Abstract
Plasmodium vivax in southern Mexico exhibits different infectivities to 2 local mosquito vectors, Anopheles pseudopunctipennis and Anopheles albimanus. Previous work has tied these differences in mosquito infectivity to variation in the central repeat motif of the malaria parasite's circumsporozoite (csp) gene, but subsequent studies have questioned this view. Here we present evidence that P. vivax in southern Mexico comprised 3 genetic populations whose distributions largely mirror those of the 2 mosquito vectors. Additionally, laboratory colony feeding experiments indicate that parasite populations are most compatible with sympatric mosquito species. Our results suggest that reciprocal selection between malaria parasites and mosquito vectors has led to local adaptation of the parasite. Adaptation to local vectors may play an important role in generating population structure in Plasmodium. A better understanding of coevolutionary dynamics between sympatric mosquitoes and parasites will facilitate the identification of molecular mechanisms relevant to disease transmission in nature and provide crucial information for malaria control.
Keywords: malaria, Plasmodium vivax, microsatellites, coevolution
Introduction
Worldwide, 30–40 Anopheles mosquito species are capable of transmitting malaria (Kiszewski et al. 2004), but the susceptibility of a mosquito to malaria infection and its ability to successfully transmit the disease is highly variable among species (Billingsley and Sinden 1997; Alavi et al. 2003) and even among genotypes within a species (Lambrechts et al. 2005). Coadaptation of vectors and parasites has been proposed to explain these differences (Cohuet et al. 2006). Local adaptation, whereby parasites exhibit greater infectivity on local as opposed to foreign hosts, is predicted when the parasite has an evolutionary advantage over the coevolving host, such as a shorter generation time, larger population size, or higher recombination rate (Kaltz and Shykoff 1998; Thompson 1999; Kawecki and Ebert 2004; Brandt et al. 2007). In order to be transmitted to humans, the malaria parasite must complete a series of developmental stages within the mosquito vector, where it encounters a number of population bottlenecks (reviewed in Abraham and Jacobs-Lorena 2004). Parasite adaptations that optimize transmission success on local hosts might contribute to the mosaic of malaria parasite–vector compatibilities observed in nature.
In southern Mexico, Plasmodium vivax parasites exhibit different compatibilities with respect to 2 local mosquito vectors, Anopheles pseudopunctipennis and Anopheles albimanus. Previous studies (Gonzalez-Ceron et al. 1999; Rodriguez et al. 2000) have suggested an association between mosquito compatibility and the central repeat motif of the malaria circumsporozoite (csp) gene that encodes a major surface protein of malaria sporozoites. The csp variants differ from each other in the amino acid composition of the central repetitive region (Rosenberg et al. 1989; Kain et al. 1992). Two variants, termed VK210 and VK247, are circulating in southern Mexico where their distributions roughly mirror those of the vectors: VK210 is found with An. albimanus on the coast and VK247 co-occurs with An. pseudopunctipennis in the foothills (Rodriguez et al. 2000). The vectors have largely nonoverlapping geographic distributions (Manguin et al. 1995; Rubio-Palis and Zimmerman 1997), although both mosquitoes and csp variants are found in the town of Tapachula, situated between the 2 ecoregions (elevation: 120–220 m).
Despite the apparent association between csp variants and mosquito compatibility (Gonzalez-Ceron et al. 1999; Rodriguez et al. 2000), the circum-sporozoite protein (CSP) is unlikely to control the observed compatibility phenotypes. Subsequent studies indicate that destruction of the incompatible parasite occurs prior to the expression of the CSP: VK210 parasites were unable to successfully traverse the An. pseudopunctipennis peritrophic membrane (Gonzalez-Ceron et al. 2007), and VK247 parasites in An. albimanus were destroyed during midgut invasion and early oocyst development (Gonzalez-Ceron et al. 2001). Further, in Columbia the reverse pattern was observed: An. albimanus fed on blood infected with VK247 parasites had a higher number of salivary gland sporozoites than those fed on VK210-infected blood (Gonzalez et al. 2001).
It is well known that underlying genetic population structure can result in spurious associations between candidate loci and a phenotype (Falush et al. 2003). If P. vivax in southern Mexico is divided into genetically isolated populations, then the association between csp variants and mosquito compatibility may simply reflect this. In this study, we surveyed P. vivax using a panel of recently developed microsatellites (MSs) to assess the presence of population structure. We found that P. vivax in our sample area comprised 3 genetic populations and that each population optimally infected the sympatric mosquito species. This study highlights the potentially misleading effect of population structure on association analysis and the importance of employing a coevolutionary framework to study epidemiologically relevant traits such as infectivity.
Materials and Methods
DNA Samples, Genome-wide Amplification, and Microsatellite Typing
Plasmodium vivax DNA samples were extracted from preexisting anonymous blood spots on Whatman No. 2 filter paper collected from Chiapas State, Mexico, between 1997 and 2005 (Gonzalez-Ceron et al. 1999). A total of 230 DNA samples were analyzed for the current study. MS loci were selected from approximately 160 genome-wide MSs identified by the Carlton laboratory. We used 16 Mexican isolates to subscreen 50 of these loci. Twenty-six loci were rejected either because they were monomorphic or because results were difficult to interpret due to the presence of stutter peaks and/or imperfect repeats. Primers and a description of the remaining 24 MS loci are given in supplementary table 1 (Supplementary Material online).
