Abstract
The development of selective type 5 metabotropic glutamate receptor (mGlu5) antagonists, such as 2-methyl-6-(phenylethynyl)-pyridine (MPEP) and 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]-pyridine (MTEP), has revealed an important role for these receptors in various disorders of the nervous system including depression, anxiety, epilepsy, Parkinson's disease, drug addiction, and alcoholism. In this study, we used microarray technology to examine changes in gene expression induced by repeated administration of the mGlu5 antagonists MPEP and MTEP. Male Wistar rats (n=5 per treatment group) were administered MPEP (10 mg/kg), MTEP (10 mg/kg) or vehicle intraperitoneally twice daily for 5 days. Approximately 30 min following the final drug administration, rats were sacrificed and frontal cortices were then dissected and examined for changes in gene expression by cDNA microarray analysis. Changes in gene expression with P-values less than 0.01 were considered to be statistically significant. The expression of 63 genes was changed by both MPEP and MTEP, with 58 genes down-regulated and 5 genes up-regulated. Quantitative PCR verified the magnitude and direction of change in expression of 9 of these genes (r2=0.556, p=0.017). Pathway analysis revealed that many of the biological processes altered by repeated MPEP and MTEP treatment were related to ATP synthesis, hydrolase activity, and signaling pathways associated with mitogen-activated protein kinase (MAPK). Our results demonstrate diverse effects of MPEP and MTEP gene expression in the frontal cortex, and these results may help elucidate the mechanisms by which these compounds produce beneficial effects in animal models of various disorders of the central nervous system.
Keywords: microarray, frontal cortex, glutamate, mGlu5, antagonist, gene expression, pathway analysis, hierarchical cluster analysis
1. Introduction
Glutamate is the predominant excitatory amino acid neurotransmitter in the brain. Receptors for glutamate receptors are classified as either ionotropic glutamate receptors, such as the N-methyl-D-aspartate (NMDA), α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and kainate receptors, or as metabotropic glutamate receptors. These metabotropic glutamate receptors are further classified into subcategories based on their pharmacological and signal transduction properties. Group I (mGlu1 and mGlu5) are positively coupled to phosphoinositide (PI) hydrolysis and intracellular Ca2+ mobilization, while Group II (mGlu2, mGlu3) and Group III (mGlu4, mGlu6, mGlu7, and mGlu8) are negatively coupled to adenylyl cyclase activity (Conn and Pin, 1997; Coutinho and Knopfel, 2002; Gerber et al., 2007; Hermans and Challiss, 2001). Stimulation of mGlu5 receptors results in activation of numerous signal transduction molecules including protein kinase C (PKC), mitogen-activated protein kinase (MAPK), Ca2+/calmodulin-dependent protein kinase (CaMK), extracellular signal-regulated kinase (ERK), c-Jun N-terminal kinase (JNK), and the phosphoinositide-3-kinase-Akt-mammalian target of rapamycin (PI3K-Akt-mTOR) pathway (Hou and Klann, 2004; Liu et al., 2006; Peavy et al., 2001; Peavy and Conn, 1998; Yang et al., 2006). These enzymes can in turn activate DNA transcription factors such as cyclic adenosine monophosphate response element binding protein (CREB), nuclear factor κB (NFκB), c-Jun, and Elk-1 (Choe and Wang, 2001a; b; Liu et al., 2006; Mao and Wang, 2002, 2003a,b; Yang et al., 2006).
The development of tools for investigating the specific function of individual metabotropic glutamate receptor subtypes in the brain has revealed important roles for these receptors in various disorders of the central nervous system. For example, mice lacking the mGlu5 receptor gene do not self-administer cocaine and are indifferent to its locomotor stimulant effects (Chiamulera et al., 2001), display abnormalities in the induction of synaptic plasticity associated with learning and memory (Jia et al., 1998) and lack certain physiological responses to stress (Brodkin et al., 2002a). Pharmacological studies in animals using systemically active mGlu5 antagonists such as 2-methyl-6-(phenylethynyl)-pyridine (MPEP) and 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]-pyridine (MTEP) (Cosford et al., 2003; Gasparini et al., 1999) have revealed potential beneficial effects of mGlu5 antagonists in the treatment of disorders such as depression, anxiety, epilepsy, Parkinson's disease, alcoholism and drug addiction (Conn, 2003; Gass and Olive, 2008; Kenny and Markou, 2004; Lea and Faden, 2006; Olive, 2005; Palucha and Pilc, 2007; Slassi et al., 2005; Spooren and Gasparini, 2004; Swanson et al., 2005; Witkin et al., 2007).
Given that mGlu5 receptors regulate the activity of numerous DNA transcription factors, it is likely that repeated blockade of these receptors results in changes in the expression of numerous genes. Therefore, the aim of the present study was to use microarray technology to identify specific genes whose expression is altered by mGluR receptor antagonism. The frontal cortex was chose as the target region since it contains moderate to high levels of expression of this receptor (Romano et al., 1995; Shigemoto and Mizuno, 2000; Shigemoto et al., 1993) and is involved in the manifestations of the aforementioned CNS disorders in which mGlu5 antagonists have shown potential beneficial effects (Charney, 2003; Dougherty and Rauch, 2007; Kalivas et al., 2005; Kellinghaus and Luders, 2004; Owen, 2004; Schoenbaum et al., 2006).
2. Methods
2.1. Animals
Male Wistar rats (Charles River, ∼250-300 g upon arrival) were housed individually in standard polycarbonate cages in a temperature and humidity controlled environment on a 12:12 light-dark cycle (lights off at 0600 h). Animals had access to food and water ad libitum. All experimental procedures were conducted in accordance with the National Research Council's Guidelines for the Care and Use of Mammals in Neuroscience and Behavioral Research (2003), and with the approval of an Institutional Animal Care and Use Committee. Rats remained undisturbed for at least three days following arrival in the colony room.
2.2. Drug treatment
Prior to the initiation of treatment, rats were handled and weighed to habituate them to handling procedures. Animals (n=5 per group) were then randomly assigned to be administered either MPEP (10 mg/kg, Sigma-Aldrich), MTEP (10 mg/kg; Alexis Biochemicals), or vehicle (20% w/v 2-hydroxypropyl-β-cyclodextrin, dissolved in sterile saline; Sigma-Aldrich). Drugs were administered twice daily (at approximately 0900 and 1400 h) for a total of 5 consecutive days via the intraperitoneal (i.p.) route in a volume of 1 ml/kg. The doses of MPEP and MTEP administered were chosen based on previous reports that they produce full occupancy of mGlu5 receptors in the rat brain for at least 60 min (Anderson et al., 2002, 2003). Peak concentration of these compounds in the brain fall in the high nanomolar to very low micromolar range following systemic administration at similar doses (Cosford et al., 2003; Nagel et al., 2007), and thus likely retain selectivity for mGlu5 over off-target proteins (Cosford et al., 2003; Gasparini et al., 1999).
2.3. Tissue preparation
In order to reduce variability of the results, the same investigator performed all of the dissections of the frontal cortex. Approximately 30 min following the last drug or vehicle administration, animals were euthanized by decapitation and brains were rapidly removed and washed twice in ice-cold phosphate buffered saline (PBS, pH=7.4) prior to dissection of the frontal cortex. Following removal of the olfactory bulb and underlying cortex and tubercles, the frontal cortex was then dissected by making a cut from approximately 1 mm anterior to bregma on the dorsal surface of the brain towards the rostroventral surface of the orbital cortex. Brain tissue was placed into 300 μl of ice-cold Trizol reagent (Invitrogen) for isolation of total RNA.
2.4. RNA isolation
Total RNA was extracted using Trizol reagent according to the manufacturer's directions. RNA from the frontal cortex was isolated from each rat individually and was not pooled within treatment groups. RNA was then further purified by elution through an RNeasy minicolumn (Qiagen). Total RNA integrity and concentration were determined by an Agilent 2100 Bioanalyzer, and RNA samples were stored at -70°C until further analysis.
