Abstract
Despite substantial work, the phylogeny of malaria parasites remains debated. The matter is complicated by concerns about patterns of evolution in potentially strongly selected genes as well as the extreme AT bias of some Plasmodium genomes. Particularly contentious has been the position of the most virulent human parasite Plasmodium falciparum, whether grouped with avian parasites or within a larger clade of mammalian parasites. Here, we study 3 classes of rare genomic changes, as well as the sequences of mitochondrial ribosomal RNA (rRNA) genes. We report 3 lines of support for a clade of mammalian parasites: 1) we find no instances of spliceosomal intron loss in a hypothetical ancestor of P. falciparum and the avian parasite Plasmodium gallinaceum, suggesting against a close relationship between those species; 2) we find 4 genomic mitochondrial indels supporting a mammalian clade, but none grouping P. falciparum with avian parasites; and 3) slowly evolving mitochondrial rRNA sequences support a mammalian parasite clade with 100% posterior probability. We further report a large deletion in the mitochondrial large subunit rRNA gene, which suggests a subclade including both African and Asian parasites within the clade of closely related primate malarias. This contrasts with previous studies that provided strong support for separate Asian and African clades, and reduces certainty about the historical and geographic origins of Plasmodium vivax. Finally, we find a lack of synapomorphic gene losses, suggesting a low rate of ancestral gene loss in Plasmodium.
Keywords: apicomplexan evolution, phylogenetic methods, rare genomic characters, parasite evolution
Introduction
Appropriate molecular markers and methods for reconstruction of Plasmodium phylogeny have long been subjects of debate. A very early molecular study compared the GC content of Plasmodium genomes across species (McCutchan et al. 1984). The first individual locus to be analyzed, the small subunit (SSU) ribosomal RNA (rRNA), eventually fell under suspicion due to the unusual presence of multiple copies in Plasmodium and their apparent concerted evolution (Corredor and Enea 1993). A second widely studied molecule, the circumsporozoite surface protein gene, may also not be an appropriate marker due to its apparent evolution under strong diversifying/balancing selection (Hughes 1991). In general, the extreme AT richness of the Plasmodium nuclear and chloroplast genomes provides unique challenges in sequence and phylogenetic analysis (e.g., Dávalos and Perkins 2008).
In particular, the evolutionary position of the most virulent human parasite Plasmodium falciparum and the closely related chimpanzee parasite Plasmodium reichenowi has been the subject of debate. Some of the best evidence to date comes from 2 particularly thorough studies of mitochondrial cytochrome b and nuclear SSU rRNA and from a recent analysis of 4 genes from the 3 organellar genomes, each of which has placed these species as sister to other mammalian parasites (Qari et al. 1996; Perkins and Schall 2002; Martinsen et al. 2007). However, an alternative clade grouping P. falciparum and P. reichenowi with avian parasites also continues to receive support (Waters et al. 1991; Escalante and Ayala 1994; Escalante et al. 1995, 1997, 1998; McCutchan et al. 1996; Leclerc et al. 2004). Recently, 1 study went so far as to argue that currently available data are insufficient to resolve the question (Hagner et al. 2007).
In cases of difficult to resolve relationships, the use of so-called rare genomic characters/changes can provide an alternative source of phylogenetic signal (Rokas and Holland 2000). Given their lower rate of change, such characters might maintain phylogenetic signal after some sequence characters have saturated. Here we study 3 alternative sources of data: 1) loss/gain of spliceosomal introns; 2) mitochondrial genomic indels; and 3) nuclear gene losses, as well as the sequences of mitochondrial rRNA genes. Our findings corroborate the existence of a clade of mammalian malaria parasites including P. falciparum and P. reichenowi. In addition, a ∼100-bp deletion including the terminal 21 bp of one fragment of the mitochondrial large subunit (LSU) ribosomal RNA gene suggests against a clade of Asian primate malaria parasites, possibly complicating the origin of the human parasite Plasmodium vivax.
Methods
Alignment and Analysis of Mitochondrial Genomes
We downloaded complete mitochondrial genome sequences for 20 Plasmodium, Haemoproteus, and Leucocytozoon species from GenBank (table 1). Overall, AT content of the sequences was much lower than for the nuclear genomes, ranging from 67.0% to 70.2%. Complete sequences were aligned in stand-alone ClustalW using default parameters. The sequence labeled Leucocytozoon sabrazesi was found to be highly divergent and was excluded from indel sequence analysis. Sequences corresponding to rRNA sequences as annotated for the Plasmodium gallinaceum sequence were extracted and concatenated. AT content for these regions was lower still, ranging from 58.2 to 59.8%. Indels were identified and analyzed by eye.
