Abstract
Plakophilins (PKPs) are armadillo family members related to the classical cadherin-associated protein p120ctn. PKPs localize to the cytoplasmic plaque of intercellular junctions and participate in linking the intermediate filament (IF)-binding protein desmoplakin (DP) to desmosomal cadherins. In response to cell–cell contact, PKP2 associates with DP in plaque precursors that form in the cytoplasm and translocate to nascent desmosomes. Here, we provide evidence that PKP2 governs DP assembly dynamics by scaffolding a DP–PKP2–protein kinase Cα (PKCα) complex, which is disrupted by PKP2 knockdown. The behavior of a phosphorylation-deficient DP mutant that associates more tightly with IF is mimicked by PKP2 and PKCα knockdown and PKC pharmacological inhibition, all of which impair junction assembly. PKP2 knockdown is accompanied by increased phosphorylation of PKC substrates, raising the possibility that global alterations in PKC signaling may contribute to pathogenesis of congenital defects caused by PKP2 deficiency.
Introduction
Armadillo family members are multifunctional proteins that play diverse roles in cell–cell adhesion and signaling. Plakophilins (PKPs) comprise a subgroup of the armadillo protein family that is related to the cadherin-associated protein p120ctn (Hatzfeld, 2007). Classically thought to be a constitutive component of the submembrane plaque in intercellular junctions, including desmosomes and the cardiac area composita (Godsel et al., 2004; Franke et al., 2006), the PKPs can also localize to the cytoplasm and nucleus (Mertens et al., 1996; Schmidt et al., 1997, 1999).
Mutations in desmosomal proteins lead to epidermal fragility and/or cardiac defects (Lai-Cheong et al., 2007). In particular, mutations in PKP2 have been described as a major causative factor for congenital cardiac arrhythmias (Gerull et al., 2004; Lai-Cheong et al., 2007). Although compromised junctional integrity is assumed to contribute to disease pathophysiology, the specific etiological mechanism is poorly understood.
PKP2 partners with several junction components, including desmoplakin (DP), with which it interacts via its N terminus (Chen et al., 2002). We recently showed that PKP2 colocalizes with cytoplasmic DP-containing precursors that form in response to cell–cell contact and subsequently translocate to nascent junctions (Godsel et al., 2005). However, the consequences of PKP2 deficiency on precursor formation and dynamics are unknown.
Here, we provide evidence that PKP2 facilitates the association of PKCα with DP, which in turn is required for the assembly competence of this junctional plaque protein. Importantly, PKP2 also regulates the availability of PKC for phosphorylation of other cellular substrates and thus may have a more global role in cellular homeostasis by serving as a scaffold for a ubiquitous signaling molecule.
Results and discussion
PKP2 is required for efficient assembly of DP into desmosomes
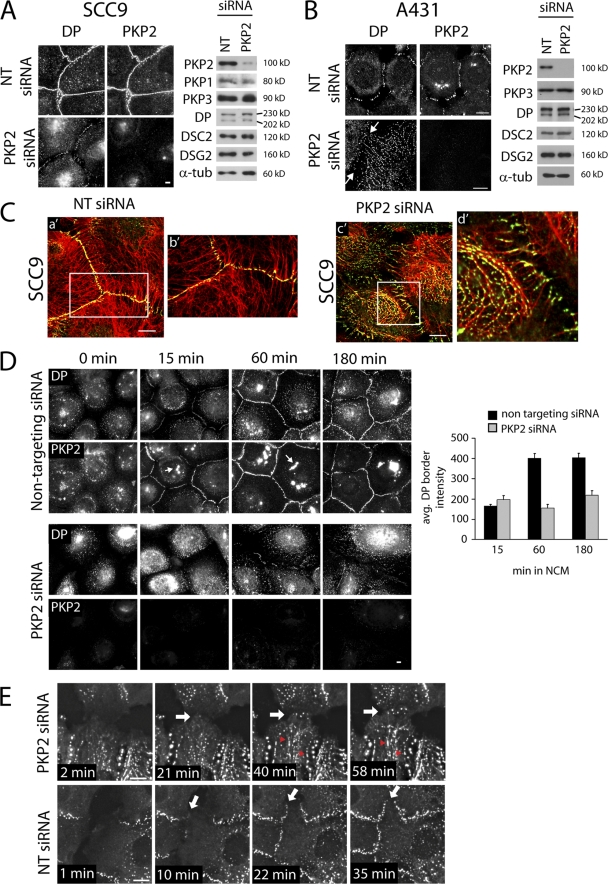
To test whether PKP2 is required for DP assembly into junctions, we introduced siRNA pools into parental or DP-GFP–expressing SCC9 and A431 epithelial cell lines (present at ≤20 and 14% of total DP, respectively), resulting in a ∼90% decrease in PKP2. Although total levels of desmosome proteins were unaffected, DP border fluorescence was severely decreased compared with control siRNA-transfected cells (Fig. 1, A and B) and restored by a silencing-resistant PKP2 (see Fig. 4 A). In addition, DP particles aligned in a striking filamentous pattern in A431 cells (Fig. 1 B), colocalizing extensively with keratin intermediate filaments (IFs; not depicted). DP particles also decorated the IF network in PKP2-deficient SCC9 cells, which appeared partially retracted, with fewer filaments extending to the membrane (Fig. 1 C). Similar results were obtained with three individual siRNAs (unpublished data), which supports the specificity of the response. DP distribution was frequently observed to be more linear in A431 than in SCC9 cells, likely reflecting a difference in keratin IF network organization in these lines. In A431 cells, IF appeared to be organized in straight radial cables compared with a more sinuous, intersecting network of bundles in SCC9s (Fig. 1 C and not depicted).