2.5. Microarray procedures
Microarray analysis was performed at the MUSC DNA Microarray and Bioinformatics Facility. Ten μg of total RNA from each tissue sample was used to generate double-stranded cDNA using a T7-(dT)24 primer (Genset) and a SuperScript cDNA synthesis kit (Invitrogen). Biotin-labeled cRNA was generated from cDNA by using a BioArray HighYield RNA Transcript labeling kit (Enzo Life Sciences) and then fragmented in 8 M sodium citrate buffer. For each sample (n=1 animal), 10 μg of fragmented cRNA was then hybridized to Affymetrix 230A GeneChip Rat Expression microarrays (which consist of 15,923 oligonucleotide (25-mer) probe sets, 11 probe pairs per sequence) using an Affymetrix Fluidics Station. GeneChip microarrays were then scanned using an Affymetrix 7G GeneArray Scanner 2500, and probe intensities were quantified using Affymetrix Microarray Suite 5.0 software with average chip intensities scaled to 150 units.
2.6. Microarray data analysis
Analysis of microarray hybridization data was performed with assistance from Creative Biolabs (Port Jefferson Station, NY). Microarray data quality was initially screened for significant deviations using (1) smoothed histograms of raw log scale probe intensities, (2) relative log expression and normalized unscaled standard error plots derived from probe-level linear model analysis (Robust Multi-Chip Average (RMA), Irizarry et al., 2003a,b), and (3) correspondence projection analysis. These data quality screening analyses revealed that 2 out of the 15 arrays (both from the MTEP treatment group) showed significant deviations from the remaining arrays, and were thus excluded from subsequent analysis. Thus, the sample sizes are n=5 for vehicle treated animals, n=5 for MPEP treated animals, and n=3 for MTEP treated animals. Data from CEL files from the remaining 13 samples were then adjusted for background, normalized, and summarized using the RMA algorithm (Irizarry et al., 2003a,b). Statistically significant changes in gene expression (p<0.01) induced by MPEP and MTEP treatment relative to vehicle-treated controls were then analyzed using the Bioconductor LIMMA software package and expressed as fold-change. This software package uses an empirical Bayes method to moderate the standard errors of the estimated log-fold changes, and results in more stable inference and improved power for studies involving smaller numbers of arrays. P-values for LIMMA analysis were corrected for multiple testing.
In order to reveal changes in expression of biological important subsets of genes, microarray data were subject to hierarchical clustering and pathway analyses. For hierarchical clustering analysis, a distance matrix containing all the pairwise distances between the genes was first calculated, and subsequently a cosine-angle distance was used to construct a dissimilarity matrix, from which clusters were visually partitioned from the complete data set based on intensity values observed in vehicle treated animals. Pathway analysis was conducted to determine whether a specified group of genes for a given biological pathway was associated with MPEP or MTEP treatment. Pathway analysis was conducted according to the method of Tian and colleagues (Tian et al., 2005). Briefly, pathways (Gene Ontology, GO, or Kyoto Encyclopedia of Genes and Genomes, KEGG) containing between 20 and 500 probe sets represented on each microarray chip were queried with the analyzed microarray data sets. To determine whether a specified pathway had a coordinated association with treatment with both MPEP and MTEP, the top 50 pathways represented by MPEP vs. vehicle treatment and the top 50 pathways represented by MTEP vs. vehicle treatment were intersected to yield the top 10 pathways common to both treatments.
2.7. Quantitive PCR procedures and data analysis
Validation of the direction and magnitude of changes in gene expression indicated by microarray analyses was performed by real-time quantitative polymerase chain reaction (qPCR). First, total RNA was converted to cDNA using an iScript Reverse Transcription kit (BioRad). Briefly, 4 μl of iScript reaction mix, 1 μl of iScript reverse transcriptase, a volume equivalent to 1 μg of total RNA, and q.s. to 20 μl with nuclease-free water were mixed in 0.25 ml PCR tubes and heated on an Applied Biosystems 9700 thermal cycler (25°C for 5 min, 42°C for 30 min, 85°C for 5 min). cDNA was then stored at -20°C until further analysis.
From the entire list of genes represented on the microarray, 9 were selected for analysis by qPCR. Pre-designed Taqman Gene Expression Assays (Applied Biosystems) primer-probe sets specific for each target gene as well as a reference gene were used. Whenever possible, pre-designed primers were used whose sequences spanned intron-exon boundaries to avoid possible contamination from genomic DNA. Primer-probe sets consisted of 20× concentrations of a 5′ FAM dye-labeled Taqman probe specific for each gene of interest, a 3′ non-fluorescent quencher, a minor groove binder, and two unlabeled gene-specific PCR primers. Target genes analyzed were Camk2b, Camk2n1, Ccnd2, Dusp6, Egr2, Grm5, Prkaa2, Ptdss1, and Strn3. Gapdh served as the reference gene, as its expression levels were not altered as determined in the analysis of microarray data. Primer-probe sets for both the target and reference genes were diluted in 2× Universal PCR Mastermix (Applied Biosystems) and nuclease-free water to a final concentration of 250 nM for the probe and 900 nM for the primers. Target and reference gene sample mixes were simultaneously loaded in triplicate (20 μl final volume) into a 96-well optical PCR plate and analyzed on a BioRad iCycler Real Time PCR system. PCR included a denaturing step (50°C for 2 min) prior to a hot start (95°C for 10 min), followed by 40 cycles with melting at 95°C for 15 sec and elongation at 60°C for 1 min. Fluorescence readings were obtained after each cycle. Melting curve analysis was performed with 0.5°C/s increases from 55°C to 95°C at the end of 40 cycles with continuous fluorescence readings to ensure that specific PCR products were obtained.
Relative gene expression was then calculated from resulting threshold cycle (CT) values, and fold change in gene expression was calculated by the 2-ΔΔCT method (Livak and Schmittgen, 2001), where fold change = 2-ΔΔCT, ΔCT = CT (target) - CT (reference), and ΔΔCT = ΔCT (MPEP or MTEP) - ΔCT (Vehicle). Pearson's correlation analysis of the fold change as detected by microarray versus that detected by qPCR was plotted using SigmaPlot (SPSS Inc.) and analyzed for statistical significance (p<0.01) using SigmaStat (SPSS Inc.).
3. Results
3.1. Microarray analysis
Only genes whose change in expression resulted in P-values less than 0.01 were considered to be statistically significant. A list of genes whose expression was altered by both MPEP and MTEP is presented in Table 1. A total of 63 genes were found to have significantly altered expression, with 5 being up-regulated and 58 being down-regulated. Biological functions of these genes included were related to biosynthesis and metabolism, cell adhesion and intercellular signaling, cell cycle control, immune system function, ion homeostasis and transport, nervous system development, nucleotide binding, modification and processing, protein kinase or phosphatase activity, protein synthesis, modification, trafficking and degradation, signal transduction, synaptic transmission, or unknown function.