The small amount of parasite DNA contained in a single blood spot was not sufficient for the multimarker typing performed here. Therefore, we used whole-genome amplification with the Qiagen Repli-g Kit (Qiagen, Valencia CA) to increase genomic DNA yield. Whole-genome–amplified DNA was washed with ethanol to remove extraneous peaks observed in preliminary analyses. Typically, 2 μl of stock DNA extracted from filter paper resulted in a 50-μl sample with a DNA concentration of 200–300 ng/μl. Loci were amplified in 10-μl reactions (1.5 mM MgCl2, 2 μM of each primer, and 0.5 U Taq polymerase) following standard protocols (Schlötterer 1998). Polymerase chain reaction (PCR) parameters were as follows: 2 min initial denaturation at 94 °C, then 42 cycles of 20 s at 94 °C, 10 s at 52 °C, 10 s at 47 °C, and 30 s at 60 °C, followed by 5 min at 60 °C. Length variation of PCR products was measured on a 16 capillary ABI3100 DNA Genetic Analyzer (Applied Biosystems, Foster City, CA). Fragment size polymorphism was analyzed using GENEMAPPER software, also from Applied Biosystems.
Estimate of Genetic Diversity
The predominant allele present in each PCR product was used to estimate allele frequencies at each locus. MS variability was measured by expected heterozygosity (He) (Nei 1978) and variance in repeat number (Vr) (Moran 1975), using the program MICROSATELLITE ANALYZER v4.05 (Dieringer and Schlotterer 2003). He and Vr were corrected for small sample sizes by multiplying n by n/(n − 1), where n is the number of chromosomes analyzed. To assess the level of multiple infections in our sample, we counted the proportion of isolates with a single allele at all 24 loci. We also measured the mean number of alleles per locus per infection.
Determination of Population Structure
We used the Bayesian model–based clustering method implemented in STRUCTURE v 2.1 (Pritchard et al. 2000) to test for the presence of population structure in our sample. The number of genetic partitions (K) was inferred by calculating the probability of the data given a certain prior value of K, [P(D|K)], over a large number of Markov chain Monte Carlo (MCMC) iterations. Genetic partitions were characterized by different allele frequencies, and individuals were probabilistically assigned to 1 or more partition according to their allele distribution. MCMC searches consisted of 500,000 “burn-in” steps followed by 1,000,000 iterations. To check for convergence of P(D|K), we performed 10 replicate runs at each K value from 1–10 under the admixture model with correlated allele frequencies. To aid in determining the most appropriate number of partitions, we used an ad hoc quantity that evaluates the second-order rate of change of the likelihood function with respect to K (Evanno et al. 2005).
We estimated population pairwise FST values for the partitions defined in the STRUCTURE analysis using θ (Weir and Cockerham 1984) in Arlequin 3.11 (Excoffier et al. 2005). The significance of pairwise FST estimates was calculated using permutation tests (N = 10,000) and Bonferroni corrections for multiple comparisons.
Principle Coordinate Analysis (PCA)
Relationships among genetic partitions were visualized using principal coordinate analysis (PCA; Gower 1966) performed in Genalex 6 (Peakall and Smouse 2006). PCA generates a set of orthogonal axes for which each successive dimension maximizes the remaining variance in the data. Input for PCA analysis consisted of individual pairwise genetic distance matrices of 1) the proportion of shared alleles (Dps,) and 2) Nei's standard genetic distance corrected for sample size (D), calculated using MsAnalyzer (Dieringer and Schlotterer 2003).
Multivariate Spatial Genetic Autocorrelation
We used multivariate spatial genetic autocorrelation (Smouse and Peakall) to test the hypothesis that P. vivax genetic structure has a spatial component that reflects the distributions of the 2 vectors, An. pseudopunctipennis and An. albimanus. We measured correlations between P. vivax pairwise geographic and pairwise squared genetic distance matrices using the program Genalex (Peakall and Smouse 2006). The autocorrelation coefficient r provides a measure of pairwise genetic similarity among isolates within a particular distance size class. We used progressively larger size classes starting with 5 km and increasing up to the maximum distance within the study area (70 km). Ten thousand random permutations of genotypes among geographic locations were used to generate a null distribution under the hypothesis of no genetic structure (Smouse and Peakall 1999; Peakell et al. 2003). In addition, 10,000 bootstrap replicates of pairwise comparisons were used to generate 95% confidence intervals (CIs) for the autocorrelation coefficient r (Peakell et al. 2003). We performed the analysis twice, both with and without samples from Tapachula.
Mosquito Infectivity Experiments
Data from 183 laboratory colony mosquito feeding experiments conducted previously at the Centro de Investigacion de Paludismo in Tapachula between 1997 and 2005 (Gonzalez-Ceron and Rodriguez 1991; Ramsey et al. 1994; Gonzalez-Ceron et al. 1999, 2007) were reanalyzed for associations between parasite population and mosquito infectivity. Only feeding experiments with no missing data were included. Both the prevalence (proportion of infected mosquitoes) and the infection intensity (mean number of oocysts) were measured.
For the analysis of infection prevalence, we used log-linear models on the number of infected mosquitoes with an offset for total number of mosquitoes. We expressed results using the relative rates (RRs) of infecting An. pseudopunctipennis compared with An. albimanus and the ratio of those relative rates (RRR). For example, to examine the effects of csp variant, we used the RRR for VK247 versus VK210:
where P(AP,VK247) is the probability of infecting An. pseudopunctipennis when the csp repeat variant was VK247, and the other notation is analogous. Values of RRR >1 in this example indicate that the RR of infecting An. pseudopunctipennis compared with An. albimanus is larger when the csp repeat variant is VK247. We explored several of these models to describe this RRR with and without additional effects for parasite genetic population. Further, we used the log-linear models to estimate several different RRRs to examine the RR of infecting An. pseudopunctipennis compared with An. albimanus for different pairs of genetic populations. To control for correlations within parasite isolate, we used the generalized estimating equation (GEE) with the working independence correlation structure. We performed Wald-type tests with t distributions and K–p degrees of freedom (dfs), where K is the number of unique parasite isolates and p is the number of parameters (Lipsitz et al. 1994).