Figure 1.
PKP2 is required for proper assembly of DP into desmosomes. (A and B) Impaired DP border localization in PKP2-deficient cultures. SCC9 cells (A, wide-field microscopy) or A431 cells (B, confocal microscopy) expressing pooled siRNAs against human PKP2 or nontargeting (NT) control, immunostained for endogenous DP and PKP2. Immunoblot analysis of SCC9 (A, right) or A431 (B, right) from NT or PKP2 siRNA-transfected cells to detect total levels of PKP2, PKP1, PKP3, DP, DSC2, DSG2, DSG3, and α-tubulin. PG protein levels were unaffected (not depicted). Arrows indicate the cell–cell border. (C) DP particles colocalize with keratin IF during PKP2 knockdown. Confocal images of SCC9 cells expressing PKP2 or NT siRNA immunostained for DP (green) and keratin (red). Panels b′ and d′ show magnified views of the boxed areas in a′ and c′, respectively. (D) DP assembly is impaired during PKP2 knockdown. Calcium switch of SCC9 cells transfected with siRNA against PKP2 or NT + siGlo, endogenous PKP2, or DP staining. DP border fluorescence intensity was measured, normalized to background, and plotted in D (right). Arrows indicate siGLO particles. Error bars represent SEM. (E) DP accumulates in a filamentous cytoplasmic pattern during PKP2 knockdown. Shown are stills from Videos 1 and 2 (available at http://www.jcb.org/cgi/content/full/jcb.200712133/DC1) depicting PKP2 or NT siRNA-transfected A431 DP-GFP cells. Arrows indicate where new borders are being formed. Red arrowheads show DP accumulation on IF. Time points indicate time lapsed from beginning of video. Bars, 10 μm.
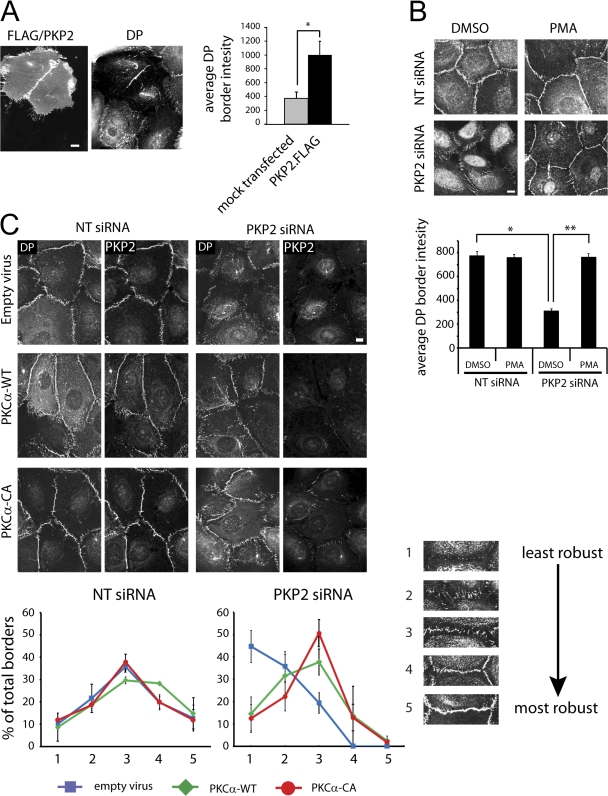
Figure 4.