Table 1. List of genes changed by MPEP and MTEP.
| GenBank | Fold Change | Fold Change | ||||
|---|---|---|---|---|---|---|
| Accession | Name | Symbol | MPEP | P value | MTEP | P value |
| Biosynthesis and Metabolism | ||||||
| NM_031720 | deiodinase, iodothyronine, type II | Dio2 | 0.792 | 0.00154 | 0.748 | 0.00084 |
| BG663460 | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit | Pafah1b1 | 0.730 | 0.00564 | 0.641 | 0.00131 |
| AA851302 | phosphatidylserine synthase 1 | Ptdss1 | 0.902 | 0.00590 | 0.879 | 0.00340 |
| Cell Adhesion and Intercellular Signaling | ||||||
| M37394 | epidermal growth factor receptor | Egfr | 0.757 | 0.00022 | 0.795 | 0.00344 |
| AI102738 | catenin (cadherin associated protein), beta 1 (beta-catenin) | Ctnnb1 | 0.724 | 0.00031 | 0.655 | 0.00009 |
| NM_012656.1 | secreted acidic cysteine rich glycoprotein | Sparc | 0.803 | 0.00100 | 0.791 | 0.00184 |
| NM_001079942.1 | semaphorin 3B (predicted) | Sema3b_pred | 1.169 | 0.00383 | 1.226 | 0.00158 |
| Cell Cycle | ||||||
| BE107226 | striatin, calmodulin binding protein 3 | Strn3 | 0.790 | 0.00027 | 0.799 | 0.00136 |
| L09752 | cyclin D2 | Ccnd2 | 0.692 | 0.00080 | 0.581 | 0.00009 |
| BF396192 | cyclin T2 (predicted) | Ccnt2_pred | 0.670 | 0.00549 | 0.631 | 0.00558 |
| Immune System Function | ||||||
| BF556820 | Casitas B-lineage lymphoma b | Cblb | 0.796 | 0.00188 | 0.783 | 0.00326 |
| Ion Homeostasis and Transport | ||||||
| M26161 | K+ voltage-gated channel, shaker-related subfamily, member 1 (Kv1.1) | Kcna1 | 0.678 | 0.00090 | 0.674 | 0.00243 |
| Nervous System Development | ||||||
| NM_152847.1 | sorting nexin family member 27 | Snx27 | 0.731 | 0.00046 | 0.720 | 0.00103 |
| Nucleotide Binding, Modification, and Processing | ||||||
| BE111095 | RNA-binding motif protein 9 (predicted) | Rbm9_pred | 0.538 | 0.00006 | 0.574 | 0.00009 |
| AI232806 | zinc finger protein 422 (predicted) | Znf422_pred | 0.729 | 0.00017 | 0.719 | 0.00041 |
| AI103040 | zinc finger protein 655 | Zfp655 | 0.803 | 0.00068 | 0.777 | 0.00069 |
| BE109033 | eukaryotic translation initiation factor 2C, member 1 (predicted) | Eif2c1_pred | 0.766 | 0.00048 | 0.738 | 0.00055 |
| BF419161 | eukaryotic translation initiation factor 4E, binding protein 2 | Eif4ebp2 | 0.814 | 0.00254 | 0.800 | 0.00396 |
| AW916745 | inhibitor of DNA binding 4 | Id4 | 0.840 | 0.00137 | 0.829 | 0.00137 |
| BI295086 | KH-type splicing regulatory protein | Khsrp | 0.658 | 0.00142 | 0.589 | 0.00069 |
| AB012232.2 | nuclear factor I/B | Nfib | 0.731 | 0.00150 | 0.706 | 0.00204 |
| NM_053633 | early growth response 2 | Egr2 | 0.667 | 0.00595 | 0.572 | 0.00169 |
| Protein kinase or phosphatase activity | ||||||
| AA858621 | CaM-kinase II inhibitor alpha | Camk2n1 | 0.742 | 0.00001 | 0.786 | 0.00041 |
| AB020480 | SNF1-like kinase | Snf1lk | 0.781 | 0.00004 | 0.833 | 0.00225 |
| NM_023991 | protein kinase, AMP-activated, alpha 2 catalytic subunit | Prkaa2 | 0.568 | 0.00018 | 0.608 | 0.00184 |
| NM_053883 | dual specificity phosphatase 6 | Dusp6 | 0.655 | 0.00071 | 0.569 | 0.00019 |
| NM_021739 | calcium/calmodulin-dependent protein kinase II beta subunit | Camk2b | 0.576 | 0.00157 | 0.594 | 0.00653 |
| BF418932 | phosphorylase kinase alpha 1 | Phka1 | 0.865 | 0.00588 | 0.835 | 0.00355 |
| NM_032062 | kalirin, RhoGEF kinase | Kalrn | 0.659 | 0.00487 | 0.600 | 0.00326 |
| Protein Synthesis, Modification, Trafficking and Degradation | ||||||
| AA946509 | proteasome 26S subunit, non-ATPase, 11 (predicted) | Psmd11_pred | 0.861 | 0.00471 | 0.766 | 0.00014 |
| Signal Trandsuction | ||||||
| AF077354 | heat shock 70 kDa protein 4 | Hspa4 | 0.647 | 0.00009 | 0.699 | 0.00177 |
| NM_031515 | Kirsten rat sarcoma viral oncogene homologue 2 | Kras2 | 0.734 | 0.00066 | 0.746 | 0.00311 |
| BM391419 | protein kinase C binding protein 1 | Prkcbp1 | 0.763 | 0.00084 | 0.785 | 0.00560 |
| BE097238 | RhoGAP involved in beta-catenin-N-cadherin and NMDA receptor signaling (predicted) | RICS_pred | 0.612 | 0.00204 | 0.566 | 0.00197 |
| Synaptic Transmission | ||||||
| NM_017215.2 | solute carrier family 1, member 2 (EAAT2, GLT-1) | Slc1a2 | 0.874 | 0.00299 | 0.797 | 0.00275 |
| Unknown Function | ||||||
| AA964250 | ankyrin repeat domain 52 (predicted) | Ankrd52_pred | 0.594 | 0.00464 | 0.542 | 0.00409 |
| BE118720 | jumonji domain containing 3 (predicted) | Jmjd3_pred | 0.680 | 0.00030 | 0.688 | 0.00131 |
| AW253339 | huntingtin interacting protein 1 (predicted) | Hip1 | 0.769 | 0.00042 | 0.759 | 0.00096 |
| BE120831 | breakpoint cluster region (predicted) | Bcr_pred | 0.657 | 0.00225 | 0.645 | 0.00459 |
| BI294227 | LOC685082 (similar to zinc finger protein 318 isoform 1) | LOC685082 | 0.814 | 0.00035 | 0.819 | 0.00149 |
| BI295883 | LOC691918 (similar to centrosomal protein 27 kDA Cep27) | LOC691919 | 1.232 | 0.00166 | 1.251 | 0.00287 |
| BI282024 | LOC686587 (similar to 40S ribosomomal protein S4. X isoform) | LOC686587 | 1.193 | 0.00448 | 1.203 | 0.00329 |