For the analysis of mean oocyst number, we also used a GEE with a working identity matrix in order to account for the correlation within parasite isolate. As with infection prevalence, we used the t distribution on the Wald test with K–p dfs. We compared means 1) among genetic population within each mosquito and 2) between mosquitoes for each genetic population. To test for significant interaction between mosquito species and parasite genotype, we again used the GEE method with an F distribution and K–p dfs. GEE analyses were carried out in R 2.5.1 using the gee package (Carey 2007; R Development Core Team 2007).
Results
Parasite Samples and Genetic Diversity
Parasite samples from a total of 98 localities within a 100-km radius in Chiapas, Mexico, were included in this study (supplementary table 2, Supplementary Material online). MSs loci were located on 9 of the 14 chromosomes (supplementary table 2, Supplementary Material online). Multiple alleles were scored at a given locus if minor peaks were more than one-third the height of the predominant peak (Imwong et al. 2007). Using this criterion, the majority of our samples appear to be single infections. The mean number of alleles per locus per infection was 1.01 (standard deviation ± 0.01). Out of 234 typed samples, 197 (84%) carried a single allele at each locus. Among the 37 putatively mixed infections, 33 (89%) showed multiple alleles at only 1–2 loci out of 24, suggesting that they represent mutations within infection rather than multiple infections (Imwong et al. 2007). The 4 isolates with >2 loci showing multiple alleles were assumed to be mixed infections and were removed from further analyses. He for the 24 MS loci ranged from 0.12 to 0.87. Three markers with low He (0.12–0.21) were retained on the assumption that even markers with low mutation rates would contribute information for the estimation of demographic processes.
Highly Differentiated Plasmodium vivax Populations
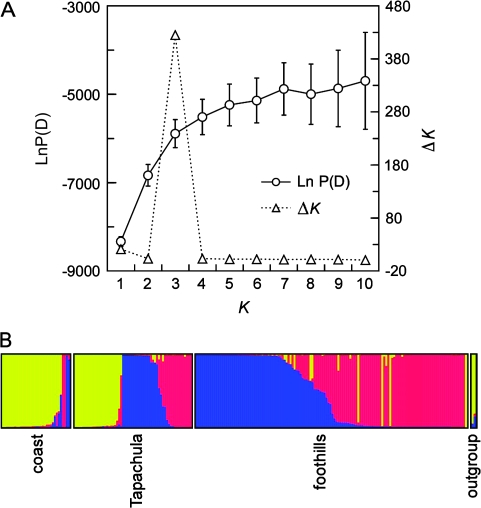
Our data showed that P. vivax in southern Mexico exhibits population structure and restricted gene flow in an area of less than 100 km2. STRUCTURE analysis indicated that P. vivax comprised 3 genetic subpopulations. The estimated logarithm of the probability of the data [P(D|K)] plateaued at K = 5–6 (fig. 1A). However, the K estimator derived from the rate of change of K (ΔK) (Evanno et al. 2005) indicated that the best estimate of K was 3. One population, hereafter termed c1, includes the majority of coastal samples (88%), as well as outgroup isolates 045V from Brazil and Sal1 from El Salvador (fig. 1B). The other 2 populations, hereafter termed f1 and f2, predominate in the foothills and included a number of individuals with mixed ancestry (fig. 1B).
FIG. 1.—
Population structure of Plasmodium vivax in southern Mexico. (A) Plot of the log probability of the data [ln P(D)] and the second-order rate of change constant (ΔK) for K = 1–10. A peak for ΔK at K = 3 suggests that 3 populations best fit the data. (B) Proportional membership of isolates in the 3 clusters identified by the STRUCTURE program. Each isolate is represented by a thin vertical line. Colors identify genetic clusters: yellow, c1; blue, f1; red, f2. Isolates are organized by geographical origin.
Significant genetic differentiation was detected among all 3 populations (FST = 0.279–0.364). He, median Vr, number of alleles, and number of private alleles were all highest in c1 (table 1), even though this sample size was smaller than the other 2 (58 vs. 83 and 89). Populations f1 and f2 showed similar levels of genetic diversity for all measures. These results suggest that the coastal parasite population may be either older or larger than the 2 foothill populations. The distributions of both mosquitoes extend beyond our study area (Manguin et al. 1995; Molina-Cruz et al. 2004), and it is likely that the parasite populations are more extensive than what we have sampled here.
Table 1.
Variability Measures for 24 MS Loci in Plasmodium vivax from Southern Mexico
| Population | n | He | Vr | Mean No. of Alleles | Mean No. of Private Alleles |
| c1 | 59.2 | 0.71 | 15.7 | 7.9 | 3.6 |
| f1 | 82.7 | 0.32 | 6.7 | 4.3 | 0.3 |
| f2 | 88.4 | 0.43 | 9.8 | 4.4 | 0.4 |
| Total | 230.4 | 0.61 | 14.4 | 9.1 | — |
NOTE.—n, mean effective number of chromosomes; He, mean heterozygosity among 24 MS markers; Vr, median variance in repeat number.
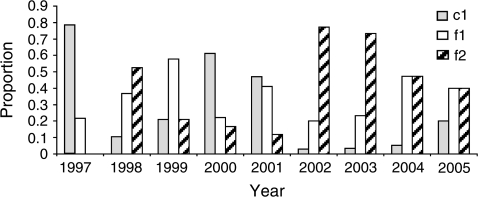
We observed temporal fluctuations in the proportional representation of the P. vivax populations in our sample (fig. 2). The proportion of c1 isolates was significantly lower during the period of 2002–2005 as compared with 1997–2001 (z = −2.62, P = 0.008). The opposite pattern was seen for f2, which comprised a significantly higher proportion of the sampled isolates during 2002–2005 as compared with 1997–2001 (z = −3.14, P = 0.0009). The proportion of f1 isolates remained relatively unchanged throughout the sampling period.
FIG. 2.—
Temporal variation in the incidence of Plasmodium vivax populations c1, f1, and f2 in parasite samples from Chiapas, Mexico, during 1997–2005.