PKP2, PMA, or PKCα restores DP incorporation into desmosomes. (A) PKP2 reexpression enhances DP border localization during PKP2 knockdown. Silencing-resistant FLAG-tagged PKP2 was coexpressed with PKP2 or nontargeting (NT) siRNA in SCC9 cells. Cells were stained for DP (right), FLAG, and PKP2 (using the same secondary antibody; left). Intensity of DP border fluorescence at borders between pairs of transfected cells or pairs of untransfected cells is shown on the far right. *, P < 0.001. (B) PKC activation rescues DP border localization during PKP2 knockdown. PKP2 siRNA or NT siRNA cells were treated with 15 nM PMA for 30 min and stained for DP. DP border fluorescence quantitation is shown on the bottom. **, P < 0.001. (C) PKC expression rescues DP border localization during PKP2 knockdown. SCC9 cells transfected with PKP2 or NT siRNA were infected with wild-type PKCα or constitutive active PKCα retrovirus and stained for PKP2 and DP. (C, top) Overexpression of PKCα rescues DP border localization. (C, bottom) DP localization at cell–cell borders was assessed for robustness by taking into account the percentage of occupied border, border continuity, and fluorescence intensity. Borders were scored on a graded scale of 1–5 and plotted as percentages of total borders counted. (C, bottom right) Representative borders scored from 1 to 5. 5 represents the most mature border, with most continuous and intense DP fluorescence, and 1 represents the least mature border, with minimal to no DP fluorescence. Graphs represent mean values from three independent experiments. NT and PKP2 siRNA experiments were performed at same time but separated in the graphs for clarity. Error bars represent SEM. Bars, 10 μm.
To test whether PKP2 knockdown affects the kinetics of DP incorporation into forming junctions, a calcium switch was performed. DP accumulation at SCC9 cell–cell borders was reduced by ∼60% at 1 h and ∼40% at 3 h (Fig. 1 D), whereas E-cadherin and plakoglobin (PG) were minimally effected (not depicted). To enhance the temporal resolution of analysis, A431 DP-GFP cells were transfected with PKP2 siRNA, and live cell imaging of cells coming into contact after scratch wounding was performed. DP border fluorescence appeared within minutes of cell–cell contact in both control and PKP2 siRNA-transfected cells (Fig. 1 E; Videos 1–3, available at http://www.jcb.org/cgi/content/full/jcb.200712133/DC1). However, at 20–30 min, a striking accumulation of DP in an IF-like filamentous pattern was observed (Fig. 1 E, top; and Videos 1 and 3). This time frame corresponds to DP appearance in the cytoplasm as small particles or dots in control cells (Fig. 1 E, bottom; and Video 2; Godsel et al., 2005). DP filamentous accumulation also correlates temporally with the point at which the border fluorescence intensity in PKP2 knockdown cells falls off relative to controls (Fig. 1 D). Although in certain cells a beads-on-a-string type of pattern was observed before the beginning of the experiment (Videos 1 and 3), DP fluorescence intensity along these tracks increased in response to cell–cell contact. These results suggest that PKP2 is required for continued accumulation of DP at borders and the maturation of junctions during desmosome assembly, possibly by regulating DP's cytoplasmic localization.
DPser2849 and PKC activity are required for proper DP border localization
The filamentous distribution of DP observed in PKP2-deficient cells was reminiscent of that exhibited by a phosphorylation-deficient point mutant, DPS2849G, which tends to be sequestered along IF and exhibits delayed assembly kinetics (Godsel et al., 2005). Ser2849 falls within a PKA/PKC consensus phosphorylation site in the C terminus of the DP IF-binding domain. Mutation at this site has been shown to enhance DP interactions with IF 10-fold in yeast two-hybrid assays (Stappenbeck et al., 1994; Meng et al., 1997; Fontao et al., 2003; Godsel et al., 2005). Previous work demonstrated that PKC activation can stimulate desmosome formation in the absence of adherens junctions (van Hengel et al., 1997) as well as DP recruitment to cell–cell borders in SCC cells cultured in low calcium (Sheu et al., 1989), both conditions that are normally inhibitory to desmosome formation. Coupled with the similarity between the PKP2-deficient and DPS2849G-expressing cells, this suggested that PKP2 may be important for PKC-dependent signaling during assembly.