| BI288731 | RGD1562686 (similar to genetic suppressor element, predicted) | RGD1562686_pred | 0.763 | 0.00002 | 0.762 | 0.00007 |
| BI304125 | RGD1563001 (predicted) | RGD1563001_pred | 0.801 | 0.00126 | 0.768 | 0.00099 |
| BE116947 | similar to RIKEN cDNA 1600012F09 (predicted) | RGD1306613_pred | 0.895 | 0.00572 | 0.854 | 0.00132 |
| AA955579 | cDNA clone UI-R-E1-fa-b-09-0-UI | 0.756 | 0.00004 | 0.699 | 0.00002 | |
| BE107173 | cDNA clone UI-R-BS1-ayq-f-12-0-UI | 0.780 | 0.00004 | 0.845 | 0.00420 | |
| AA818910 | cDNA clone UI-R-A0-as-a-09-0-UI | 0.678 | 0.00007 | 0.696 | 0.00060 | |
| AI229240 | cDNA clone RBRDF82 | 0.759 | 0.00025 | 0.751 | 0.00064 | |
| BF550404 | cDNA clone UI-R-A1-eg-c-09-0-UI | 0.733 | 0.00049 | 0.744 | 0.00236 | |
| BE095660 | cDNA clone UI-R-BO1-apo-f-12-0-UI | 0.683 | 0.00106 | 0.704 | 0.00546 | |
| BM391905 | cDNA clone UI-R-DM1-ckc-b-23-0-UI | 0.875 | 0.00202 | 0.868 | 0.00384 | |
| BG373799 | cDNA clone UI-R-CV1-bsk-a-10-0-UI | 0.814 | 0.00234 | 0.796 | 0.00308 | |
| BF393051 | cDNA clone UI-R-CA0-bgp-f-01-0-UI | 0.842 | 0.00258 | 0.763 | 0.00020 | |
| AI234119 | cDNA clone RLUCV43 | 1.145 | 0.00415 | 1.162 | 0.00546 | |
| BI288873 | cDNA clone UI-R-DK0-cdf-h-07-0-UI | 0.882 | 0.00416 | 0.859 | 0.00303 | |
| AI137837 | cDNA clone UI-R-C0-ig-e-11-0-UI | 1.196 | 0.00420 | 1.265 | 0.00169 | |
| AI175666 | cDNA clone ROVBD47 | 0.847 | 0.00672 | 0.792 | 0.00178 | |
| BF290416 | cDNA clone RGIHV60 | 0.868 | 0.00779 | 0.828 | 0.00303 | |
| AI177753 | cDNA clone RPLCF68 | 0.823 | 0.00344 | 0.801 | 0.00370 | |
| BI282077 | cDNA clone UI-R-Y0-mk-e-10-0-UI | 0.784 | 0.00016 | 0.819 | 0.00269 | |
| BE106526 | cDNA clone UI-R-BO1-asp-a-08-0-UI | 0.825 | 0.00016 | 0.870 | 0.00639 | |
| BF555171 | cDNA clone UI-R-E0-ct-a-05-0-UI | 0.755 | 0.00667 | 0.630 | 0.00046 | |
| AI103026 | DD6A4-12(7)-1 mRNA partial sequence | 0.716 | 0.00183 | 0.609 | 0.00021 | |
pred = predicted
3.2. Quantitative PCR analysis
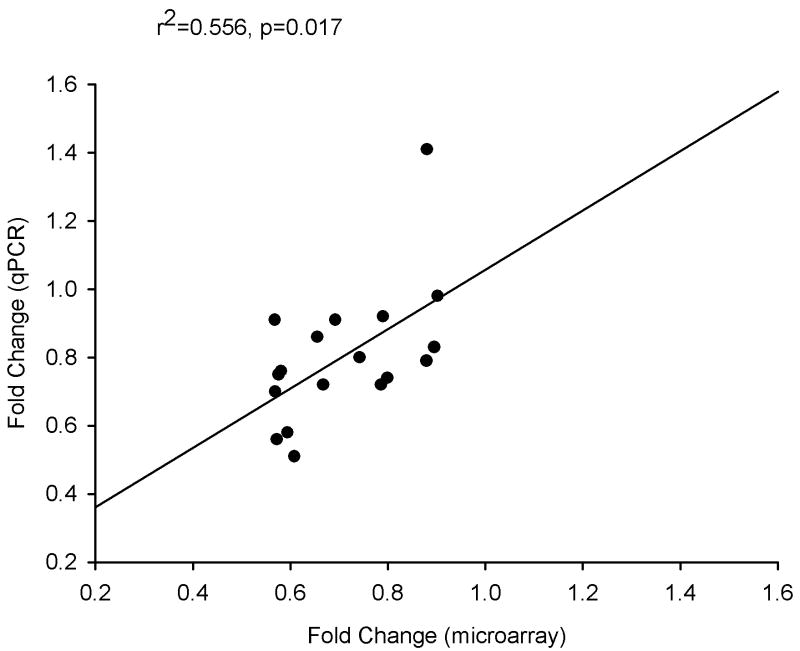
The magnitude and direction of changes in expression of 9 genes produced by treatment with MPEP and/or MTEP was verified by qPCR. Target genes analyzed were Camk2b, Camk2n1, Ccnd2, Dusp6, Egr2, Grm5, Prkaa2, Ptdss1, and Strn3, with Gapdh serving as the reference gene. A statistically significant correlation between the fold-change induced by drug treatment as measured by microarray analysis as compared to that measured by qPCR (r2=0.556, p=0.017). This correlation is depicted in Figure 1, and the results of the qPCR analysis are listed in Table 3.
Fig. 1.
Correlation between the fold-change in expression of 9 genes induced by MPEP or MTEP treatment as revealed by microarray analysis versus qPCR. A statistically significant correlation coefficient was found (r2=0.556, p=0.017).
Table 3.
Results of qPCR analysis of 9 selected genes from microarray findings.
| Gene Symbol | Treatment | Fold Change (mean ± SEM) |
|---|---|---|
| Camk2b | MPEP | 0.75 ± 0.06 |
| MTEP | 0.58 ± 0.14 | |
| Camk2n1 | MPEP | 0.80 ± 0.14 |
| MTEP | 0.58 ± 0.17 | |
| Ccnd2 | MPEP | 0.91 ± 0.47 |
| MTEP | 0.76 ± 0.16 | |
| Dusp6 | MPEP | 0.86 ± 0.10 |
| MTEP | 0.70 ± 0.08 | |
| Egr2 | MPEP | 0.72 ± 0.14 |
| MTEP | 0.56 ± 0.05 | |
| Grm5 | MPEP | 1.41 ± 0.90 |
| MTEP | 0.83 ± 0.25 | |
| Prkaa2 | MPEP | 0.91 ± 0.09 |
| MTEP | 0.51 ± 0.06 | |
| Ptdss1 | MPEP | 0.98 ± 0.09 |
| MTEP | 0.79 ± 0.19 | |
| Strn3 | MPEP | 0.92 ± 0.16 |
| MTEP | 0.74 ± 0.09 |
3.3. Hierarchical clustering and pathway analyses
Figure 2 shows a heat map and dendrogram of the 63 genes altered by both MPEP and MTEP. A total of six main clusters were identified visually based on the heat map signal intensity in vehicle-treated animals. Clusters 1, 3 and 4 consisted of genes with relatively high levels of expression, cluster 2 consisted of genes with moderate levels of expression, and clusters 5 and 6 consisted mainly genes whose expression levels were low in vehicle-treated animals. Several of these clusters contained genes of related function. For example, Cluster 1 contained genes encoding nucleotide binding proteins including Eif2c1_pred, Nfib, and Khrsp. Cluster 3 contained genes related to cell adhesion and intracellular signaling such as Sparc and Ctnnb1. Cluster 4 contained two genes specifically related to CaM kinase function (Camk2n1 and Camk2b). Cluster 5 contained genes related to protein kinase activity including Prkaa2, Snf1lk and Phka1. Finally, Cluster 6 contained genes encoding nucleotide binding proteins including Zpf422_pred and Zfp655.
Fig. 2.
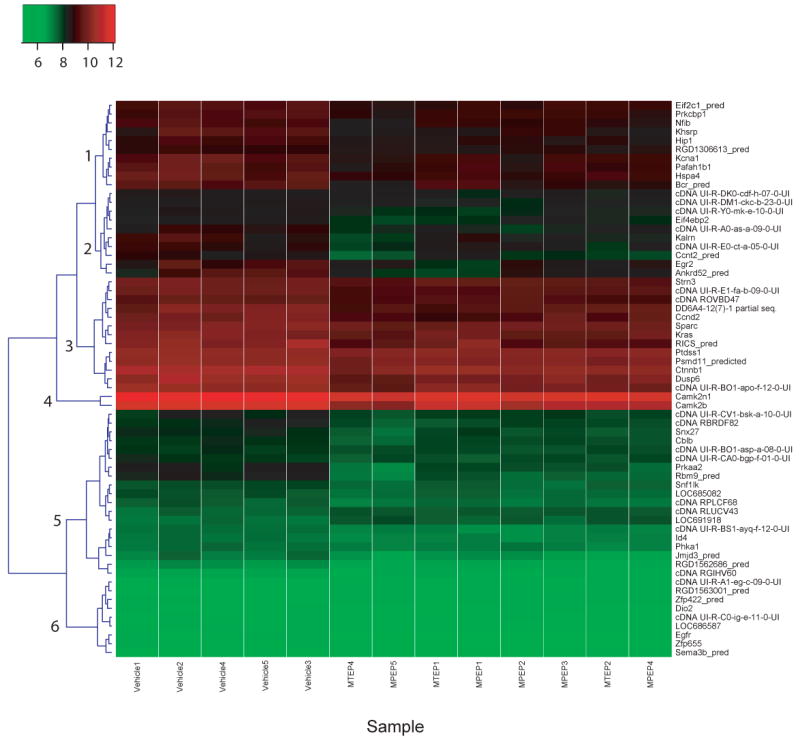
Heat map and dendrogram showing clusters of genes based on signal intensity of each individual gene in vehicle-treated animals. Each column represents the signal intensity (see color code at inset) of each individual biological sample. Visually identified clustered are numbered along the left side of the dendrogram.
