-10, and active TGF-β from R & D Systems (lower detection limits: 15.6, 15.6, 3.9, 31.2, 6.25, 31.2, 15.6, 31.2, and 31.2 pg/ml, respectively).
Airway Resistance and Compliance Measurements.
Airway resistance was measured by using the flexiVent system (Scireq Scientific Respiratory Equipment). Briefly, the mice were anesthetized, a tracheotomy was performed, and a cannula was inserted. A positive end-expiratory pressure of 0.2 kPa was established. Saline aerosol followed by β-methylcholine (Sigma–Aldrich; 25–100 mg/ml) established control baseline. Aerosols were generated with an ultrasonic nebulizer (DeVilbiss UltraNeb 2000) and delivered to the inspiratory line of the FlexiVent. Each aerosol was delivered for 20 seconds during which time regular ventilation was maintained. Five measurements were made at 25-second intervals, and three peak responses were compared to the mean response of the saline aerosol.
Lung Histopathologic Changes.
Hematoxylin and eosin or periodic acid Schiff (PAS) staining was performed (30).
Microarray Data Analysis.
Whole-lung RNA was extracted by using TRIzol Reagent (Invitrogen Life Technologies). Microarray hybridization to mouse expression array (MOE430 2.1) was performed by the Affymetrix Gene Chip Core facility at Cincinnati Children's Hospital Medical Center (19).
Chemotaxis Assays.
Chemotaxis was assessed by using eosinophils obtained from CD2-IL-5 transgenic mice as described (31). Cells (1.5 × 106) were either untreated or treated with anti-CCR3 or an isotype-matched antibody control (50 μg/ml at 4°C for 30 min) (R & D Systems). Thereafter, the cells were washed and placed in the upper chamber, and 30% BALF (in HBSS) from WT or Il13ra1−/− mice was placed in the lower chamber. After 3 h, total eosinophils in the lower chamber were assessed by using a hemacytometer.
Statistical Analysis.
Data were analyzed by ANOVA followed by the Tukey post hoc test using GraphPad Prism 4. Data are presented as mean ± SD, and values of P < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments.
We thank Tatyana Orekov and Drs. Simon P. Hogan, Gurjit Khurana Hershey, Marsha Wills-Karp, and Carine Blanchard for assistance and/or helpful discussions. This work was supported by National Institutes of Health Grants P01 HL-076383 (to M.E.R. and F.D.F.) and R01 AI057803 (to M.E.R.), a fellowship award (to A.M.) from the Machiah Foundation of the Jewish Community Endowment Fund, the generous support of the Alexander M. and June L. Maisin Foundation, and the Kanbar Charitable Trust.
Footnotes
Conflict of interest statement: F.D.F. is a consultant for Amgen, Abbott, Plexxikon, CSI, Schering–Plough, Centocor, and Peptimmune and has received research support from Abbott, Amgen, Centocor, and Plexxikon. M.E.R. is a consultant for Merck and Ception Therapeutics.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802465105/DCSupplemental.
References
- 1.Wynn TA. IL-13 effector functions. Annu Rev Immunol. 2003;21:425–456. doi: 10.1146/annurev.immunol.21.120601.141142. [DOI] [PubMed] [Google Scholar]
- 2.Elias JA, Lee CG, Zheng T, Shim Y, Zhu Z. Interleukin-13 and leukotrienes: An intersection of pathogenetic schema. Am J Respir Cell Mol Biol. 2003;28:401–404. doi: 10.1165/rcmb.F264. [DOI] [PubMed] [Google Scholar]
- 3.Mattes J, et al. IL-13 induces airways hyperreactivity independently of the IL-4R alpha chain in the allergic lung. J Immunol. 2001;167:1683–1692. doi: 10.4049/jimmunol.167.3.1683. [DOI] [PubMed] [Google Scholar]
- 4.Mentink-Kane MM, Wynn TA. Opposing roles for IL-13 and IL-13 receptor alpha 2 in health and disease. Immunol Rev. 2004;202:191–202. doi: 10.1111/j.0105-2896.2004.00210.x. [DOI] [PubMed] [Google Scholar]
- 5.Fichtner-Feigl S, et al. Induction of IL-13 triggers TGF-beta1-dependent tissue fibrosis in chronic 2,4,6-trinitrobenzene sulfonic acid colitis. J Immunol. 2007;178:5859–5870. doi: 10.4049/jimmunol.178.9.5859. [DOI] [PubMed] [Google Scholar]
- 6.Fichtner-Feigl S, Strober W, Kawakami K, Puri RK, Kitani A. IL-13 signaling through the IL-13alpha2 receptor is involved in induction of TGF-beta1 production and fibrosis. Nat Med. 2006;12:99–106. doi: 10.1038/nm1332. [DOI] [PubMed] [Google Scholar]