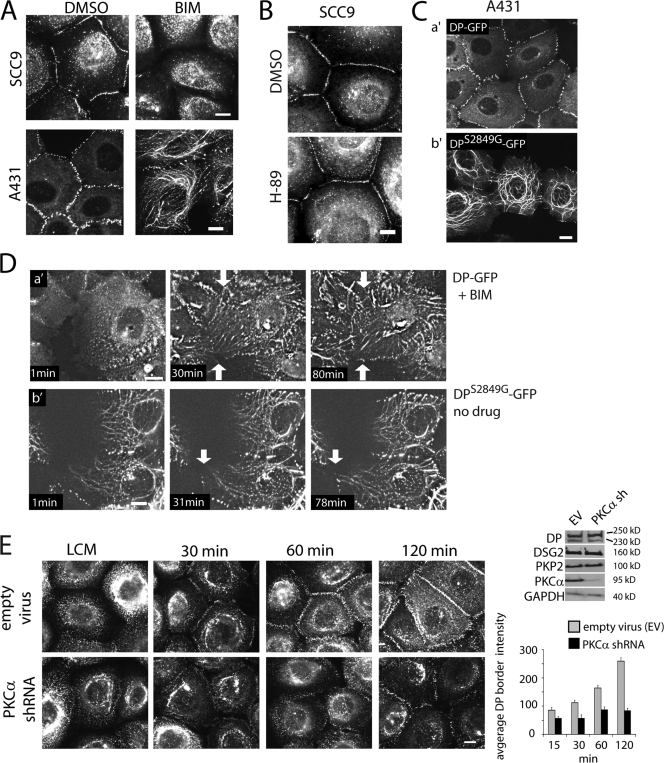
Supporting this possibility, treatment of cells with the PKC inhibitor bisindolylmaleimide I (BIM) severely impaired DP border localization in SCC9 cells (Fig. 2 A, left), whereas PKA inhibition by H-89 had no effect (Fig. 2 B), which suggests that PKC is more important than PKA in regulating DP incorporation into junctions. BIM treatment induced a filamentous DP pattern in A431 cells (Fig. 2 A) similar to that observed in PKP2-deficient cells (Fig. 1 B). Furthermore, when A431 cells expressing wild-type DP-GFP were treated with BIM and analyzed by live cell imaging, DP particles that formed in the cytoplasm underwent a dramatic filamentous alignment, recapitulating the phenotype of DPS2849G and PKP2 knockdown (Fig. 2, C and D; and Videos 4 and 5, available at http://www.jcb.org/cgi/content/full/jcb.200712133/DC1).
Figure 2.
DPser2849 and PKC activity are required for proper DP border localization. (A, left) PKC inhibition impairs DP border localization and induces a filamentous pattern. SCC9 cells treated with 12.5 μM BIM or DMSO for 30 min and immunostained for endogenous DP. (A, right) BIM-treated A431 DP-GFP cells. (B) PKA inhibition does not affect DP border localization. SCC9 cells in low calcium treated with 10 μM H-89 or DMSO and switched to high calcium for 3 h, immunostained for endogenous DP. Quantitative analysis of border fluorescence intensity confirmed that DP localization was comparable in H-89–treated cells. (C) Mutation of Ser2849 results in IF alignment (Godsel et al., 2005). Fixed A431 cells expressing DP-GFP (a′) or DPS2849G-GFP (b′) for comparison with D. (D, a′) Selected stills from Video 4 (available at http://www.jcb.org/cgi/content/full/jcb.200712133/DC1) of A431 DP-GFP cells treated with 12.5 μM BIM. The drug was added at beginning of video and maintained throughout the 90-min time course. (D, b′) Video 5 of untreated A431 DPS2849G-GFP. Arrows indicate areas where new cell–cell contacts occur. Note the variability in the DPS2849G-GFP pattern ranging from particulate to continuous filaments (compare with C). (E) Impaired DP border localization during PKCα knockdown. Calcium switch of SCC9 cells infected with PKCα small hairpin RNA retrovirus or empty virus immunostained for DP. DP border fluorescence was measured, normalized to background, and plotted on the right. Immunoblot analysis of PKC-deficient cells for DP, PKP2, DSG2, PKCα, or GAPDH loading control. Error bars represent SEM. Bars, 10 μm.
