Abstract
Background
Dried blood spots (DBSs) are an attractive alternative to plasma for HIV-1 drug resistance testing in resource-limited settings. We recently showed that HIV-1 can be efficiently genotyped from DBSs stored at −20°C for prolonged periods (0.5–4 years). Here, we evaluated the efficiency of genotyping from DBSs stored at 4°C for 1 year.
Methods
A total of 40 DBSs were prepared from residual diagnostic specimens collected from HIV subtype B-infected persons and were stored with desiccant at 4°C. Total nucleic acids were extracted after 1 year using a modification of the Nuclisens assay. Resistance testing was performed using the ViroSeq HIV-1 assay and an in-house nested RT–PCR method validated for HIV-1 subtype B that amplifies a smaller (1 kb) pol fragment.
Results
Using the ViroSeq assay, only 23 of the 40 (57.5%) DBS specimens were successfully genotyped; 22 of these specimens had plasma viraemia >10 000 RNA copies/mL. When the specimens were tested using the in-house assay, 38 of the 40 DBSs (95%) were successfully genotyped. Overall, resistance genotypes generated from the DBSs and plasma were highly concordant.
Conclusions
We show that drug resistance genotyping from DBSs stored at 4°C with desiccant is highly efficient but requires the amplification of small pol fragments and the use of an in-house nested PCR protocol with quality-controlled reagents. These findings suggest that 4°C may represent a suitable temperature for long-term storage of DBSs.
Keywords: resistance testing, ViroSeq assay, 903 filter paper
Introduction
The availability of clinical specimens that can be easily collected, stored and transported is advantageous in areas that lack appropriate infrastructure for blood processing. Whole blood samples collected by finger stick and dried onto a filter paper [dried blood spots (DBSs)] represent an attractive alternative to the conventional collection of blood in tubes. DBSs have been extensively used for HIV-1 antibody testing,1 molecular diagnostics2 and virus load (VL) quantification3–5 and are now considered a convenient alternative to plasma for HIV-1 drug resistance testing. We recently found that drug resistance genotypes generated from DBSs were similar to those derived from plasma in antiretroviral-naive and -experienced patients.6,7 We also found that resistance testing from DBSs can be as sensitive as with plasma when using a genotypic assay that amplifies a large (1.8 kb) pol fragment.6 The high success of amplification was noted using DBSs stored at −20°C in the presence of desiccant, suggesting that −20°C may represent a feasible storage temperature for DBSs. Although these findings were encouraging, storage at −20°C is not always possible in resource-limited settings where 4°C or room temperature may represent a more realistic alternative. Studies on HIV-1 VL determinations using DBSs have reported the efficient amplification of short HIV-1 RNA sequences from filter papers stored at 4°C or room temperature.3,8,9 However, genotypic assays usually rely on the amplification of large pol fragments and are thought to be particularly sensitive to HIV-1 RNA degradation that may occur under suboptimal storage conditions. Here, we report on the efficiency of amplification and drug resistance genotyping from DBSs stored at 4°C for 1 year.
Methods
Preparation and storage of DBSs
We prepared a total of 40 DBSs from residual diagnostic specimens collected from highly antiretroviral-experienced HIV-1 subtype B-infected persons. A more detailed description of the study population including antiretroviral drug treatment has been reported elsewhere.6 Drug resistance genotypes were available for the matched plasma specimens.6 The median plasma VL in these patients was 13 680 RNA copies/mL (range 518–676 694, Versant HIV-1 RNA 3.0 Assay, Bayer HealthCare Diagnostic Division, Tarrytown, NY, USA) (Table 1). DBSs were prepared by pipetting 50 µL of whole blood onto pre-marked circles on 903 filter paper cards (Schleicher & Schuell, Keene, NH, USA). Cards were dried overnight at room temperature (25°C), placed in a gas-impermeable, sealable plastic bag (Fisher Scientific Company, Pittsburgh, PA, USA) containing a silica gel desiccant (Mini Pax Sorbent, Multisorb technologies, Buffalo, NY, USA) and stored at 4°C until they were shipped to CDC in dry ice. Upon arrival, individual plastic bags were put in a larger bag containing three extra desiccants and stored at 4°C. All desiccants were evaluated for humidity at 6 months; 21 of the 40 bags had evidence of humidity as indicated by the colour indicator in the desiccant. Therefore, a new desiccant was added to each individual bag after allowing the spots to equilibrate at room temperature for 30 min. None of the specimens had mould after 12 months of storage at 4°C.