Table 2 lists the results of pathway analysis of the effects of MPEP and MTEP on gene expression. This list was generated by intersecting the top 50 biological pathways representing genes changed by MPEP treatment with the top 50 pathways representing genes changed by MTEP treatment. These pathways were: ATP synthesis, TGF-beta signaling pathway, Wnt signaling pathway, muscle development, phosphoric ester hydrolase activity, phosphoric monoester hydrolase activity, hydrolase activity, acting on acid anhydrides, hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides, neuropeptide signaling pathway, and MAPK signaling pathway.
Table 2.
Pathway analysis of effects of MPEP and MTEP treatment on gene expression in the rat frontal cortex.
| ID | Name |
|---|---|
| KEGG:00193 | ATP synthesis |
| KEGG:04350 | TGF-beta signaling pathway |
| KEGG:04310 | Wnt signaling pathway |
| GO:0007517 | muscle development |
| GO:0042578 | phosphoric ester hydrolase activity |
| GO:0016791 | phosphoric monoester hydrolase activity |
| GO:0016817 | hydrolase activity, acting on acid anhydrides |
| GO:0016818 | hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides |
| GO:0007218 | neuropeptide signaling pathway |
| KEGG:04010 | MAPK signaling pathway |
4. Discussion
Utilizing microarray technology, the present study demonstrates that repeated administration of mGlu5 receptor antagonists MPEP and MTEP produces changes in the expression of a multitude of genes in the rat frontal cortex. The functions of these genes were not confined to synaptic transmission or signal transduction pathways known to be linked to mGlu5 receptor function, but instead represented a host of biological functions including regulation of biosynthesis and metabolism, cell-cell adhesion and communication, cell cycle control, and protein, protein kinase and nucleotide binding activity. In addition, expression of genes involved in immune system function, ion homeostasis and transport, and nervous system development was also altered. Our data suggest that mGluR5 receptors may regulate a host of cellular functions other than modulatory synaptic transmission by glutamate. A statistically significant correlation between the fold-change observed by microarray analysis and the fold-change of 9 genes selected for analysis by qPCR was observed, indicating a degree of reliability in the changes in gene expression as detected by microarray analysis. However, additional studies examining whether these alterations in gene expression result in changes in actual protein synthesis are needed to further confirm these findings.
The doses of MPEP and MTEP chosen for the present study were each 10 mg/kg i.p.. It has previously been shown that this dose of MPEP produces rapid and full occupancy of mGlu5 receptors in the rat brain following systemic administration (Anderson et al., 2003). In contrast, only a 3 mg/kg i.p. dose of MTEP has been shown to be required to produce full occupancy of brain mGlu5 receptors. However, Anderson and colleagues also noted differences in the clearance rates of MPEP and MTEP from the brain. For example, approximately 4 hours following systemic administration MPEP or MTEP, these investigators found that mGlu5 receptor occupancy had declined to approximately 40-50% in MPEP rats, whereas in MTEP treated rats mGlu5 receptor occupancy had declined to approximately 5-10% (Anderson et al., 2003). Therefore, while MTEP appears to be a more potent mGlu5 antagonist, it also appears to be shorter acting in the brain. We therefore attempted to compensate for this by increasing the dose of MTEP to 10 mg/kg so as to obtain roughly equal durations of mGlu5 receptor occupancy following each injection. Recent microdialysis studies have shown that extracellular levels of both MPEP and MTEP are in the high nanomolar to low micromolar range following systemic administration of similar doses (Nagel et al., 2007), and therefore likely retain their selectivity for mGlu5 receptors over other off-target proteins (Cosford et al., 2003; Gasparini et al., 1999). However, the receptor occupancy study (Anderson et al., 2003) and the microdialysis study (Nagel et al., 2007) were performed following acute administration of MPEP or MTEP, and it is unknown how the receptor occupancy, extracellular levels or pharmacokinetics of MPEP or MTEP are altered following repeated administration twice daily, as was performed in the present study.
Various studies examining the behavioral effects of MPEP and MTEP in animal models of CNS disorders have demonstrated positive effects after acute systemic administration (c.f., Brodkin et al., 2002b; Chiamulera et al., 2001; Pietraszek et al., 2005; Spooren et al., 2000; Tatarczynska et al., 2001; Tessari et al., 2004; Varty et al., 2005). However, some investigators have reported that multiple dosing procedures with these compounds produces increases efficacy in the anxiolytic, antidepressant or anti-addictive effects as compared with acute administration (Backstrom et al., 2004; Cowen et al., 2005; Klodzinska et al., 2004; Li et al., 2006; Nordquist et al., 2007; Pilc et al., 2002). This raises the possibility that changes in gene expression may underlie some of the behavioral effects of MPEP and/or MTEP following repeated exposure. Thus, a multiple dosing schedule was utilized in the present study to determine effects of these compounds on changes in gene expression.
The expression of potassium voltage-gated channel shaker-related subfamily member 1 (Kcna1), also termed Kv1.1, was significantly down-regulated by both mGlu5 antagonists, suggesting that these compounds may alter neuronal excitability and activity in the frontal cortex (Homayoun and Moghaddam, 2006) by modulating potassium channel expression.
Repeated administration of both MPEP and MTEP was found to decrease the expression of numerous signaling proteins that are known to be downstream of mGlu5 receptors, including proteins involved to calcium second messenger and MAPK-related signaling such as CaM kinase II inhibitor alpha (Camk2n1), protein kinase AMP activated alpha 2 catalytic subunit (Prkaa2), also termed protein kinase A Cα2, calcium/calmodulin dependent protein kinase II beta (Camk2b), also termed CaMKIIβ, and striatin/calmodulin-binding protein 3 (Strn3) The mGluR5 antagonists also decreased the expression of protein kinase C binding protein 1 (Prkcbp1) and the predicted sequence of RhoGAP involved in beta-catenin-N-cadherin and NMDA receptor signaling (RICS_pred). Many of these proteins are postsynaptic molecular substrates that underlie synaptic plasticity, which may explain the deleterious effects of mGlu5 antagonists on some forms of learning and memory (Simonyi et al., 2005). Despite the well-established fact that mGlu5 receptors are coupled to PKC signaling, we did not observe changes in the expression of any PKC isoforms.
The expression of the gene encoding Casitas B-lineage lymphoma b (Cblb) was significantly downregulated by both MPEP and MTEP treatment. mGlu5 receptors are constitutively expressed on lymphocytes (Pacheco et al., 2004), and it is therefore possible that MPEP and MTEP may have immunomodulatory properties, although we are unaware of the existence of any such reports on this topic in the literature.
Both MPEP and MTEP altered the expression of genes encoding proteins involved in biosynthesis such as phosphatidylserine synthase 1 (Ptdss1) and platelet-activating factor acetylhydrolase isoform Ib alpha subunit (Pafah1b1), cell-cell adhesion and intercellular communication proteins such as epidermal growth factor receptor (Egfr) and beta-catenin (Ctnnb1), and cell cycle control proteins such as cyclin D2 (Ccnd2) and the predicted sequence for cyclin T2 (Ccnt2-pred). These compounds also down-regulated the expression of CNS development proteins such as sorting nexin family member 27 (Snx27), and transcription factors and DNA-binding proteins such as zinc finger proteins 422 (Znf422_pred), and 655 (Zfp655), nuclear factor I/B (Nfib), inhibitor of DNA binding 4 (Id4), and eukaryotic translation initiating factor 4E binding protein 2 (Eif4ebp2) and the predicted sequences of eukaryotic translation initiating factor 2C member 1 (Eif2c1_pred) and RNA-binding motif protein 9 (Rbm9_pred). These findings suggest that some of the observed effects of mGlu5 antagonists on gene expression may be mediated by down-regulation of the expression of various transcription factors and nucleotide binding proteins.