Table 1.
Complete Mitochondrial Genomes and Species Abbreviations
| Parasite | Host | Abbreviation | GenBank Accession Number |
| Plasmodium of primates | |||
| Plasmodium cynomolgi | Macaques | Pcynm | AY800108.1 |
| Plasmodium falciparum | Homo sapiens | Pfalc | AY282930.1 |
| Plasmodium fragile | Macaca sinica and Macaca radiata | Pfrag | AY722799.1 |
| Plasmodium gonderi | Mangabey monkeys | Pgond | AY800111.1 |
| Plasmodium knowlesi | Macaca irus | Pknow | AY722797.1 |
| Plasmodium reichenowi | Pan troglodytes | Preic | AJ251941.1 |
| Plasmodium simiovale | M. sinica | Psimv | AY800109.1 |
| Plasmodium simium | Alouatta fuscus | Psimi | AY722798.1 |
| Plasmodium sp. DAJ-2004 | Mandrillus leucophaeus | P-DAJ | AY800112.1 |
| Plasmodium vivax | H. sapiens | Pviva | NC_007243.1 |
| Plasmodium of rodents | |||
| Plasmodium berghei | Grammomys surdaster | Pberg | AF014115.1 |
| Plasmodium chabaudi | Thamnomys rutilans | Pchab | AF014116.1 |
| Plasmodium yoelii | T. rutilans | Pyoel | M29000.1 |
| Plasmodium of birds | |||
| Plasmodium gallinaceum | Gallus gallus | Pgall | AB250690.1 |
| Plasmodium juxtanucleare | Domestic fowl | Pjuxt | AB250415.1 |
| Plasmodium relictum | Zenaida macroura | Preli | AY733090.1 |
| Haemoproteus | |||
| Haemoproteus sp. jb1.JA27 | Meliphaga lewinii | H-jb1 | AY733086.1 |
| Haemoproteus sp. jb2.SEW5141 | Lichenostomus frenatus | H-jb2 | AY733087.1 |
| Leucocytozoon | |||
| Leucocytozoon caulleryi | G. gallus | Lcaul | AB302215.1 |
| Leucocytozoon sabrazesi | G. gallus | Lsabr | AB299369.1 |
NOTE.—Representative hosts are drawn from a combination of sources (Garnham 1966; Perkins and Schall 2002; Martinsen et al. 2007).
Phylogenetic Analysis of rRNA Sequences
We constructed a concatamer of 14 pieces of the highly fragmented rRNA genes from the mitochondrial long and short ribosomal subunits. We performed Neighbor-Joining (NJ) and Bayesian inference analyses. The NJ analysis was performed on MEGA 4.0, using a Kimura 2-parameters model with gamma distributed rates (gamma parameter = 1) and complete deletion. A total of 5,000 replicates were carried out for the bootstrap analysis. The phylogeny by Bayesian inference was performed using the MrBayes3.1.2 software. We assumed a general time-reversible model with a gamma-shaped distribution of rates. We ran 1,000,000 tree generations with a sample frequency of 100, and the first 100,000 were discarded before inferring the tree with the highest posterior probability. Leucocytozoon species were used as outgroups. We repeated the same analysis using smaller groups of 4 species to show that the signal holds even with scant taxon sampling. We used the following groups: P. falciparum–P. gallinaceum–Leucocytozoon caulleryi–P. vivax and P. falciparum–P. gallinaceum–L. caulleryi–Plasmodium yoelii. Results from NJ and Bayesian inferences were highly consistent.
Loss of Ancestral Genes in Plasmodium
We downloaded the genomes and annotations of Theileria parva (GenBank accession numbers AAGK01000001–AAGK01000009), Toxoplasma gondii (http://www.toxodb.org), and P. falciparum (http://www.plasmodb.org) and the genome assemblies for P. vivax and P. gallinaceum (http://www.plasmodb.org). We identified putative orthologs between T. parva and T. gondii by reciprocal searches using stand-alone BlastP. We then scored presence/absence in P. falciparum by BlastP searches of the T. parva representative against the predicted P. falciparum proteome. For each case of apparent gene loss in P. falciparum, we next performed TBlastN searches using the T. parva representative against the P. gallinaceum and P. vivax genome assemblies in order to identify potential synapomorphic losses. For all searches, we used a cutoff E value of 10−10.