- 7.Grunig G, et al. Requirement for IL-13 independently of IL-4 in experimental asthma. Science. 1998;282:2261–2263. doi: 10.1126/science.282.5397.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perkins C, Wills-Karp E, Finkelman FD. IL-4 induces IL-13-independent allergic airway inflammation. J Allergy Clin Immunol. 2006;118:410–419. doi: 10.1016/j.jaci.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 9.McKenzie GJ, Fallon PG, Emson CL, Grencis RK, McKenzie AN. Simultaneous disruption of interleukin (IL)-4 and IL-13 defines individual roles in T helper cell type 2-mediated responses. J Exp Med. 1999;189:1565–1572. doi: 10.1084/jem.189.10.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ramalingam TR, et al. Unique functions of the type II interleukin 4 receptor identified in mice lacking the interleukin 13 receptor alpha1 chain. Nat Immunol. 2008;9:25–33. doi: 10.1038/ni1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershey GK. IL-13 receptors and signaling pathways: An evolving web. J Allergy Clin Immunol. 2003;111:677–690. doi: 10.1067/mai.2003.1333. quiz 691. [DOI] [PubMed] [Google Scholar]
- 12.Chiaramonte MG, et al. Regulation and function of the interleukin 13 receptor alpha 2 during a T helper cell type 2-dominant immune response. J Exp Med. 2003;197:687–701. doi: 10.1084/jem.20020903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wood N, et al. Enhanced interleukin (IL)-13 responses in mice lacking IL-13 receptor alpha 2. J Exp Med. 2003;197:703–709. doi: 10.1084/jem.20020906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tabata Y, et al. Allergy-driven alternative splicing of IL-13 receptor alpha2 yields distinct membrane and soluble forms. J Immunol. 2006;177:7905–7912. doi: 10.4049/jimmunol.177.11.7905. [DOI] [PubMed] [Google Scholar]
- 15.McKenzie AN, Zurawski G. Interleukin-13: Characterization and biologic properties. Cancer Treat Res. 1995;80:367–378. doi: 10.1007/978-1-4613-1241-3_15. [DOI] [PubMed] [Google Scholar]
- 16.Chomarat P, Banchereau J. Interleukin-4 and interleukin-13: Their similarities and discrepancies. Int Rev Immunol. 1998;17:1–52. doi: 10.3109/08830189809084486. [DOI] [PubMed] [Google Scholar]
- 17.Wills-Karp M. Interleukin-13 in asthma pathogenesis. Immunol Rev. 2004;202:175–190. doi: 10.1111/j.0105-2896.2004.00215.x. [DOI] [PubMed] [Google Scholar]
- 18.Finkelman FD, et al. IL-4 is required to generate and sustain in vivo IgE responses. J Immunol. 1988;141:2335–2341. [PubMed] [Google Scholar]
- 19.Zimmermann N, et al. Dissection of experimental asthma with DNA microarray analysis identifies arginase in asthma pathogenesis. J Clin Invest. 2003;111:1863–1874. doi: 10.1172/JCI17912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finkelman FD, et al. Anti-cytokine antibodies as carrier proteins. Prolongation of in vivo effects of exogenous cytokines by injection of cytokine-anti-cytokine antibody complexes. J Immunol. 1993;151:1235–1244. [PubMed] [Google Scholar]
- 21.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–964. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 22.Anthony RM, Rutitzky LI, Urban JR, Jr, Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat Rev Immunol. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuperman DA, et al. Dissecting asthma using focused transgenic modeling and functional genomics. J Allergy Clin Immunol. 2005;116:305–311. doi: 10.1016/j.jaci.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 24.Chupp GL, et al. A chitinase-like protein in the lung and circulation of patients with severe asthma. N Engl J Med. 2007;357:2016–2027. doi: 10.1056/NEJMoa073600. [DOI] [PubMed] [Google Scholar]
- 25.McCoy KD, et al. Natural IgE production in the absence of MHC class II cognate help. Immunity. 2006;24:329–339. doi: 10.1016/j.immuni.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 26.Mathew A, et al. Signal transducer and activator of transcription 6 controls chemokine production and T helper cell type 2 cell trafficking in allergic pulmonary inflammation. J Exp Med. 2001;193:1087–1096. doi: 10.1084/jem.193.9.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavett SH, et al. Interleukin-4 receptor blockade prevents airway responses induced by antigen challenge in mice. Am J Physiol. 1997;272:L253–L261. doi: 10.1152/ajplung.1997.272.2.L253. [DOI] [PubMed] [Google Scholar]
- 28.Khodoun M, et al. Differences in expression, affinity, and function of soluble(s) IL-4Ralpha and sIL-13Ralpha2 suggest opposite effects on allergic responses. J Immunol. 2007;179:6429–6438. doi: 10.4049/jimmunol.179.10.6429. [DOI] [PubMed] [Google Scholar]
- 29.Morris SC, et al. IL-4 induces in vivo production of IFN-gamma by NK and NKT cells. J Immunol. 2006;176:5299–5305. doi: 10.4049/jimmunol.176.9.5299. [DOI] [PubMed] [Google Scholar]
- 30.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM, Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35:337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fulkerson PC, et al. Negative regulation of eosinophil recruitment to the lung by the chemokine monokine induced by IFN-gamma (Mig, CXCL9) Proc Natl Acad Sci USA. 2004;101:1987–1992. doi: 10.1073/pnas.0308544100. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.