Of particular interest was the observation that both MPEP and MTEP decreased the expression of the gene encoding the glutamate transporter solute carrier family 1 member 2 (Slc1a2), also known as excitatory amino acid transporter 2 (EAAT2) or glutamate transporter 1 (GLT-1). This sodium-dependent glutamate transporter is localized to presynaptic terminals and regulates extracellular levels of glutamate (Shigeri et al., 2004). One might hypothesize that the ability of repeated administration of MPEP and MTEP to decrease the expression of EAAT2/GLT-1 might result in increased extracellular levels of glutamate. While it has been reported that local application of MPEP in vitro (Thomas et al., 2001) or in vivo (de Novellis et al., 2003; Mills et al., 2001; Thomas et al., 2001) actually reduces extracellular levels of glutamate, to our knowledge no studies to date have measured changes in extracellular glutamate in response to repeated administration of mGlu5 antagonists, as was performed in the present study.
Pathway analysis revealed that many of the common biological pathways whose components were altered by both MPEP and MTEP were related to biosynthesis (ATP synthesis, hydrolase activity), muscle development, or various intracellular signaling pathways (TGF-beta, Wnt, neuropeptide, and MAPK signaling). Surprisingly, the expression of the gene encoding mGlu5 (Grm5) was not altered by repeated administration of MPEP or MTEP, and this lack of change in expression was confirmed by qPCR. These data indicate a substantial degree of resilience in expression this receptor in the frontal cortex in response to repeated administration of the antagonists MPEP or MTEP (10 mg/kg each twice daily for 5 days). These data are consistent with that of a previous report, which showed that repeated treatment of MTEP (2 mg/kg/day for 12 days) to rats produced no changes in mGlu5 mRNA expression in the cingulate cortex, dorsal striatum or nucleus accumbens, as measured by in situ hybridization (Cowen et al., 2005). However, these authors did note a 25% reduction in mGlu5 mRNA expression in the olfactory tubercle induced by repeated MTEP administration, suggesting that the mechanisms regulating the expression of mGlu5 in response to repeated antagonist treatment may be brain region-specific.
In summary, we have demonstrated that repeated administration of the mGlu5 antagonists MPEP and MTEP resulted in changes in the expression of 63 genes, with a predominant trend towards inhibition of gene expression since 58 genes were down-regulated and only 5 were up-regulated. Genes whose expression was altered by these mGluR5 antagonists encode proteins known to be involved in biosynthesis and metabolism, cell-cell adhesion and communication, cell cycle control, immune system function, ion homeostasis and transport, nervous system development, transcription factors and DNA-binding proteins, protein kinase or phosphatase activity, protein synthesis, modification and trafficking, signal transduction, and synaptic transmission. These data suggest that mGlu5 antagonists can alter the expression of a wide array of genes that is much larger than previously characterized. Given that mGlu5 receptors are coupled to numerous signal transduction cascades, and are expressed on cells of the immune system, we speculate that ability of MPEP and MTEP to alter the expression of genes related to intracellular signaling cascades (i.e., MAPK) and immune system function are primarily related to the pharmacological actions of these compounds. In contrast, given the observed changes in expression of numerous genes not previously associated with mGlu5 function (i.e., biosynthesis and metabolism, cell-cell adhesion, and cell cycle control), we speculate that these changes may be secondary to the ability of these compounds to alter gene expression via specific transcription factors or nucleotide binding proteins.
Acknowledgments
This study was supported by Public Health Service grant AA013852 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA, Bethesda, MD) to MFO, and an institutional training grant AA007474 (JTG). The authors wish to thank Dr. Jacqueline McGinty for her kind donation of the microarray chips, Victor Fresco of the MUSC DNA Microarray and Bioinformatics Facility for assistance in microarray hybridization procedures, and Robert Lee of Creative Biolabs Inc. for assistance in the analysis of the microarray data.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson JJ, Bradbury MJ, Giracello DR, Chapman DF, Holtz G, Roppe J, King C, Cosford ND, Varney MA. In vivo receptor occupancy of mGlu5 receptor antagonists using the novel radioligand [3H]-methoxy-5-(pyridin-2-ylethynyl)pyridine) Eur J Pharmacol. 2003;473:35–40. doi: 10.1016/s0014-2999(03)01935-6. [DOI] [PubMed] [Google Scholar]
- Anderson JJ, Rao SP, Rowe B, Giracello DR, Holtz G, Chapman DF, Tehrani L, Bradbury MJ, Cosford ND, Varney MA. [3H]Methoxymethyl-3-[(2-methyl-1,3-thiazol-4-yl)ethynyl]pyridine binding to metabotropic glutamate receptor subtype 5 in rodent brain: in vitro and in vivo characterization. J Pharmacol Exp Ther. 2002;303:1044–1051. doi: 10.1124/jpet.102.040618. [DOI] [PubMed] [Google Scholar]
- Backstrom P, Bachteler D, Koch S, Hyytia P, Spanagel R. mGluR5 antagonist MPEP reduces ethanol-seeking and relapse behavior. Neuropsychopharmacology. 2004;29:921–928. doi: 10.1038/sj.npp.1300381. [DOI] [PubMed] [Google Scholar]
- Brodkin J, Bradbury M, Busse C, Warren N, Bristow LJ, Varney MA. Reduced stress-induced hyperthermia in mGluR5 knockout mice. Eur J Neurosci. 2002a;16:2241–2244. doi: 10.1046/j.1460-9568.2002.02294.x. [DOI] [PubMed] [Google Scholar]
- Brodkin J, Busse C, Sukhoff SJ, Varney MA. Anxioltyic-like activity of the mGluR5 antagonist MPEP: a comparison with diazepam and buspirone. Pharmacol Biochem Behav. 2002b;73:359–366. doi: 10.1016/s0091-3057(02)00828-6. [DOI] [PubMed] [Google Scholar]
- Charney DS. Neuroanatomical circuits modulating fear and anxiety behaviors. Acta Psychiatr Scand Suppl. 2003;417:38–50. doi: 10.1034/j.1600-0447.108.s417.3.x. [DOI] [PubMed] [Google Scholar]
- Chiamulera C, Epping-Jordan MP, Zocchi A, Marcon C, Cottiny C, Tacconi S, Corsi M, Orzi F, Conquet F. Reinforcing and locomotor stimulant effects of cocaine are absent in mGluR5 null mutant mice. Nat Neurosci. 2001;4:873–874. doi: 10.1038/nn0901-873. [DOI] [PubMed] [Google Scholar]
- Choe ES, Wang JQ. Group I metabotropic glutamate receptor activation increases phosphorylation of cAMP response element-binding protein, Elk-1, and extracellular signal-regulated kinases in rat dorsal striatum. Mol Brain Res. 2001a;94:75–84. doi: 10.1016/s0169-328x(01)00217-0. [DOI] [PubMed] [Google Scholar]
- Choe ES, Wang JQ. Group I metabotropic glutamate receptors control phosphorylation of CREB, Elk-1 and ERK via a CaMKII-dependent pathway in rat striatum. Neurosci Lett. 2001b;313:129–132. doi: 10.1016/s0304-3940(01)02258-3. [DOI] [PubMed] [Google Scholar]