Results
Spliceosomal Intron Loss
Following previous studies utilizing spliceosomal intron losses and gains as phylogenetic characters (Venkatesh et al. 1999; Nguyen et al. 2005; Roy and Gilbert 2005a; Zheng et al. 2007; Roy and Irimia 2008), we studied spliceosomal intron loss and gain in Plasmodium. We previously showed that intron loss/gain is very rare in Plasmodium, with 2,185 shared introns but only 27 intron gain/losses in conserved coding regions between P. falciparum and the rodent parasite P. yoelii (Roy and Hartl 2006). In all, 26 of these 27 introns were also found to be shared with the avian parasite P. gallinaceum; no homologous P. gallinaceum sequence could be found for the remaining intron (Roy and Hartl 2006). Four pairs of adjacent P. falciparum introns are absent in P. yoelii, suggesting their concerted loss by recombination with a reverse transcribed copy of an mRNA (Frugoli et al. 1998; Niu et al. 2005; Roy and Gilbert 2005b), yielding a total of 22 (multiple) intron gain/loss events in conserved regions (table 2).
Table 2.
Summary of Intron Loss/Gain Pattern for 27 Observed Loss/Gain Events from a Previous Study (Roy and Penny 2006)
| Plasmodium gallinaceum | Plasmodium falciparum | Plasmodium yoelii | Events (Introns) |
| + | + | − | 15 (19) |
| + | − | + | 7 (7) |
| ? | − | + | 1 (1) |
| − | − | + | 0 (0) |
NOTE.—Cases of intron absence at multiple adjacent positions in P. yoelii are consistent with coincident loss of multiple introns and are treated as single events.
We sought to test the hypothesis that P. falciparum arose via lateral transfer from an avian host. Assuming a P. gallinaceum–P. falciparum affinity, P. falciparum has experienced 7 loss events since the P. gallinaceum–P. falciparum divergence (shared P. gallinaceum–P. yoelii introns absent in P. falciparum), but none before the divergence (no P. yoelii-specific introns). If the P. gallinaceum–P. yoelii divergence and the P. gallinaceum–P. falciparum divergence occurred at times Tgy and Tgf in the past (Tgy > Tgf), respectively, the probability that all 7 losses in P. falciparum postdate the P. gallinaceum–P. falciparum divergence is P = (Tgf/Tgy)7, assuming constant rate of loss. P < 0.05 for Tgf/Tgy < 0.65, suggesting that the P. gallinaceum–P. falciparum divergence is not recent relative to the P. gallinaceum–P. yoelii divergence. Assuming that the 15 introns (or intron pairs) that are absent in P. yoelii are all due to intron loss, the probability of observing 15 losses in P. yoelii in Tgy but no losses in the P. falciparum–P. gallinaceum ancestor over Tgy − Tgf My, is [Tgy/(2Tgy − Tgf)]15 assuming constant rate of loss. P < 0.05 for Tgf/Tgy < 0.78, again suggesting against a recent P. falciparum–P. gallinaceum divergence. This situation is not eased by attributing some of the shared P. falciparum–P. gallinaceum introns to intron gain because in this case the lack of intron gains in P. yoelii over a longer time period is unexpected.
If avian (e.g., P. gallinaceum) and most mammalian (e.g., P. yoelii) parasites diverged coincident with their hosts (bird–mammal divergence ∼315 MYA, Reisz and Müller 2004), these arguments suggest that P. falciparum is unlikely to have diverged from P. gallinaceum less than 205–246 MYA (i.e., 0.65 × 315–0.78 × 315 MYA). Assuming ancient host switching, the P. yoelii–P. gallinaceum divergence could be more recent; however, unless this divergence too reflects a transfer between mammals and bird ancestors, it cannot postdate the mammalian radiation ∼210 MYA (monotreme–therian divergence; Hugall et al. 2007), suggesting against a P. gallinaceum–P. falciparum divergence less than 136–164 MYA (i.e., 0.65 × 210–0.78 × 210 MYA). Each of these dates is well before the estimated radiation of birds ∼115 MYA (Hugall et al. 2007), suggesting against an avian–mammalian transfer leading to P. falciparum.