Table 1.
Frequency of amplification of HIV-1 pol from plasma or DBSs stored at −20°C for 6 months or at 4°C for 12 months
| Specimen ID | Plasma VL (log10 copies/mL) | CD4 cells/mm3 | Plasmaa | DBS (−20°C, 6 months, ViroSeq assay)b | DBS (4°C, 12 months, ViroSeq assay) | DBS (4°C, 12 months, in-house assay) |
|---|---|---|---|---|---|---|
| 1 | 676 694 | 32 | + | + | + | + |
| 2 | 266 612 | 195 | + | + | + | + |
| 3 | 214 330 | 195 | + | + | + | + |
| 4 | 191 674 | 48 | + | + | + | + |
| 5 | 170 289 | 663 | + | + | + | + |
| 6 | 90 758 | 324 | + | + | + | + |
| 7 | 63 261 | 169 | + | + | + | + |
| 8 | 51 598 | 324 | + | + | + | + |
| 9 | 52 342 | 457 | + | + | + | + |
| 10 | 49 805 | 450 | + | + | + | + |
| 11 | 37 010 | 152 | + | + | + | + |
| 13 | 30 267 | 525 | + | + | + | + |
| 14 | 28 742 | 378 | + | + | + | + |
| 16 | 23 352 | 221 | + | + | + | + |
| 17 | 23 321 | 168 | + | + | + | + |
| 18 | 21 896 | 391 | + | + | + | + |
| 19 | 17 563 | 289 | + | + | + | + |
| 20 | 16 359 | 609 | + | + | n.s | + |
| 21 | 14 622 | 483 | + | + | + | + |
| 22 | 13 952 | 396 | + | + | + | + |
| 23 | 13 407 | 224 | + | + | + | + |
| 24 | 12 452 | 374 | + | + | − | + |
| 25 | 12 307 | 465 | + | + | + | + |
| 26 | 12 264 | 1320 | + | + | + | + |
| 27 | 11 999 | 81 | + | + | − | + |
| 28 | 11 663 | 336 | + | + | n.s | + |
| 29 | 10 998 | 442 | + | + | n.s. | − |
| 30 | 9955 | 342 | + | + | − | + |
| 31 | 8316 | 288 | + | + | − | + |
| 32 | 4717 | + | + | − | + | |
| 33 | 4349 | 990 | + | + | − | + |
| 34 | 4213 | 1080 | + | + | − | + |
| 36 | 3880 | 434 | + | + | n.s. | + |
| 37 | 3455 | 468 | + | + | − | + |
| 38 | 2240 | 154 | + | + | + | + |
| 39 | 1929 | 416 | + | + | − | + |
| 42 | 1254 | 858 | + | + | − | n.s. |
| 44 | 1045 | 360 | + | + | − | + |
| 45 | 929 | 63 | + | + | n.s. | + |
| 46 | 518 | 240 | + | + | − | + |
n.s., HIV-1 RT and protease were amplified but failed to generate full-length sequences.
aPlasma genotypes were generated using the Viroseq assay except for 1, 3, 18, 32, 33, 39 and 42 (TrueGene assay).