PKCα is the only calcium-dependent classical PKC that is expressed in keratinocytes (Denning, 2004), localizes to desmosomes (Jansen et al., 2001; Garrod et al., 2005), and modulates their sensitivity to calcium depletion and adhesive state (Wallis et al., 2000; Kimura et al., 2006). However, the potential mechanistic role of this specific kinase in desmosome assembly is unknown. To test whether PKCα is required for DP assembly into borders, we infected cells with retrovirus-encoded PKCα small hairpin RNA and performed a calcium switch. DP border localization was dramatically impaired (67% decrease) in PKCα-deficient cells compared with control cells (Fig. 2 E), whereas total desmosome protein levels were unaffected (Fig. 2 E, right). These studies demonstrate that PKCα regulates DP assembly into desmosomes, possibly through modulation of DP phosphorylation at Ser2849 and regulating its association with IF.
PKP2 regulates PKC signaling by recruiting it to DP
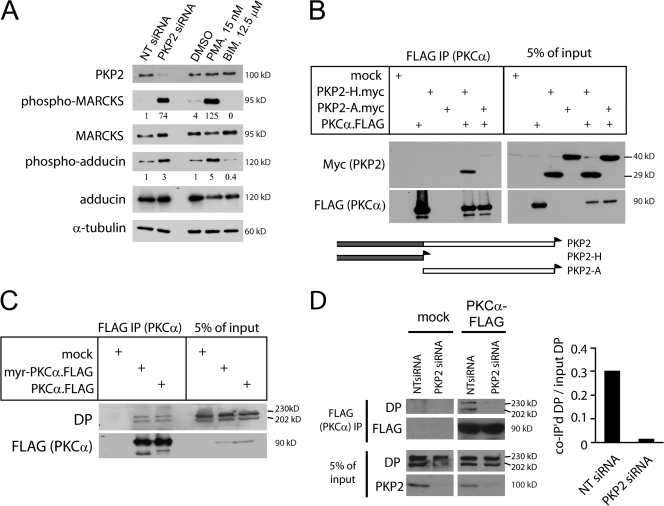
Based on the similarity in DP behavior under PKP2- and PKC-deficient conditions, we set out to test whether the loss of PKP2 affects PKC signaling. Lysates from cells transfected with PKP2 siRNA were probed with phospho-specific antibodies against known PKC substrates. Relative levels of phosphorylated myristoylated alanine-rich PKC substrate (MARCKS) and adducin were increased by >70-fold and threefold, respectively, in PKP2 siRNA-transfected cells (Fig. 3 A). MARCKS phosphorylation was increased in PKP2-deficient cells even when cultured in low calcium (unpublished data), which suggests that cell–cell contact is not required for PKP2 regulation of PKC signaling. However, PKCα phosphorylation at Thr638 was not consistently increased (unpublished data), which suggests that total cellular levels of PKC activity are not affected by PKP2 knockdown. Based on these findings, we hypothesized that PKP2 serves as a scaffold that recruits PKC locally to control the proper assembly and behavior of DP precursors, and that in its absence, PKC is free to phosphorylate other substrates. Consistent with this idea, PKP2 and the PKP2 head domain but not the central armadillo repeats coprecipitated with PKCα-FLAG (Fig. 3 B). Endogenous DP also coprecipitated with PKCα, raising the possibility that the three proteins form a cytoplasmic complex (Fig. 3 C). Importantly, the level of DP that coprecipitated with PKCα in PKP2-deficient cells was reduced by 95%, which is consistent with the idea that PKP2, directly or indirectly, serves as a scaffold for PKC (Fig. 3 D).
Figure 3.
PKP2 regulates PKC signaling. (A) Enhanced PKC substrate phosphorylation during PKP2 knockdown. SCC9 cell lysates from nontargeting (NT) or PKP2 siRNA-transfected cells or cells treated with DMSO, 15 nM PMA, or 12.5 μM BIM for 30 min were probed for PKC substrates phospho-MARCKS or -adducin, total MARCKS, adducin, PKP2, or α-tubulin (loading control). Densitometry numbers below the blots represent ratios of phosphoprotein normalized to total protein from treatment samples relative to the NT siRNA sample. (B) PKCα coimmunoprecipitates the PKP2 N-terminal head but not the central armadillo repeat domain. FLAG-tagged PKCα and myc-tagged PKP2-Head or -Arm were cotransfected into HEK293 cells and subjected to anti-FLAG IP. Blots were probed with anti-myc or anti-FLAG antibodies. (bottom) A schematic of PKP2 constructs. 5% input blots represent 5% of triton lysate from which IP was performed, removed before IP for immunoblot analysis. (C) PKCα coimmunoprecipitates endogenous DP in HEK293 cells. FLAG-tagged wild-type PKCα or myristylated (constitutively active) PKCα were transfected into HEK293 cells and subjected to FLAG IP. Precipitates and lysates were probed for endogenous DP or FLAG. Data are representative of three independent experiments. (D) PKP2 is required for PKC–DP interaction. PKCα-FLAG was immunoprecipitated from HEK293 cells transfected with PKP2 or NT siRNA. Endogenous DP, PKP2, and FLAG were probed. Data are representative of three independent experiments. Densitometry (right) reveals 95% reduction in DP coimmunoprecipitated with PKCα.