Notably, shifts in rates of intron loss/gain are by no means impossible, as very large rate shifts have been observed within apicomplexan parasites (Roy and Penny 2006); however, to explain the data in terms of changes in intron loss rates through Plasmodium history requires 2 independent shifts, 1 each to explain the greater loss rates in P. yoelii and in P. falciparum relative to that in the putative P. falciparum–P. gallinaceum ancestor. In total, these findings complicate the hypothesis of a recent P. falciparum–P. gallinaceum divergence.
Alignment of Complete Mitochondrial Genomes
We aligned publicly available full-length mitochondrial genome sequences from 16 Plasmodium species, as well as 2 species of Haemoproteus (bird and reptile parasites closely related to Plasmodium) and 2 species of Leucocytozoon (a speciose genus of avian parasites; table 1). Genomes show more moderate AT contents than for Plasmodium nuclear and apicoplast genomes (80.6% and 86.9%, respectively in P. falciparum), ranging from 67.0 to 70.2% AT. The genomes are highly conserved, with 83.8% and 99.9% conservation between species pairs across the entire genome at ungapped positions.
Analysis of Mitochondrial Indels
We identified indels for which either 1) there were clearly only 2 lengths, suggesting a single indel event and/or 2) 1 long indel (>20 bp) was shared among 2 or more sequences. In total, we identified 22 phylogenetically informative indels. Supporting their phylogenetic utility, the majority of indels supported well-established groups, including P. falciparum–P. reichenowi (5 indels), other primate parasites (1 indel), rodent parasites (4 indels), the 2 Haemoproteus species (2 indels), mammalian parasites other than P. falciparum–P. reichenowi (2 indels), and a clade containing Plasmodium floridense and Plasmodium mexicanum (1 indel). Only 1 indel showed clear homoplasy, grouping all nonrodent Plasmodium parasites.
Consistent with the existence of a clade of all mammalian parasites, 4 indels (as well as 1 borderline case) supported this clade, whereas no characters supported the alternative clade grouping of P. falciparum–P. reichenowi with bird and/or lizard parasites. Very few characters yielded insights to the grouping of bird parasites, with only a single character which groups bird Plasmodium (but not Haemoproteus) parasites.
Shared Deletion in LS1
Intriguingly, we also identified a ∼100-bp deletion including the last 21 bp of the LS1 gene fragment of the LSU (fig. 1) that was shared between 2 studied African primate parasites (Plasmodium gonderi and an unknown parasite taken from a mandrill) and 2 Asian primate parasites (Plasmodium cynomolgi and Plasmodium simiovale). The region is otherwise conserved across all species, consistent with this deletion representing a single deletion event. This is in contrast to evidence from cytochrome b sequences strongly supporting separate clades of Asian and African parasites (Perkins and Schall 2002).
FIG. 1.—
A long deletion spanning the 3′ end of the LS1 rRNA gene suggests a clade grouping sampled African primate parasites (Plasmodium gonderi and a species isolated from a mandrill [P-DAJ]) with some Asian parasites (Plasmodium cynomolgi and Plasmodium semiovale) within the clade of closely related primate parasites (bold). The deletion spans both the 3′ terminus of the LS1 gene (first line of the alignment) and downstream intergenic sequence (subsequent lines of the alignment). The alignment corresponds to positions 5140–5314 in the Plasmodium falciparum genome sequence (GenBank accession number AJ276844.1).
Phylogenetic Analysis of Mitochondrial rRNA Gene Sequences
Next, we extracted regions of the alignment corresponding to the highly fragmented mitochondrial rRNA genes and analyzed ungapped positions. In all, 14/15 fragments of the small and large rRNA subunits show very high levels of sequence conservation. The other LSU fragment, LS2, appears to evolve largely by microsatellite indels and was excluded from the analysis.