bAmplification from these DBSs stored at −20°C has been reported elsewhere.6
Nucleic acid extraction
Total nucleic acids were extracted from one spot after 1 year of storage at 4°C using a modification of the Nuclisens assay described previously.7 Briefly, a whole spot containing 50 µL of blood was cut with scissors and added into 9 mL of Nuclisens lysis buffer (bioMérieux, Inc., Durham, NC, USA). Special care was taken to avoid contact of the scissors with the blood spots; scissors and forceps were sprayed with 70% ethanol after each use and wiped dry. After 2 h of incubation with lysis buffer at room temperature under gentle rotation, the supernatant was clarified by centrifugation at 250 g for 5 min and then transferred to a clean 15 mL conical tube. Nucleic acids were then extracted following the manufacturer’s instructions, resuspended in 45 µL of elution buffer and stored at −80°C until use.
Drug resistance testing
Resistance testing from DBSs was performed using the ViroSeq HIV-1 Genotyping System (Abbott Molecular, Des Plains, IL, USA) and an in-house reverse transcriptase (RT)-nested PCR method described previously.7 The ViroSeq assay amplifies a 1.8 kb pol fragment and has a sensitivity of detection of 2000 RNA copies/mL plasma. The in-house assay amplifies a 1023 bp fragment of HIV-1 pol comprising amino acids 15–99 of the protease and 1–256 of the RT. This assay has been validated only for HIV-1 subtype B viruses and has a sensitivity of detection of 1000 RNA copies/mL of plasma.7 Genotypes were interpreted using the Stanford Genotypic Resistance Interpretation Algorithm (version 4.2.6) available at http://hivdb.stanford.edu/pages/algs/HIVdb.html.
Results
When the ViroSeq assay was used, only 23 of the 40 (57.5%) DBS specimens stored at 4°C were successfully genotyped; 22 of the successful amplifications were from specimens that had plasma viraemia >10 000 RNA copies/mL (Table 1). An additional five specimens produced amplicons that did not generate full-length RT and protease sequences. Visual inspection of the desiccant bags after 6 months of storage at 4°C showed evidence of humidity in 21 of the 40 specimens. Of these 21 specimens, only those with plasma VL higher than 14 000 RNA copies/mL (12 specimens) were successfully genotyped by the ViroSeq assay at 12 months (data not shown).
As we previously noted using parallel DBSs stored at −20°C, resistance genotypes generated from DBSs and matched plasma specimens were highly concordant. Of the 163 drug resistance mutations identified in these 23 plasma sequences, 158 were also found in sequences generated from DBSs. Of the five mutations absent in DBSs, three were protease mutations, two were major (V82A and I54M) and one minor (L10IT), and two were polymorphisms at codon 333 of the RT (G333E and G333D) (data not shown). We also compared the efficiency of amplification seen in these DBSs with that previously seen in DBSs prepared in parallel from the same patients and stored at −20°C for 6 months.6 Under storage at −20°C, all of the 40 DBS specimens were successfully amplified and genotyped (Table 1).
The ViroSeq assay amplifies a large (1.8 kb) pol fragment in a single round of PCR and may be particularly sensitive to the degradation of HIV-1 nucleic acids that may occur during long-term storage. We, therefore, assessed if the rate of amplification from DBSs stored at 4°C could be improved by using an in-house assay that amplifies a smaller (1023 bp) fragment and uses a nested PCR step.7 Using this method, viruses from 38 of the 40 DBSs (95%) were successfully genotyped (Table 1). Despite the use of a different assay to genotype viruses from these 38 DBS specimens, resistance genotypes were highly concordant with those generated from plasma using the ViroSeq assay; 275 of the 291 mutations found in plasma viruses were also found in viruses from DBSs (data not shown).