Restricted Gene Flow Between Plasmodium vivax Populations
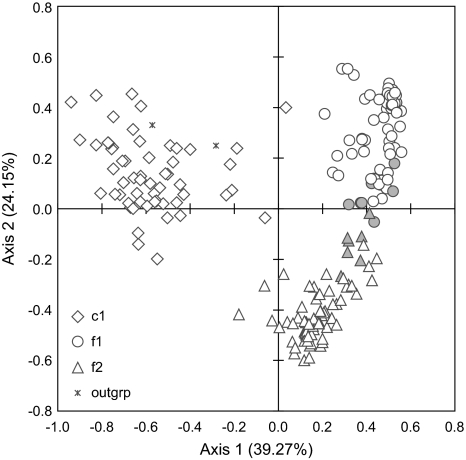
To better characterize the degree of genetic relatedness among P. vivax populations, we performed principal coordinates analysis (PCA). Results based on Dps and D did not differ greatly from each other; therefore, we present results for Dps only (fig. 3). The 3 genetic populations showed little overlap. Axis 1 primarily separated c1 from f1 and f2, whereas axis 2 further separated f2 from f1. We observed a cline rather than discreet clusters for f1 and f2 isolates. The dispersion of samples along a line suggests mixed ancestry (Patterson et al. 2006), and this is confirmed by the central location of admixed f1/f2 individuals along the cline (fig. 3).
FIG. 3.—
PCAs of the Plasmodium vivax populations. Plot of the first 2 PCA axes based on the proportion of shared alleles (Dps). The percentage of variation explained by each axis is indicated. Individuals with mixed f1/f2 ancestry (membership >0.20 in each) are shown as gray.
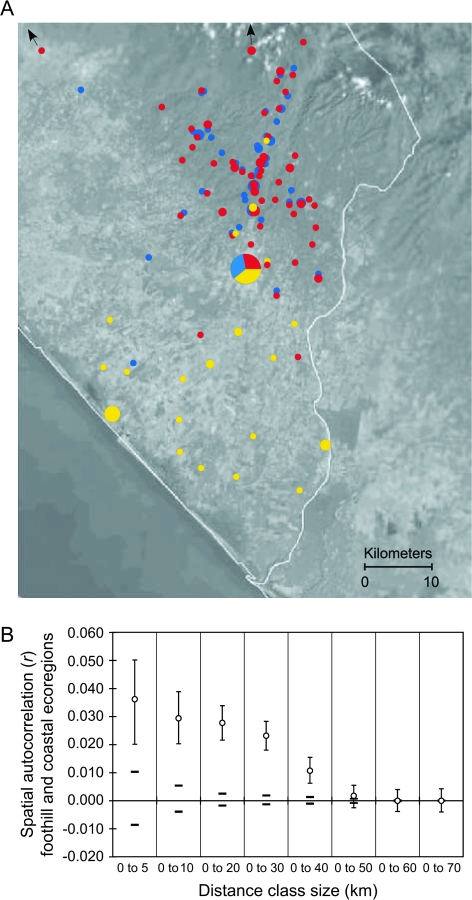
We detected significant positive spatial structure extending up to 40 km (fig. 4). Results did not differ greatly for analyses including Tapachula isolates; therefore, we present only the results from the non-Tapachula samples here. In the presence of positive structure, r, the autocorrelation coefficient, should decrease with increasing distance class size (Peakell et al. 2003) and the distance class for which r is no longer significant can be taken as the upper limit of positive structure (Double et al. 2005). There was no evidence of significant structure within either the coastal or the foothill ecoregions when analyzed separately (data not shown) despite the presence of 2 parasite populations in the foothills. The mean distance between coastal and foothill sites is 34.3 km (SD ± 10.2), and the observed positive autocorrelation extending to 40 km is likely to reflect restricted gene flow between parasites collected from the coastal and foothill ecoregions.
FIG. 4.—
Spatial genetic structure of Plasmodium vivax in southern Mexico. (A) Study area in southern Chiapas State, Mexico. Circle size is proportional to frequency in the sample, with the exception of Tapachula (n = 135). Individuals with mixed ancestry are shown as a single color corresponding to the population with the highest membership proportion. Yellow, c1; blue, f1; red, f2. (B) Spatial genetic autocorrelation (r) across multiple distance classes. Samples from the town of Tapachula were not included. Ninety-five percent CIs for estimates of r based on 1,000 bootstrap replicates are shown as error bars, and 95% CIs based on 1,000 random permutations for the null hypothesis of no spatial autocorrelation are shown by the bold dashes surrounding r = 0.
Enhanced Infectivity of Sympatric Mosquito Species
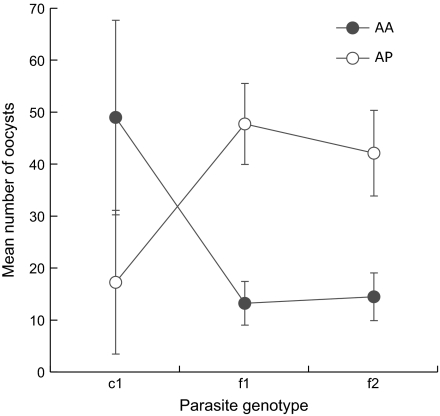
There was a clear phenotypic separation between c1 and f1/f2 populations with respect to mean oocyst number in the 2 mosquito species, with each parasite population being more compatible with the sympatric mosquito (fig. 5). The main effects of mosquito species and parasite population were both significant (P = 8.32 × 10−04 and P = 0.0016, respectively). The mean number of oocysts was significantly different for c1 compared with f1/f2 within each vector. No differences between f1 and f2 were observed in either vector. Most importantly in terms of coevolutionary dynamics, the interaction between mosquito species and parasite genetic population had a highly significant effect (P = 1.191 × 10−08; fig. 5).