- Conn PJ. Physiological roles and therapeutic potential of metabotropic glutamate receptors. Ann N Y Acad Sci. 2003;1003:12–21. doi: 10.1196/annals.1300.002. [DOI] [PubMed] [Google Scholar]
- Conn PJ, Pin JP. Pharmacology and functions of metabotropic glutamate receptors. Annu Rev Pharmacol Toxicol. 1997;37:205–237. doi: 10.1146/annurev.pharmtox.37.1.205. [DOI] [PubMed] [Google Scholar]
- Cosford ND, Tehrani L, Roppe J, Schwieger E, Smith ND, Anderson J, Bristow L, Brodkin J, Jiang X, McDonald I, Rao S, Washburn M, Varney MA. 3-[(2-Methyl-1,3-thiazol-4-yl)ethynyl]-pyridine: a potent and highly selective metabotropic glutamate subtype 5 receptor antagonist with anxiolytic activity. J Med Chem. 2003;46:204–206. doi: 10.1021/jm025570j. [DOI] [PubMed] [Google Scholar]
- Coutinho V, Knopfel T. Metabotropic glutamate receptors: electrical and chemical signaling properties. Neuroscientist. 2002;8:551–561. doi: 10.1177/1073858402238514. [DOI] [PubMed] [Google Scholar]
- Cowen MS, Djouma E, Lawrence AJ. The metabotropic glutamate 5 receptor antagonist 3-[(2-methyl-1,3-thiazol-4-yl)ethynyl)-pyridine reduces ethanol self-administration in multiple strains of alcohol-preferring rats and regulates olfactory glutamatergic systems. J Pharmacol Exp Ther. 2005;315:590–600. doi: 10.1124/jpet.105.090449. [DOI] [PubMed] [Google Scholar]
- de Novellis V, Marabese I, Palazzo E, Rossi F, Berrino L, Rodella L, Bianchi R, Rossi F, Maione S. Group I metabotropic glutamate receptors modulate glutamate and g-aminobutyric acid release in the periaqueductal grey of rats. Eur J Pharmacol. 2003;462:73–81. doi: 10.1016/s0014-2999(03)01342-6. [DOI] [PubMed] [Google Scholar]
- Dougherty DD, Rauch SL. Brain correlates of antidepressant treatment outcome from neuroimaging studies in depression. Psychiatr Clin North Am. 2007;30:91–103. doi: 10.1016/j.psc.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Gasparini F, Lingenhohl K, Stoehr N, Flor PJ, Heinrich M, Vranesic I, Biollaz M, Allgeier H, Heckendorn R, Urwyler S, Varney MA, Johnson EC, Hess SD, Rao SP, Sacaan AI, Santori EM, Velicelebi G, Kuhn R. 2-Methyl-6-(phenylethynyl)-pyridine (MPEP), a potent, selective and systemically active mGlu5 receptor antagonist. Neuropharmacology. 1999;38:1493–1503. doi: 10.1016/s0028-3908(99)00082-9. [DOI] [PubMed] [Google Scholar]
- Gass JT, Olive MF. Glutamatergic substrates of drug addiction and alcoholism. Biochem Pharmacol. 2008;75:218–265. doi: 10.1016/j.bcp.2007.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber U, Gee CE, Benquet P. Metabotropic glutamate receptors: intracellular signaling pathways. Curr Opin Pharmacol. 2007;7:56–61. doi: 10.1016/j.coph.2006.08.008. [DOI] [PubMed] [Google Scholar]
- Hermans E, Challiss RA. Structural, signalling and regulatory properties of the group I metabotropic glutamate receptors: prototypic family C G-protein-coupled receptors. Biochem J. 2001;359:465–484. doi: 10.1042/0264-6021:3590465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homayoun H, Moghaddam B. Bursting of prefrontal cortex neurons in awake rats is regulated by metabotropic glutamate 5 (mGlu5) receptors: rate-dependent influence and interaction with NMDA receptors. Cereb Cortex. 2006;16:93–105. doi: 10.1093/cercor/bhi087. [DOI] [PubMed] [Google Scholar]
- Hou L, Klann E. Activation of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin signaling pathway is required for metabotropic glutamate receptor-dependent long-term depression. J Neurosci. 2004;24:6352–6361. doi: 10.1523/JNEUROSCI.0995-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003a;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jia Z, Lu Y, Henderson J, Taverna F, Romano C, Abramow-Newerly W, Wojtowicz JM, Roder J. Selective abolition of the NMDA component of long-term potentiation in mice lacking mGluR5. Learn Mem. 1998;5:331–343. [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Volkow N, Seamans J. Unmanageable motivation in addiction: a pathology in prefrontal-accumbens glutamate transmission. Neuron. 2005;45:647–650. doi: 10.1016/j.neuron.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Kellinghaus C, Luders HO. Frontal lobe epilepsy. Epileptic Disord. 2004;6:223–239. [PubMed] [Google Scholar]
- Kenny PJ, Markou A. The ups and downs of addiction: role of metabotropic glutamate receptors. Trends Pharmacol Sci. 2004;25:265–272. doi: 10.1016/j.tips.2004.03.009. [DOI] [PubMed] [Google Scholar]
- Klodzinska A, Tatarczynska E, Chojnacka-Wojcik E, Nowak G, Cosford ND, Pilc A. Anxiolytic-like effects of MTEP, a potent and selective mGlu5 receptor antagonist, does not involve GABAA signaling. Neuropharmacology. 2004;47:342–350. doi: 10.1016/j.neuropharm.2004.04.013. [DOI] [PubMed] [Google Scholar]
- Lea PM, Faden AI. Metabotropic glutamate receptor subtype 5 antagonists MPEP and MTEP. CNS Drug Rev. 2006;12:149–166. doi: 10.1111/j.1527-3458.2006.00149.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Need AB, Baez M, Witkin JM. Metabotropic glutamate 5 receptor antagonism is associated with antidepressant-like effects in mice. J Pharmacol Exp Ther. 2006;319:254–259. doi: 10.1124/jpet.106.103143. [DOI] [PubMed] [Google Scholar]
- Liu F, Zhang G, Hornby G, Vasylyev D, Bowlby M, Park K, Gilbert A, Marquis K, Andree TH. The effect of mGlu5 receptor positive allosteric modulators on signaling molecules in brain slices. Eur J Pharmacol. 2006;536:262–268. doi: 10.1016/j.ejphar.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Interactions between ionotropic and metabotropic glutamate receptors regulate cAMP response element-binding protein phosphorylation in cultured striatal neurons. Neuroscience. 2002;115:395–402. doi: 10.1016/s0306-4522(02)00400-1. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Metabotropic glutamate receptor 5-regulated Elk-1 phosphorylation and immediate early gene expression in striatal neurons. J Neurochem. 2003a;85:1006–1017. doi: 10.1046/j.1471-4159.2003.01750.x. [DOI] [PubMed] [Google Scholar]
- Mao L, Wang JQ. Phosphorylation of cAMP response element-binding protein in cultured striatal neurons by metabotropic glutamate receptor subtype 5. J Neurochem. 2003b;84:233–243. doi: 10.1046/j.1471-4159.2003.01256.x. [DOI] [PubMed] [Google Scholar]
- Mills CD, Xu GY, McAdoo DJ, Hulsebosch CE. Involvement of metabotropic glutamate receptors in excitatory amino acid and GABA. release following spinal cord injury in rat. J Neurochem. 2001;79:835–848. doi: 10.1046/j.1471-4159.2001.00630.x. [DOI] [PubMed] [Google Scholar]
- Nagel J, Greco S, Klein KU, Eilbacher B, Valastro B, Tober C, Danysz W. Program No 711.19. 2007 Neuroscience Meeting Planner. San Diego, CA: Society for Neuroscience; 2007. Microdialysis reveals different brain PK of the selective mGluR5 negative modulators MTEP and MPEP. 2007. Online. [Google Scholar]
- Nordquist RE, Durkin S, Jaeschke G, Spooren W. Stress-induced hyperthermia: effects of acute and repeated dosing of MPEP. Eur J Pharmacol. 2007;568:199–202. doi: 10.1016/j.ejphar.2007.04.034. [DOI] [PubMed] [Google Scholar]