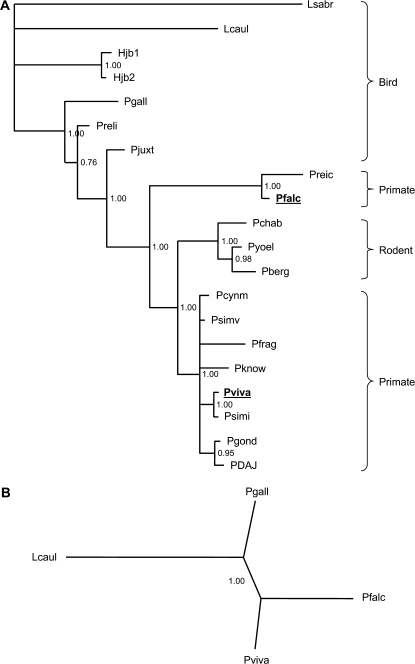
We concatenated the 14 remaining rRNA gene fragments, comprising 1,306 ungapped positions (somewhat fewer than the 2,334 bp recently analyzed by Martinsen et al. [2007]). Across the entire concatenated region, species pairs showed between 93.4% and 100.0% sequence identity at ungapped positions. We reconstructed the phylogeny from this alignment using standard protocols (see Methods). The results are given in figure 2a. The mammalian clade is strongly supported with 100% posterior probability.
FIG. 2.—
Phylogenetic tree of Plasmodium parasites and relatives reconstructed by Bayesian methods from 14 mitochondrial rRNA genes. (a) Complete reconstruction using 20 genomes gives 100% posterior support for a mammalian malaria clade. (b) Reconstruction using only 4 representative genomes gives 100% posterior probability for grouping Plasmodium falciparum with Plasmodium yoelii.
The clarity of the signal supporting the mammalian clade is underscored by its recovery with strong Bayesian posterior probability (again, 100%) when only a few taxa are used: quartets of taxa including P. gallinaceum, P. falciparum, L. caulleryi, and either P. vivax or P. yoelii, both recovered the grouping of P. falciparum with other mammalian parasites (fig. 2b). These findings suggest that mitochondrial rRNA genes may be of use in resolving relationships within genera for other groups of apicomplexans.
Nuclear Gene Losses
Finally, we studied apparent gene losses in Plasmodium. We performed reciprocal BlastP searches to identify putative ortholog pairs between the distantly related apicomplexans T. gondii and T. parva. We then Blasted the T. parva representative against the predicted P. falciparum proteome. In all, 68/1,331 genes lacked Blast hits (E value > 10−10), suggesting loss in P. falciparum. TBlastN searches of these genes against the P. yoelii and P. gallinaceum genomes indicated absence in both genomes for most genes (56/68), presence in both genomes in 4 cases (apparent lineage-specific loss in P. falciparum), and 8 cases of differential presence between the 2 species, which are possible synapomorphic gene losses (7 genes were present in P. gallinaceum and 1 in P. vivax). However, upon further inspection, most of these cases were found to have borderline Blast hits in both species, suggesting that they are not true synapomorphic gene losses. In total then, we found very little phylogenetic signal from gene losses, with only 2 single informative genes, 1 each supporting the 2 alternative relationships.
Discussion
We used rare genomic changes to study phylogeny of Plasmodium and Haemoproteus parasites. Three separate analyses—loss/gain events of spliceosomal introns, mitochondrial indels, and rRNA genes from mitochondrial genomes—each support the existence of a clade of mammalian parasites including primate and rodent parasites along with the more distant P. falciparum and P. reichenowi. These results corroborate that of previous studies—1 of nuclear SSU rRNA sequences (Qari et al. 1996), 1 of cytochrome b (Perkins and Schall 2002), and a very recent multilocus analysis (Martinsen et al. 2007)—that used dense species sampling, and underscore that the alternative signal which groups P. falciparum–P. reichenowi with avian parasites likely reflects an artifact of long-branch attraction and problems associated with extreme base composition (Dávalos and Perkins 2008).
Interestingly, we also discovered a substantial mitochondrial deletion of the 3′ region of a LS1 rRNA gene fragment, which was found in 2 African primate parasites as well as a subset of Asian primate parasites. This deletion of an otherwise highly conserved region seems unlikely to be homoplastic, indicating that these 4 species form a clade, in contrast to previous studies suggesting separate Asian and African clades (Perkins and Schall 2002).
This finding has implications for the origin of P. vivax. The geographic and historical origins of P. vivax pose something of a mystery. On the one hand, the very high incidence of the Duffy negative blood type, which confers resistance to P. vivax, in western Africa but not in Asia suggests a long history of P. vivax in Africa; on the other hand, several other factors, including haplotype analysis, reconstructions of P. vivax demographic history, and host–parasite cophylogeny reconstruction, suggest an Asian origin (Mu et al. 2005, and references therein). As the current results undermine the finding of an Asian clade, they weaken the support for an Asian origin of P. vivax. Notably, grouping of the 2 Asian macaque parasites with the 2 African parasites (P. gonderi and a parasite of unknown species taken from a mandrill) could possibly reduce the number of required host switches to explain the cophylogeny of parasites and hosts (Mu et al. 2005). However, it is important not to discount the possibility of frequent movement of parasites and hosts over large geographic distances, in which case relationships between species based on geographic range may not be informative about point of origin of a given species.