FIG. 5.—
Comparison of the infection intensities of 3 parasite populations in 2 mosquito species. Anopheles pseudopunctipennis (AP) and Anopheles albimanus (AA) were challenged with blood infected with Plasmodium vivax isolates belonging to 1 of 3 populations: c1, f1, and f2. Error bars represent 95% CIs. Data are from 183 laboratory colony feeding experiments conducted between 1997 and 2005. A strong interaction between parasite genetic background and mosquito species can be seen.
We also examined the RR of infecting An. pseudopunctipennis compared with An. albimanus for the CSP VK247 variant over the analogous RR for the VK210 variant, termed the ratio of relative rate (RRR) of VK247 versus VK210. We first modeled this RRR without accounting for parasite population, giving an RRR of 8.82 (95% CI = 5.33–14.62), indicating elevated infectivity in An. pseudopunctipennis for VK247 parasites. A separate analysis for each parasite population (c1, f1, f2) shows that this effect is confined to c1 (RRR = 106.74); however, the CI is unreliable as there is only one c1/VK247 isolate in our sample (table 2). Removing this isolate does not greatly alter the overall RR (8.75, 95% CI = 5.31–14.43). Further examination of the RR of infecting An. pseudopunctipennis compared with An. albimanus for each parasite population/CSP variant combination revealed that the overall RRR is being driven by the low RR for c1/VK210 (table 2). Within populations f1 and f2, CSP variant had no effect on infection prevalence.
Table 2.
The RR of Infecting Anopheles pseudopunctipennis Compared with Anopheles Albimanus (e.g., an RR of 5 Means that the Rate of Infection is 5 Times Larger for An. pseudopunctipennis than for An. albimanus)
| Genetic Population/csp Variant | Isolates (n) | Mosquitoes (n) | 95% CI | ||
| RR | Lower | Upper | |||
| c1/VK247 | 1 | 35 | 12.500 | NaN | NaN |
| c1/VK210 | 36 | 1081 | 0.117 | 0.056 | 0.244 |
| f1/VK247 | 66 | 2006 | 5.025 | 4.050 | 6.235 |
| f1/VK210 | 7 | 200 | 4.500 | 1.295 | 15.636 |
| f2/VK247 | 54 | 1546 | 6.269 | 4.627 | 8.494 |
| f2/VK210 | 19 | 616 | 4.495 | 2.472 | 8.174 |
| c1/ALL | 37 | 1116 | 0.170 | 0.075 | 0.384 |
| f1/ALL | 73 | 2206 | 4.872 | 3.921 | 6.055 |
| f2/ALL | 73 | 2162 | 5.679 | 4.294 | 7.512 |
NOTE.—NaN, not a number.
When CSP variant is excluded from the model, the RRR for f1 versus c1 is 28.64 (95% CI = 12.33–66.55) and for f2 versus c1 is 33.39 (95% CI = 14.12–78.99). In contrast, the RRR for f1 versus f2 does not differ from 1 (RRR = 0.858, 95% CI = 0.602–1.222). These data support the hypothesis that c1 is better adapted to infect An. albimanus and f1 and f2 are better adapted to infect An. pseudopunctipennis.
Discussion
Adaptation of a parasite to its local host is predicted when the parasite is under stronger selection pressure and/or has a greater evolutionary potential (e.g., larger population sizes, shorter generation times, and higher mutation rates) than the host (Gandon et al. 1996; Gandon and Michalakis 2002; Brandt et al. 2007). We have identified distinct P. vivax genetic populations in southern Mexico and have demonstrated that they differ markedly from each other in their ability to successfully infect local mosquito vectors. Further, we showed that sympatric mosquito–parasite combinations are more compatible than allopatric ones, constituting evidence for local adaptation (Woolhouse et al. 2002). Our data support the view that asymmetries in the coevolutionary arms race between mosquito and parasite favor local adaptation of the parasite. Selection pressure is predicted to be greater for the parasite than for the mosquito because successful host exploitation is essential for parasite survival, whereas the cost of infection incurred by the mosquito (Lyimo and Koella 1992; Hogg and Hurd 1997) must be balanced against the cost of maintaining resistance mechanisms (Hurd et al. 2006). Additionally, in low transmission regions such as southern Mexico, where infection incidence among wild-caught mosquitoes is <0.01%, most hosts may never encounter a parasite.
Although 3 parasite populations were identified, 2—f1 and f2—are not distinguishable from each other with regard to mosquito infectivity, geographic distribution, or genetic diversity. One possible explanation for the identification of 2 populations in the foothills is that one may constitute a more recent immigration to the region and that not enough time has elapsed for complete admixture to have occurred between them. The distribution of f1/f2 haplotypes in the PCA plot as 2 widely separated foci connected by a smaller number of haplotypes is suggestive of this scenario (fig. 3), as is the absence of f2 in the 1997 collection (fig. 2), although we cannot completely rule out the possibility that f1/f2 constitute a single parasite population.
Both ecological and evolutionary factors associated with the mosquitoes may act to promoted divergence and restrict gene flow among parasite lineages adapted to different vectors. Anopheles albimanus and An. pseudopunctipennis differ in their ecological requirements, resulting in minimal overlap in both space and time. Anopheles albimanus is coastal and is found mainly at elevations <100 m. Anopheles pseudopunctipennis prefers higher elevations and is often the only vector above 600 m (Rubio-Palis and Zimmerman 1997). Although their distributions overlap in and around Tapachula where all 3 parasite populations were collected, we found little evidence for recombination between c1 and f1/f2 isolates (fig. 1B). Anopheles albimanus densities peak during the rainy season (May–November) coinciding with a peak in coastal malaria infections. In contrast, An. pseudopunctipennis densities and the number of malaria infections in the foothills peak during the dry season (December–April) and are associated with pools forming in dry river beds (Savage et al. 1990) that are eliminated by heavy rains during the rainy season (Fernandez-Salas et al. 1994).