PKC activation restores DP desmosome incorporation
If PKP2-dependent recruitment of a limiting pool of active PKC to DP is required for proper regulation of DP–IF interactions, then global activation of PKC might bypass the need for PKP2. Indeed, PMA treatment restored DP borders in PKP2 siRNA-treated cells that had been reduced to 40% of the control (Fig. 4 B). This observation suggests that hyperactivation of PKC leads to an increased probability that DP will be properly phosphorylated by PKC, facilitating its incorporation into desmosomes. Furthermore, ectopic expression of wild-type PKCα and, even more so, constitutively active PKCα in PKP2-deficient cells shifted the proportion of borders from those that exhibit weakly localized DP to those that appear more robust, continuous, and mature (Fig. 4 C). The fact that restoration was not complete may reflect a requirement for PKC-independent structural and/or signaling functions contributed by PKP2 in desmosome assembly or maintenance.
Collectively, these data support a role for PKP2 in coordinating signals required for assembling the DP-rich desmosome plaque (Fig. 5, A and B). In particular, DP behavior during the later phases of assembly is aberrant in PKP2-deficient cells. Instead of coalescing into assembly competent particles (Godsel et al., 2005), DP and/or DP-containing particles appear to coat filaments all along their length and fail to efficiently translocate to cell–cell interfaces. The data are consistent with a model whereby cell contact triggers a signal (possibly PKC activation; van Hengel et al., 1997), and PKP2, either directly or in conjunction with other scaffolding proteins, is required for communicating this signal by recruiting activated PKC to DP, which in turn phosphorylates DP at Ser2849. Although the molecular mechanism is not yet known, it seems possible that the observed reduction in affinity for IF exhibited by phosphorylated DP may be caused by alterations in the conformation of adjacent regions making up the IF-binding domain. A more dynamic association with IF may be necessary for ultimately promoting plaque precursor assembly while maintaining precursors locally concentrated near the cortex for proper delivery to junctions.
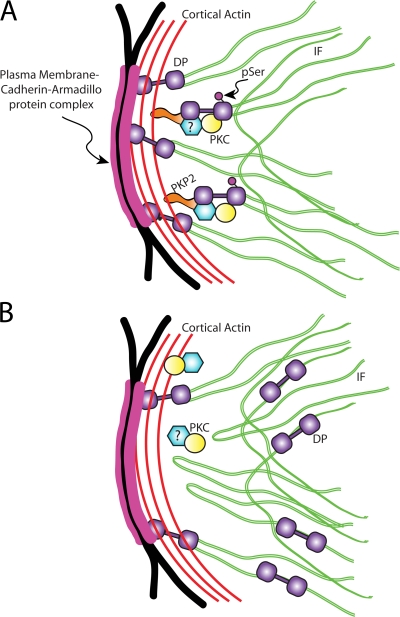
Figure 5.
Model for role of PKP2 in PKC-regulated DP assembly. (A) Wild type. PKP2 recruits PKC to DP cytoplasmic complexes, where phosphorylation of DP at Ser2849 modulates its interaction with IF and allows DP assembly into cell–cell junctions. (B) PKP2 deficiency. PKC is no longer recruited to DP complexes and is free to phosphorylate other substrates, leading to aberrant accumulation of DP along IF and impaired DP assembly.
Somewhat surprisingly, although PKP2 appears at borders earlier than DP during the assembly process (unpublished data), the loss of PKP2 did not completely abrogate DP clustering at cell borders during assembly (Fig. 1), which suggests that its initial accumulation does not require PKP2. PG and/or PKP3 may partially compensate for the loss of PKP2 and stabilize DP near the membrane, particularly in tissues where PKP2 is not the primary PKP expressed. However, as the nonmembrane-bound assembly competent dots that appear in the cytoplasm after cell contact contain PKP2 but not PKP3 or PG, the role of PKP2 at this intermediate stage may be more critical (Godsel et al., 2005). DP border intensity is reduced at steady state in all cultured cell types tested, which suggests that compensatory mechanisms are insufficient for proper maturation and/or stability of junctions.