We also studied gene loss among Plasmodium species but found only 2 phylogenetically informative gene losses. This is perhaps surprising in light of the paradigm of reductive evolution occurring among parasites, particularly intracellular parasites (Andersson and Kurland 1998; Darby et al. 2007). Interestingly, this lack of clear ongoing gene loss echoes the lack of intron loss in Plasmodium evolution (Roy and Hartl 2006; Nguyen et al. 2007). The forces driving reductive genome evolution in parasitic species, and the limits thereof, remain important questions in understanding the biology of eukaryotic parasites.
Concluding Remarks
We report that a variety of alternative genomic characters corroborate the finding of a clade comprising mammalian malaria parasites but not the finding of a clade of Asian primate parasites. These results represent another example of the utility of rare genomic characters in phylogenetic analysis as a complementary approach to resolving controversial phylogenetic questions.
Acknowledgments
SWR thanks Martine Zilversmit for critical comments on a very early draft of the manuscript and for general tutoring in apicomplexan phylogenetics. MI was funded by the Spanish Ministerio of Educación y Ciencia, through the FPI grant (BFU2005-00252), and SWR by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/DHHS. We thank Eugene Koonin and Jordi Garcia-Fernàndez and their groups for intellectual support and stimulation, for financial support, and for fostering environments of open intellectual exploration in their respective groups.
References
- Andersson SG, Kurland CG. Reductive evolution of resident genomes. Trends Microbiol. 1998;6:263–268. doi: 10.1016/s0966-842x(98)01312-2. [DOI] [PubMed] [Google Scholar]
- Corredor V, Enea V. Plasmodial ribosomal RNA as phylogenetic probe: a cautionary note. Mol Biol Evol. 1993;10:924–926. doi: 10.1093/oxfordjournals.molbev.a040039. [DOI] [PubMed] [Google Scholar]
- Darby AC, Cho NH, Fuxelius HH, Westberg J, Andersson SG. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 2007;23:211–220. doi: 10.1016/j.tig.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Dávalos LM, Perkins SL. Saturation and base composition bias explain phylogenomic conflict in Plasmodium. Genomics. 2008 doi: 10.1016/j.ygeno.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci USA. 1994;91:11373–11377. doi: 10.1073/pnas.91.24.11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–626. doi: 10.1093/oxfordjournals.molbev.a040241. [DOI] [PubMed] [Google Scholar]
- Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci USA. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante AA, Goldman IF, De Rijk P, De Wachter R, Collins WE, Qari SH, Lal AA. Phylogenetic study of the genus Plasmodium based on the secondary structure-based alignment of the small subunit ribosomal RNA. Mol Biochem Parasitol. 1997;90:317–321. doi: 10.1016/s0166-6851(97)00121-7. [DOI] [PubMed] [Google Scholar]
- Frugoli JA, McPeek MA, Thomas TL, McClung CR. Intron loss and gain during evolution of the catalase gene family in angiosperms. Genetics. 1998;149:355–365. doi: 10.1093/genetics/149.1.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnham PCC. Malaria parasites and other Haemosporidia. Oxford: Blackwell; 1966. [Google Scholar]
- Hagner S, Misof B, Maier W, Kampen H. Bayesian analysis of new and old malaria parasite DNA sequence data demonstrates the need for more phylogenetic signal to clarify the descent of Plasmodium falciparum. Parasitol Res. 2007;101:493–503. doi: 10.1007/s00436-007-0499-6. [DOI] [PubMed] [Google Scholar]
- Hugall AF, Foster R, Lee MS. Calibration choice, rate smoothing, and the pattern of tetrapod diversification according to the long nuclear gene RAG-1. Syst Biol. 2007;56:543–563. doi: 10.1080/10635150701477825. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Circumsporozoite protein genes of malaria parasites (Plasmodium spp.): evidence for positive selection on immunogenic regions. Genetics. 1991;127:345–353. doi: 10.1093/genetics/127.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc MC, Hugot JP, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. Parasitol. 2004;129:677–684. doi: 10.1017/s0031182004006146. [DOI] [PubMed] [Google Scholar]