Anopheles albimanus and An. pseudopunctipennis are not closely related but rather belong to nonsister subgenera Nyssorhynchus and Anopheles, respectively (Krzywinski et al. 2001). They likely diverged >90 to 106 MYA, the estimated divergence time for sister subgenera Cellia and Anopheles (Krzywinski et al. 2006). Thus, these 2 mosquitoes present potentially divergent environments to developing malaria parasites. As such, adaptations for successful transmission in one species may not be advantageous in the other. The strong interaction between parasite genetic background and mosquito species that we observed in oocyst number supports this view.
A limitation of this study is its dependence on laboratory mosquito colonies that are likely to have greatly reduced genetic diversity compared with natural mosquito populations, due to selection and genetic drift (Norris et al. 2001). On the other hand, we have shown that the infectivity phenotype is consistent across a range of parasite genotypes collected during a 7-year period. Further infection experiments with recently established mosquito colonies are needed to directly address the contribution of the mosquito genotype.
Although the effects of artificial selection (i.e., drug pressure) on Plasmodium genetic diversity have been extensively studied (Wootton et al. 2002; Roper et al. 2004; Nash et al. 2005; Laufer et al. 2006; Nair et al. 2007), this is to our knowledge the first study to examine the effects of natural selection. Here we have identified strong local adaptation of malaria parasites to their vectors on a small geographic scale, resulting in distinct parasite populations. The previously observed association between csp alleles and mosquito species in southern Mexico can be attributed to the confounding effects of population structure. The presence of only a single c1/VK247 isolate in our sample was surprising given that CSP repeat variants are segregating freely in f1 and f2 and that c1 is more genetically diverse than f1 or f2. Nonetheless, the overall geographical distribution of CSP variants strongly suggests that CSP does not play a role in adaptation to these local vectors. Vector-mediated population structure may also explain the differences in susceptibility to csp variants observed for Anopheles aquasalis and Anopheles darlingi in Brazil (Silva et al. 2006).
Local adaptation and vector–parasite compatibilities may play an important role in generating population structure in Plasmodium, especially in Central and South America, where extensive genetic subdivision on small geographic scales has been observed for both Plasmodium falciparum and P. vivax. (Anderson et al. 2000; Machado et al. 2004; Ferreira et al. 2007). Local adaptation to vectors offers a possible explanation for the paradoxical finding of high genetic diversity coupled with strong linkage disequilibrium in P. vivax in Brazil (Ferreira et al. 2007), as well as the absence of a relationship between geographic and genetic distance observed for P. falciparum, also in Brazil (Machado et al. 2004). Spatial heterogeneity in the prevalence and distribution of lineages has also been reported for avian malaria on an even smaller geographic scale than this study (Wood et al. 2007). We have shown that genetically distinct P. vivax populations differ from each other in terms of vector compatibility, but the mechanism and genes responsible for these differences remain unknown. We have, however, identified the relevant components for a genetic cross (c1 vs. f1/f2) and subsequent genome-wide scan for loci under selection, a necessary next step to identify parasite genes contributing to vector–parasite compatibility.
Supplementary Materials
Supplementary tables 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
This work was supported by Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health. Marco A. Sandoval and Frida Santillan provided technical assistance with sample collection and csp genotyping. We thank intramural editor Brenda Rae Marshall for assistance.
References
- Abraham EG, Jacobs-Lorena M. Mosquito midgut barriers to malaria parasite development. Insect Biochem Mol Biol. 2004;34:667–671. doi: 10.1016/j.ibmb.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Alavi Y, Arai M, Mendoza J, et al. (11 co-authors) The dynamics of interactions between Plasmodium and the mosquito: a study of the infectivity of Plasmodium berghei and Plasmodium gallinaceum, and their transmission by Anopheles stephensi, Anopheles gambiae and Aedes aegypti. Int J Parasitol. 2003;33:933–943. doi: 10.1016/s0020-7519(03)00112-7. [DOI] [PubMed] [Google Scholar]
- Anderson TJC, Haubold B, Williams JT, et al. (16 co-authors) Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–1482. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- Billingsley PF, Sinden RE. Determinants of malaria-mosquito specificity. Parasitol Today. 1997;13:297–301. [Google Scholar]
- Brandt M, Fischer-Blass B, Heinze J, Foitzik S. Population structure and the co-evolution between social parasites and their hosts. Mol Ecol. 2007;16:2063–2078. doi: 10.1111/j.1365-294X.2007.03300.x. [DOI] [PubMed] [Google Scholar]
- Carey VJ. 2007 gee: Generalized Estimation Equation solver. Ported to R by T. Lumley and B. Ripley. R package Version 4. 13–13. [Google Scholar]
- Cohuet A, Osta MA, Morlais I, Awono-Ambene PH, Michel K, Simard F, Christophides GK, Fontenille D, Kafatos FC. Anopheles and Plasmodium: from laboratory models to natural systems in the field. EMBO Rep. 2006;7:1285–1289. doi: 10.1038/sj.embor.7400831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieringer D, Schlotterer C. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Mol Ecol Notes. 2003;3:167–169. [Google Scholar]