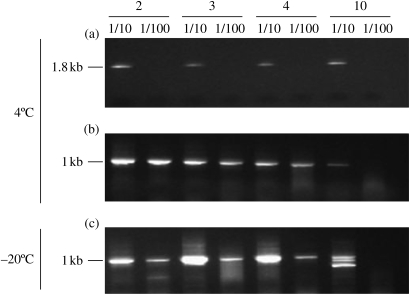
To investigate if the decreased amplification success rate observed with the ViroSeq assay was due to insufficient or degraded HIV nucleic acids, we performed serial dilutions of RNA extracts from high VL specimens and tested them by both the in-house and the ViroSeq assays (Figure 1). Although both assays have comparable sensitivity, amplification signals were only maintained when the shorter pol fragment was amplified by the in-house method. Figure 1 shows that such signals were consistently lost when the larger pol fragment was amplified using the ViroSeq assay, suggesting the possibility of nucleic acid degradation during storage at 4°C. Such a possibility was further supported by parallel testing of serial dilutions of RNA extracts prepared from specimens stored at −20°C. Figure 1(c) shows that amplification signals generated from these specimens using the in-house assay were generally stronger than those obtained from the same DBS specimens stored at 4°C (Figure 1b).
Figure 1.
Agarose gel showing the HIV-1 pol amplification signals obtained from DBS specimens stored at 4 or −20°C. Two serial 10-fold dilutions (1/10 and 1/100) of specimens 2 (266 612 RNA copies/mL), 3 (214 330 RNA copies/mL), 4 (191 674 RNA copies/mL) and 10 (49 805 RNA copies/mL) were prepared in Nuclisens elution buffer and were tested in parallel using the ViroSeq assay (1.8 kb) or the in-house RT-nested PCR method (1023 bp). (a) DBS specimens stored at 4°C and amplified using the ViroSeq assay. (b) DBS specimens stored at 4°C and amplified using the in-house method. (c) DBS specimens stored at −20°C and amplified using the in-house method.
Discussion
Our study on a small number of specimens suggests that storage at 4°C may represent a feasible alternative to −20°C for long-term storage of DBSs. However, we noted that the efficient genotyping from these DBS specimens required the use of an in-house nested PCR assay and that the ViroSeq assay failed to genotype a substantial proportion of specimens. The results with the ViroSeq assay differed from our previous findings with this assay showing a high success of genotyping from DBSs stored at −20°C and suggest that some degradation of RNA may have occurred during long-term storage at 4°C possibly due to suboptimal storage temperature, humidity or both. Such a possibility was suggested by the weaker amplification signals observed in RNA extracts from specimens stored at 4°C compared with those stored at −20°C upon serial dilutions. Although different efficiencies of RNA extraction could partially explain these differences, we found that a high proportion of the specimens stored at 4°C had evidence of humidity at 6 months, whereas none of the specimens stored at −20°C did. High humidity conditions are thought to be detrimental to resistance testing from DBSs given the extreme sensitivity of HIV nucleic acids to degradation in the presence of humidity.10 Nonetheless, we showed that it is possible to overcome potential losses in HIV-1 RNA integrity and efficiently genotype from DBSs stored at 4°C by using an in-house nested PCR protocol that amplifies a smaller fragment using stringent, quality-controlled reagents. These results are encouraging and expand on our earlier findings showing the efficient genotyping of HIV-1 pol from DBSs stored at −20°C for up to 4 years.6,7 A high success of genotyping has also been noted from DBSs stored for 3 months at 37°C and 85% humidity in the presence of desiccant.10 Under these extreme conditions, viruses from ∼85% of the DBS specimens could be genotyped by using in-house nested PCR assays that amplified small (700 bp), overlapping pol sequences.10 The use of nested PCR protocols and/or the amplification of small fragments may likely result in higher amplification success rates from DBSs stored under less optimal conditions. Such a possibility was supported by our findings, showing a better maintenance of amplification efficiencies using the in-house method upon serial dilution of RNA extracts. Despite having comparable sensitivity, amplifications were rapidly lost when a larger fragment was amplified using the ViroSeq assay.