- Olive MF. mGlu5 receptors: neuroanatomy, pharmacology, and role in drug addiction. Curr Psychiatry Rev. 2005;1:197–214. [Google Scholar]
- Owen AM. Cognitive dysfunction in Parkinson's disease: the role of frontostriatal circuitry. Neuroscientist. 2004;10:525–537. doi: 10.1177/1073858404266776. [DOI] [PubMed] [Google Scholar]
- Pacheco R, Ciruela F, Casado V, Mallol J, Gallart T, Lluis C, Franco R. Group I metabotropic glutamate receptors mediate a dual role of glutamate in T cell activation. J Biol Chem. 2004;279:33352–33358. doi: 10.1074/jbc.M401761200. [DOI] [PubMed] [Google Scholar]
- Palucha A, Pilc A. Metabotropic glutamate receptor ligands as possible anxiolytic and antidepressant drugs. Pharmacol Ther. 2007;115:116–147. doi: 10.1016/j.pharmthera.2007.04.007. [DOI] [PubMed] [Google Scholar]
- Peavy RD, Chang MS, Sanders-Bush E, Conn PJ. Metabotropic glutamate receptor 5-induced phosphorylation of extracellular signal-regulated kinase in astrocytes depends on transactivation of the epidermal growth factor receptor. J Neurosci. 2001;21:9619–9628. doi: 10.1523/JNEUROSCI.21-24-09619.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peavy RD, Conn PJ. Phosphorylation of mitogen-activated protein kinase in cultured rat cortical glia by stimulation of metabotropic glutamate receptors. J Neurochem. 1998;71:603–612. doi: 10.1046/j.1471-4159.1998.71020603.x. [DOI] [PubMed] [Google Scholar]
- Pietraszek M, Sukhanov I, Maciejak P, Szyndler J, Gravius A, Wislowska A, Plaznik A, Bespalov AY, Danysz W. Anxiolytic-like effects of mGlu1 and mGlu5 receptor antagonists in rats. Eur J Pharmacol. 2005;514:25–34. doi: 10.1016/j.ejphar.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Pilc A, Klodzinska A, Branski P, Nowak G, Palucha A, Szewczyk B, Tatarczynska E, Chojnacka-Wojcik E, Wieronska JM. Multiple MPEP administrations evoke anxiolytic- and antidepressant-like effects in rats. Neuropharmacology. 2002;43:181–187. doi: 10.1016/s0028-3908(02)00082-5. [DOI] [PubMed] [Google Scholar]
- Romano C, Sesma MA, McDonald CT, O'Malley K, van den Pol AN, Olney JW. Distribution of metabotropic glutamate receptor mGluR5 immunoreactivity in rat brain. J Comp Neurol. 1995;355:455–469. doi: 10.1002/cne.903550310. [DOI] [PubMed] [Google Scholar]
- Schoenbaum G, Roesch MR, Stalnaker TA. Orbitofrontal cortex, decision-making and drug addiction. Trends Neurosci. 2006;29:116–124. doi: 10.1016/j.tins.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigemoto R, Mizuno N. Metabotropic glutamate receptors - immunocytochemical and in situ hybridization analysis. In: Ottersen OP, Storm-Mathisen J, editors. Handbook of Chemical Neuroanatomy: Metabotropic Glutamate Receptors: Immunocytochemical and In Situ Hybridization Analyses. Vol. 18. Elsevier; London: 2000. pp. 63–98. [Google Scholar]
- Shigemoto R, Nomura S, Ohishi H, Sugihara H, Nakanishi S, Mizuno N. Immunohistochemical localization of a metabotropic glutamate receptor, mGluR5, in the rat brain. Neurosci Lett. 1993;163:53–57. doi: 10.1016/0304-3940(93)90227-c. [DOI] [PubMed] [Google Scholar]
- Shigeri Y, Seal RP, Shimamoto K. Molecular pharmacology of glutamate transporters, EAATs and VGLUTs. Brain Res Rev. 2004;45:250–265. doi: 10.1016/j.brainresrev.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Simonyi A, Schachtman TR, Christoffersen GR. The role of metabotropic glutamate receptor 5 in learning and memory processes. Drug News Perspect. 2005;18:353–361. doi: 10.1358/dnp.2005.18.6.927927. [DOI] [PubMed] [Google Scholar]
- Slassi A, Isaac M, Edwards L, Minidis A, Wensbo D, Mattsson J, Nilsson K, Raboisson P, McLeod D, Stormann TM, Hammerland LG, Johnson E. Recent advances in non-competitive mGlu5 receptor antagonists and their potential therapeutic applications. Curr Top Med Chem. 2005;5:897–911. doi: 10.2174/1568026054750236. [DOI] [PubMed] [Google Scholar]
- Spooren W, Gasparini F. mGlu5 receptor antagonists: a novel class of anxiolytics? Drug News Perspect. 2004;17:251–257. doi: 10.1358/dnp.2004.17.4.829052. [DOI] [PubMed] [Google Scholar]
- Spooren WP, Vassout A, Neijt HC, Kuhn R, Gasparini F, Roux S, Porsolt RD, Gentsch C. Anxiolytic-like effects of the prototypical metabotropic glutamate receptor 5 antagonist 2-methyl-6-(phenylethynyl)pyridine in rodents. J Pharmacol Exp Ther. 2000;295:1267–1275. [PubMed] [Google Scholar]
- Swanson CJ, Bures M, Johnson MP, Linden AM, Monn JA, Schoepp DD. Metabotropic glutamate receptors as novel targets for anxiety and stress disorders. Nat Rev Drug Discov. 2005;4:131–144. doi: 10.1038/nrd1630. [DOI] [PubMed] [Google Scholar]
- Tatarczynska E, Klodzinska A, Chojnacka-Wojcik E, Palucha A, Gasparini F, Kuhn R, Pilc A. Potential anxiolytic- and antidepressant-like effects of MPEP, a potent, selective and systemically active mGlu5 receptor antagonist. Br J Pharmacol. 2001;132:1423–1430. doi: 10.1038/sj.bjp.0703923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessari M, Pilla M, Andreoli M, Hutcheson DM, Heidbreder CA. Antagonism at metabotropic glutamate 5 receptors inhibits nicotine- and cocaine-taking behaviours and prevents nicotine-triggered relapse to nicotine-seeking. Eur J Pharmacol. 2004;499:121–133. doi: 10.1016/j.ejphar.2004.07.056. [DOI] [PubMed] [Google Scholar]
- Thomas LS, Jane DE, Gasparini F, Croucher MJ. Glutamate release inhibiting properties of the novel mGlu5 receptor antagonist 2-methyl-6-(phenylethynyl)-pyridine (MPEP): complementary in vitro and in vivo evidence. Neuropharmacology. 2001;41:523–527. doi: 10.1016/s0028-3908(01)00091-0. [DOI] [PubMed] [Google Scholar]
- Tian L, Greenberg SA, Kong SW, Altschuler J, Kohane IS, Park PJ. Discovering statistically significant pathways in expression profiling studies. Proc Natl Acad Sci U S A. 2005;102:13544–13549. doi: 10.1073/pnas.0506577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varty GB, Grilli M, Forlani A, Fredduzzi S, Grzelak ME, Guthrie DH, Hodgson RA, Lu SX, Nicolussi E, Pond AJ, Parker EM, Hunter JC, Higgins GA, Reggiani A, Bertorelli R. The antinociceptive and anxiolytic-like effects of the metabotropic glutamate receptor 5 (mGluR5) antagonists, MPEP and MTEP, and the mGluR1 antagonist, LY456236, in rodents: a comparison of efficacy and side-effect profiles. Psychopharmacology. 2005;179:207–217. doi: 10.1007/s00213-005-2143-4. [DOI] [PubMed] [Google Scholar]
- Witkin JM, Marek GJ, Johnson BG, Schoepp DD. Metabotropic glutamate receptors in the control of mood disorders. CNS Neurol Disord Drug Targets. 2007;6:87–100. doi: 10.2174/187152707780363302. [DOI] [PubMed] [Google Scholar]
- Yang L, Mao L, Chen H, Catavsan M, Kozinn J, Arora A, Liu X, Wang JQ. A signaling mechanism from Gαq-protein-coupled metabotropic glutamate receptors to gene expression: role of the c-Jun N-terminal kinase pathway. J Neurosci. 2006;26:971–980. doi: 10.1523/JNEUROSCI.4423-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]