- Double M, Peakall R, Beck N, Cockburn A. Dispersal, philopatry, and infidelity: dissecting local genetic structure in Superb Fairy-wrens (Malurus cyaneus) Evolution. 2005;59:625–635. [PubMed] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin. Version. 3.0: an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Salas I, Rodriguez MH, Roberts DR, Rodriguez MC, Wirtz RA. Bionomics of adult Anopheles pseudopunctipennis (Diptera: culicidae) in the Tapachula foothills area of southern Mexico. J Med Entomol. 1994;31:663–670. doi: 10.1093/jmedent/31.5.663. [DOI] [PubMed] [Google Scholar]
- Ferreira M, Karunaweera N, da Silva-Nunes M, da Silva NS, Wirth D, Hartl D. Population structure and transmission dynamics of Plasmodium vivax in rural Amazonia. J Infect Dis. 2007;195:1218–1226. doi: 10.1086/512685. [DOI] [PubMed] [Google Scholar]
- Gandon S, Capowiez Y, Dubois Y, Michalakis Y, Olivieri I. Local adaptation and gene-for-gene coevolution in a metapopulation model. Proc R Soc Lond B Biol Sci. 1996;263:1003–1009. [Google Scholar]
- Gandon S, Michalakis Y. Local adaptation, evolutionary potential and host-parasite coevolution: interactions between migration, mutation, population size and generation time. J Evol Biol. 2002;15:451–462. [Google Scholar]
- Gonzalez-Ceron L, Rodriguez MH. An enzyme-linked immunosorbent assay using detergent-soluble Plasmodium vivax antigen for seroepidemiological surveys. Trans R Soc Trop Med Hyg. 1991;85:358–361. doi: 10.1016/0035-9203(91)90289-b. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ceron L, Rodriguez MH, Chavez-Munguia B, Santillan F, Nettel JA, Hernandez-Avila JE. Plasmodium vivax: impaired escape of Vk210 phenotype ookinetes from the midgut blood bolus of Anopheles pseudopunctipennis. Exp Parasitol. 2007;115:59–67. doi: 10.1016/j.exppara.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ceron L, Rodriguez MH, Nettel JC, Villarreal C, Kain KC, Hernandez JE. Differential susceptibilities of Anopheles albimanus and Anopheles pseudopunctipennis to infections with coindigenous Plasmodium vivax variants VK210 and VK247 in southern Mexico. Infect Immun. 1999;67:410–412. doi: 10.1128/iai.67.1.410-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ceron L, Rodriguez MH, Santillan F, Chavez B, Nettel JA, Hernandez-Avila JE, Kain KC. Plasmodium vivax: ookinete destruction and oocyst development arrest are responsible for Anopheles albimanus resistance to circumsporozoite phenotype VK247 parasites. Exp Parasitol. 2001;98:152–161. doi: 10.1006/expr.2001.4626. [DOI] [PubMed] [Google Scholar]
- Gonzalez JM, Hurtado S, Arevalo-Herrera M. Variants of the Plasmodium vivax circumsporozoite protein (VK210 and VK247) in Colombian isolates. Mem Inst Oswaldo Cruz. 2001;96:709–712. doi: 10.1590/s0074-02762001000500023. [DOI] [PubMed] [Google Scholar]
- Gower JHS. Some distance properties of latent root and vector methods used in multivariate analysis. Biometrika. 1966;53:325–338. [Google Scholar]
- Hogg J, Hurd H. The effects of natural Plasmodium faliparum infection on the fecundity and mortality of Anopheles gambiae s.l. in northeast Tanzania. Parasitology. 1997;114:325–331. doi: 10.1017/s0031182096008542. [DOI] [PubMed] [Google Scholar]
- Hurd H, Taylor PJ, Adams D, Underhill A, Eggleston P. Evaluating the costs of mosquito resistance to malaria parasites. Evolution Int J Org Evolution. 2006;59:2560–2572. [PMC free article] [PubMed] [Google Scholar]
- Imwong M, Nair S, Pukrittayakamee S, et al. (14 co-authors) Contrasting genetic structure in Plasmodium vivax populations from Asia and South America. Int J Parasitol. 2007;37:1013–1022. doi: 10.1016/j.ijpara.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Kain KC, Brown AE, Webster HK, Wirtz RA, Keystone JS, Rodriguez MH, Kinahan J, Rowland M, Lanar DE. Circumsporozoite genotyping of global isolates of Plasmodium vivax from dried blood specimens. J Clin Microbiol. 1992;30:1863–1866. doi: 10.1128/jcm.30.7.1863-1866.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltz O, Shykoff JA. Local adaptation in host-parasite systems. Heredity. 1998;81:361–370. [Google Scholar]
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. [Google Scholar]
- Kiszewski A, Mellinger A, Spielman A, Malaney P, Sachs SE, Sachs J. A global index representing the stability of malaria transmission. Am J Trop Med Hyg. 2004;70:486–498. [PubMed] [Google Scholar]
- Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phyl Evol. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: insights from nuclear single-copy genes and the weight of evidence. Syst Biol. 2001;50:540–556. [PubMed] [Google Scholar]
- Lambrechts L, Halbert J, Durand P, Gouagna L, Koella J. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malaria J. 2005;4:3. doi: 10.1186/1475-2875-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufer MK, Thesing PC, Eddington ND, Masonga R, Dzinjalamala FK, Takala SL, Taylor TE, Plowe CV. Return of chloroquine antimalarial efficacy in Malawi. N Engl J Med. 2006;355:1959–1966. doi: 10.1056/NEJMoa062032. [DOI] [PubMed] [Google Scholar]
- Lipsitz SR, Fitzmaurice GM, Orav EJ, Laird NM. Performance of generalized estimating equations in practical situations. Biometrics. 1994;50:270–278. [PubMed] [Google Scholar]
- Lyimo E, Koella J. Relationship between body size of adult Anopheles gambiae s.l. and infection with the malaria parasite Plasmodium falciparum. Parasitology. 1992;104:233–237. doi: 10.1017/s0031182000061667. [DOI] [PubMed] [Google Scholar]