The main objective of our study was to evaluate the amplification success rate from DBSs stored at 4°C for a prolonged period of time. We used an in-house assay validated for HIV-1 subtype B since our study population included patients infected with subtype B viruses. It is important to note, however, that information on the performance of our assay with non-B subtypes is limited to a small number of specimens.7 Therefore, we caution that resistance testing from DBSs collected in areas with prevalent non-B subtypes will require in-house assays appropriately validated for other HIV subtypes.
The ability to collect blood samples on filter paper represents an advantage for HIV drug resistance surveillance and monitoring, particularly in areas that lack the appropriate infrastructure for plasma processing and transport. Much effort is currently underway to define the best conditions that will facilitate resistance testing from DBSs including the most appropriate storage temperature and time. The findings reported here suggest that 4°C may represent a feasible storage temperature for long-term storage of DBSs and add to the promise of DBSs as a convenient specimen type for HIV-1 drug resistance testing.
Funding
Work at the CDC was done with intramural funding. Work at Hospital Carlos III was supported by grants from the Fondo de Investigación Sanitaria (FIS, CP06/284 and PI06/1826).
Transparency declarations
None to declare.
Disclaimers
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the Public Health Service, or the Centers for Disease Control and Prevention.
References
- 1.Solomon SS, Pulimi S, Rodriguez, et al. Dried blood spots are an acceptable and useful HIV surveillance tool in a remote developing world setting. Int J STD AIDS. 2004;15:658–61. doi: 10.1177/095646240401501005. [DOI] [PubMed] [Google Scholar]
- 2.Beck IA, Drennan KD, Melvin AJ, et al. Simple, sensitive, and specific detection of human immunodeficiency virus type 1 subtype B DNA in dried blood samples for diagnosis in infants in the field. J Clin Microbiol. 2001;39:29–33. doi: 10.1128/JCM.39.1.29-33.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brambilla D, Jennings C, Aldrovandi G, et al. Multicenter evaluation of use of dried blood and plasma spot specimens in quantitative assays for human immunodeficiency virus RNA: measurement, precision, and RNA stability. J Clin Microbiol. 2003;41:1888–93. doi: 10.1128/JCM.41.5.1888-1893.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fiscus SA, Brambilla D, Grosso L, et al. Quantitation of human immunodeficiency virus type 1 RNA in plasma by using blood dried on filter paper. J Clin Microbiol. 1998;36:258–60. doi: 10.1128/jcm.36.1.258-260.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Shea S, Mullen J, Corbett K, et al. Use of dried whole blood spots for quantification of HIV-1 RNA. AIDS. 1999;13:630. doi: 10.1097/00002030-199904010-00019. [DOI] [PubMed] [Google Scholar]
- 6.Masciotra S, Garrido C, Youngpairoj Ae S, et al. High concordance between HIV-1 drug resistance genotypes generated from plasma and dried blood spots in antiretroviral-experienced patients. AIDS. 2007;21:2503–11. doi: 10.1097/QAD.0b013e3281c618db. [DOI] [PubMed] [Google Scholar]
- 7.McNulty A, Jennings C, Bennett D, et al. Evaluation of dried blood spots for HIV-1 drug resistance testing. J Clin Microbiol. 2007;45:517–21. doi: 10.1128/JCM.02016-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alvarez-Muñoz MT, Zaragoza-Rodríguez S, Rojas-Montes O, et al. High correlation of human immunodeficiency virus type-1 viral load measured in dried-blood spot samples and in plasma under different storage conditions. Arch Med Res. 2005;36:382–6. doi: 10.1016/j.arcmed.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Cassol S, Gill MJ, Pilon R, et al. Quantification of human immunodeficiency virus type 1 RNA from dried plasma spots collected on filter paper. J Clin Microbiol. 1997;35:2795–801. doi: 10.1128/jcm.35.11.2795-2801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertagnolio S, Soto-Ramirez L, Pilon R, et al. HIV-1 drug resistance surveillance using dried whole blood spots. Antiviral Ther. 2007;12:107–13. [PubMed] [Google Scholar]