- Martinsen ES, Perkins SL, Schall JJ. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): evolution of life-history traits and host switches. Mol Phylogenet Evol. 2007;47:261–273. doi: 10.1016/j.ympev.2007.11.012. [DOI] [PubMed] [Google Scholar]
- McCutchan TF, Dame JB, Miller LH, Barnwell J. Evolutionary relatedness of Plasmodium species as determined by the structure of DNA. Science. 1984;225:808–811. doi: 10.1126/science.6382604. [DOI] [PubMed] [Google Scholar]
- McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Braga EM, Krettli AU, Miller LH. Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci USA. 1996;93:11889–11894. doi: 10.1073/pnas.93.21.11889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Joy DA, Duan J, et al. (11 co-authors) Host switch leads to emergence of Plasmodium vivax malaria in humans. Mol Biol Evol. 2005;22:1686–1693. doi: 10.1093/molbev/msi160. [DOI] [PubMed] [Google Scholar]
- Nguyen H, Yoshihama M, Kenmochi N. New maximum likelihood estimators for eukaryotic intron evolution. PLoS Comput Biol. 2005;1:e79. doi: 10.1371/journal.pcbi.0010079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HD, Yoshihama M, Kenmochi N. The evolution of spliceosomal introns in alveolates. Mol Biol Evol. 2007;24:1093–1096. doi: 10.1093/molbev/msm037. [DOI] [PubMed] [Google Scholar]
- Niu D-K, Hou W-R, Li S-W. mRNA-mediated intron losses: evidence from extraordinarily large exons. Mol Biol Evol. 2005;22:1475–1481. doi: 10.1093/molbev/msi138. [DOI] [PubMed] [Google Scholar]
- Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Qari SH, Shi YP, Pieniazek NJ, Collins WE, Lal AA. Phylogenetic relationship among the malaria parasites based on small subunit rRNA gene sequences: monophyletic nature of the human malaria parasite, Plasmodium falciparum. Mol Phylogenet Evol. 1996;6:157–165. doi: 10.1006/mpev.1996.0068. [DOI] [PubMed] [Google Scholar]
- Reisz RR, Müller J. Molecular timescales and the fossil record: a paleontological perspective. Trends Genet. 2004;20:237–241. doi: 10.1016/j.tig.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Rokas A, Holland PWH. Rare genomic changes as a tool for phylogenetics. Trends Ecol Evol. 2000;15:454–459. doi: 10.1016/s0169-5347(00)01967-4. [DOI] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. Resolution of a deep animal divergence by the pattern of intron conservation. Proc Natl Acad Sci USA. 2005a;102:4403–4408. doi: 10.1073/pnas.0409891102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The pattern of intron loss. Proc Natl Acad Sci USA. 2005b;102:713–718. doi: 10.1073/pnas.0408274102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Hartl DL. Very little intron loss/gain in Plasmodium: intron loss/gain mutation rates and intron number. Genome Res. 2006;16:750–756. doi: 10.1101/gr.4845406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Irimia M. Rare genomic characters do not support Coelomata: intron loss/gain. Mol Biol Evol. 2008;25:620–623. doi: 10.1093/molbev/msn035. [DOI] [PubMed] [Google Scholar]
- Roy SW, Penny D. Large-scale intron conservation and order-of-magnitude variation in intron loss/gain rates in apicomplexan evolution. Genome Res. 2006;16:1270–1275. doi: 10.1101/gr.5410606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Ning Y, Brenner S. Late changes in spliceosomal introns define clades in vertebrate evolution. Proc Natl Acad Sci USA. 1999;96:10267–10271. doi: 10.1073/pnas.96.18.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters AP, Higgins DG, McCutchan TF. Plasmodium falciparum appears to have arisen as a result of lateral transfer between avian and human hosts. Proc Natl Acad Sci USA. 1991;88:3140–3144. doi: 10.1073/pnas.88.8.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Rogozin IB, Koonin EV, Przytycka TM. Support for the Coelomata clade of animals from a rigorous analysis of the pattern of intron conservation. Mol Biol Evol. 2007;24:2583–2592. doi: 10.1093/molbev/msm207. [DOI] [PubMed] [Google Scholar]