Our data are consistent with observations performed in PKP2-deficient mice (Grossmann et al., 2004). The authors reported a loss of DP from cell–cell junctions and the presence of cytoplasmic DP-containing aggregates that were intimately associated with desmin IF of the cardiac muscle. Along with the fact that Ser2849 phosphorylation also regulates DP–desmin IF interactions (Lapouge et al., 2006), these observations support a similar role for PKP2 in other cell types. It was proposed that mechanical defects caused by loss of junctional integrity underlie the observed cardiac rupture and embryonic lethality in PKP2 knockout mice and may also contribute to human cardiac arrhythmias associated with PKP2 mutations. The relative contributions of PKPs in different cell contexts may be different, as patients with PKP2 mutations and prominent heart defects have not been reported to exhibit loss of keratinocyte adhesion.
In addition to promoting junction assembly and maintenance by locally harnessing PKC activity, the observation that PKP2 prevents aberrant phosphorylation of PKC substrates suggests that it may have broader cellular functions. Such global changes in the distribution of active PKC could contribute to disease pathogenesis in patients with PKP2 deficiency. The PKC substrates investigated in this report and others are actin-binding proteins (Larsson, 2006). Thus, this PKP2–PKC signaling pathway could also contribute to actin remodeling that is important for desmosome assembly, an idea that is consistent with our previous observation that DP translocation is actin dependent (Godsel et al., 2005). Coupled with recently ascribed roles for p120ctn and p0071 in actomyosin contraction signaling and regulation of Rho GTPases (Hatzfeld, 2005; Wolf et al., 2006), the possibility that PKP2 scaffolds PKC suggests an emerging common function for these armadillo proteins in locally coordinating signals that direct cytoskeletal remodeling and regulate junction dynamics.
Materials and methods
DNA constructs
DP-GFP and DPS2849G-GFP (Godsel et al., 2005) and PKP2-FLAG, PKP2-H-FLAG, and PKP2-A-FLAG (Chen et al., 2002) have been described previously. PKP2-H-myc and -A-myc were created by PCR amplifying the PKP2-H and -A into pBK-CMV EcoRI and XbaI sites. PKCα-FLAG cDNAs were provided by A. Toker (Beth Israel Deaconess Medical Center, Boston, MA; Rabinovitz et al., 1999). siRNA-resistant PKP2-FLAG was made by site-directed mutagenesis (Stratagene).
Retroviral constructs were obtained as follows: pSUPER.Retro was a gift from V. Cryns (Northwestern University, Chicago, IL); pSUPER.Retro.shPKCα (Leirdal and Sioud, 2002) and LZRS-Linker have been described previously (Kinsella and Nolan, 1996); and LZRS-PKCα was constructed by cloning EcoRI-digested hPKCα cDNA (No. 65978; American Type Culture Collection) into the EcoRI sites of LZRS-Linker. Constitutively active PKCα (LZRS-PKCαΔ22-28) was constructed by deleting the pseudosubstrate domain (amino acids 22–29) by site-directed mutagenesis and was cloned into the EcoRI sites of LZRS-linker.
Nonspecific siRNA (negative control), siGLO (positive transfection control), SMARTpool, and individual siRNAs against hPKP2 (Thermo Fisher Scientific) were used for siRNA experiments.
Cell lines and transfections
SCC9, SCC9 DP-GFP, and A431 DP-GFP lines and culture conditions have been described previously (Godsel et al., 2005). HEK293 cells were grown in DME and 10% FBS. Phoenix amphotropic cells were a gift from G. Nolan (Stanford University, Stanford, CA) and were maintained in DME and 10% hI-FBS.
cDNAs were transfected using ExGen 500 (Fermentas) or Lipofectamine 2000 (Invitrogen). siRNAs (Thermo Fisher Scientific) were transfected using DharmaFECT1. Cells were analyzed 72–96 h after transfection. Retroviruses were packaged and transduced as described previously (Getsios et al., 2004).