- Machado RD, Povoa M, Calvosa VP, Ferreira M, Rossit AB, dos Santos EM, Conway D. Genetic structure of Plasmodium falciparum populations in the Brazilian Amazon region. J Infect Dis. 2004;190:1547–1555. doi: 10.1086/424601. [DOI] [PubMed] [Google Scholar]
- Manguin S, Roberts D, Peyton E, Fernandez-Salas I, Barreto M, Fernandez LR, Elgueta SR, Martinez Granaou R, Rodriguez M. Biochemical systematics and population genetic structure of Anopheles pseudopunctipennis, vector of malaria in Central and South America. Am J Trop Med Hyg. 1995;53:362–377. doi: 10.4269/ajtmh.1995.53.362. [DOI] [PubMed] [Google Scholar]
- Molina-Cruz A, De Merida AMP, Mills K, Rodriguez F, Schoua C, Yurrita MM, Molina E, Palmieri M, Black WCI. Gene flow among Anopheles albimanus populations in Central America, South America, and the Caribbean assessed by microsatellites and mitochondrial DNA. Am J Trop Med Hyg. 2004;71:350–359. [PubMed] [Google Scholar]
- Moran P. Wandering distributions and the electrophoretic profile. Theor Popul Biol. 1975;8:318–330. doi: 10.1016/0040-5809(75)90049-0. [DOI] [PubMed] [Google Scholar]
- Nair S, Nash D, Sudimack D, Jaidee A, Barends M, Uhlemann A-C, Krishna S, Nosten F, Anderson TJC. Recurrent gene amplification and soft selective sweeps during evolution of multidrug resistance in malaria parasites. Mol Biol Evol. 2007;24:562–573. doi: 10.1093/molbev/msl185. [DOI] [PubMed] [Google Scholar]
- Nash D, Nair S, Mayxay M, Newton P, Guthmann J, Nosten F, Anderson T. Selection strength and hitchhiking around two anti-malarial resistance genes. Proc R Soc Lond B Biol Sci. 2005;272:1153–1161. doi: 10.1098/rspb.2004.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris D, Shurtleff A, Touré Y, Lanzaro G. Microsatellite DNA polymorphism and heterozygosity among field and laboratory populations of Anopheles gambiae ss (Diptera: culicidae) J Med Entomol. 2001;38:336–340. doi: 10.1603/0022-2585-38.2.336. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:2074–2093. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall R, Smouse P. GenAlEx 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakell R, Ruibal M, Lindenmayer D. Spatial autocorrelation analysis offers new insights into gene flow in the Australian bush rat, Rattus fuscipes. Evolution. 2003:1182–1195. doi: 10.1111/j.0014-3820.2003.tb00327.x. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JM, Salinas E, Rodriguez MH, Beaudoin RL. Effects of transmission blocking immunity on Plasmodium vivax infections in Anopheles albimanus populations. J Parasitol. 1994;80:88–92. [PubMed] [Google Scholar]
- Rodriguez MH, Gonzalez-Ceron L, Hernandez JE, Nettel JA, Villarreal C, Kain KC, Wirtz RA. Different prevalences of Plasmodium vivax phenotypes VK210 and VK247 associated with the distribution of Anopheles albimanus and Anopheles pseudopunctipennis in Mexico. Am J Trop Med Hyg. 2000;62:122–127. doi: 10.4269/ajtmh.2000.62.122. [DOI] [PubMed] [Google Scholar]
- Roper C, Pearce R, Nair S, Sharp B, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- Rosenberg R, Wirtz RA, Lanar DE, Sattabongkot J, Hall T, Waters AP, Prasittisuk C. Circumsporozoite protein heterogeneity in the human malaria parasite Plasmodium vivax. Science. 1989;245:973–976. doi: 10.1126/science.2672336. [DOI] [PubMed] [Google Scholar]
- Rubio-Palis Y, Zimmerman R. Ecoregional classification of malaria vectors in the neotropics. J Med Entomol. 1997;34:499–510. doi: 10.1093/jmedent/34.5.499. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: A language for statistical computing. 2007 Vienna (Austria): R Foundation for Statistical Computing. Available at: http://www.R-project.org. [Google Scholar]
- Savage H, Rejmankova E, Arredondo-Jimenez J, Roberts D, Rodriguez M. Limnological and botanical characterization of larval habitats for the two primary malaria vectors, Anopheles albimanus and Anopheles pseudopunctipennis, in coastal areas of Chiapas State, Mexico. J Am Mosq Control Assoc. 1990;6:612–620. [PubMed] [Google Scholar]
- Schlötterer C. Microsatellites. In: Hoelzel AR, editor. Molecular genetic analysis of populations: a practical approach. Oxford (UK): Oxford University Press; 1998. pp. 237–261. [Google Scholar]
- da Silva AN, Santos CC, Lacerda RN, Machado RL, Póvoa MM. Susceptibility of Anopheles aquasalis and An. darlingi to Plasmodium vivax VK210 and VK247. Mem Inst Oswaldo Cruz. 2006;101:547–550. doi: 10.1590/s0074-02762006000500011. [DOI] [PubMed] [Google Scholar]
- Smouse P, Peakall R. Spatial autocorrelation analysis of individual multiallele and multilocus genetic structure. Heredity. 1999;82:561–573. doi: 10.1038/sj.hdy.6885180. [DOI] [PubMed] [Google Scholar]
- Thompson JN. Specific hypotheses on the geographic mosaic of coevolution. Am Nat. 1999;153:S1–S14. [Google Scholar]
- Weir B, Cockerham C. Estimating F statistics for the analysis of population structure. Evolution. 1984:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Wood MJ, Cosgrove CL, Wilkin TA, Knowles SCL, Day KP, Sheldon BC. Within-population variation in prevalence and lineage distribution of avian malaria in blue tits, Cyanistes caeruleus. Mol Ecol. 2007;16:3263–3273. doi: 10.1111/j.1365-294X.2007.03362.x. [DOI] [PubMed] [Google Scholar]
- Woolhouse MEJ, Webster JP, Domingo E, Charlesworth B, Levin BR. Biological and biomedical implications of the co-evolution of pathogens and their hosts. Nat Genetics. 2002;32:569–577. doi: 10.1038/ng1202-569. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su X-Z. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.