Antibodies and chemical reagents
Rabbit polyclonal antibodies used were: NW6 anti-DP; anti-MARCKS (Santa Cruz Biotechnology, Inc.), anti-phospho-MARCKS (Cell Signaling Technology); anti-PKCα/βII and rIgG (Sigma-Aldrich); anti-FLAG (Affinity BioReagents and Sigma-Aldrich); anti-GFP (Clontech Laboratories, Inc.); anti–α-adducin and anti–phospho-adducin (provided by V. Bennett (Duke University, Durham, NC; Matsuoka et al., 1998); 667 anti-PKP1 (provided by M. Hatzfeld; Martin Luther University Halle, Halle, Germany); and 2026 anti-cMyc and 1905 anti-Dsg3 (provided by J. Stanley, University of Pennsylvania, Philadelphia, PA). Mouse monoclonal antibodies used were: 12G10 anti–α-tubulin (provided by J. Frankel and E.M. Nelsen from the Developmental Studies Hybridoma Bank under the auspices of the National Institute of Child Health and Human Development and maintained by the University of Iowa, Department of Biological Sciences, Iowa City, IA); anti-GAPDH (Novus Biologicals); KSB17.2 anti-keratin, 9E10 anti-myc, and mIgG (Sigma-Aldrich); MAB6013S anti-PKP2 (BioDesign, Inc.); 1G4 anti-DP and 7G6 anti-Dsc2; 6D8 anti-Dsg2; 23E3 anti-PKP3 (provided by F. van Roy, University of Ghent, Ghent, Belgium); and chicken polyclonal 1407 anti-PG. Alexa Fluor 568 or 488–conjugated goat IgGs (Invitrogen), and HRP-conjugated goat IgGs were also used (Kirkegaard & Perry Laboratories, Inc.; Rockland Immunochemicals, Inc.). For kinase inhibition and activation, exhausted media was supplemented with BIM, PMA, or H-89 (EMD) or DMSO vehicle (Sigma-Aldrich).
Calcium switch
SCC9 cells were incubated in low-calcium medium (DME with 0.05 mM CaCl2) for 16–20 h, switched to normal growth media containing ∼1.2 mM Ca2+ to induce cell junction assembly for time periods ranging up to 3 h, and processed for immunofluorescence analysis.
Immunoprecipitation (IP) and immunoblotting
Lysates were processed for IP as described previously (Chen et al., 2002) in lysis buffer (10 mM TrisCl, pH 7.5, 1% Triton X-100, 145 mM NaCl, 5 mM EDTA, 2 mM EGTA, and protease inhibitor cocktail [Roche] ± 2% phosphatase inhibitor cocktail IV [EMD]). FLAG-tagged proteins were immunoprecipitated with anti-FLAG M2-agarose (Sigma-Aldrich). Samples were resolved by 6.5 or 7.5% SDS-PAGE and immunoblotted as described previously (Chen et al., 2002).
Immunofluorescence analysis, fixed and time-lapse image acquisition
All immunofluorescence, fixed, and time-lapse imaging procedures were performed as described previously (Godsel et al., 2005). SCC9 or A431 cells were seeded onto 0.1-mg/ml collagen I–coated (BD Biosciences) glass coverslips. For fixed immunofluorescence analysis, coverslips were washed in PBS, fixed in anhydrous methanol for 2 min at −20°C, and processed for indirect immunofluorescence. When using MAB6013, fixed cells were extracted with 0.5% Triton X-100 for 30 min at 4°C before antibody incubation. After incubation in the appropriate secondary antibody (see Antibodies and chemical reagents), coverslips were mounted in polyvinyl alcohol (Sigma-Aldrich).
Fixed cells were visualized with a microscope (DMR; Leica) using 40× PL Fluotar, NA 1.0 or 63× PL APO, NA 1.32 objectives (Leica), a charge-coupled device camera (Orca 100, model C4742-95; Hamamatsu) and MetaMorph 6.1 software (MDS Analytical Technologies) or a laser scanning confocal microscope (LSM 510; Carl Zeiss, Inc.) using a 100× Plan Apo Chromat, NA 1.4 objective and LSM 510 software (Carl Zeiss, Inc.). Images were further processed using Photoshop CS3 (Adobe) and compiled using Illustrator CS3 (Adobe).
For live imaging, 3 d after transfection, PKP2 siRNA-transfected, control siRNA with siGlo-transfected (to locate siRNA-transfected cells), or untransfected cells were seeded onto Lab-Tek chambered coverglass slides (Thermo Fisher Scientific). Cell monolayers were wounded with a 26-gauge needle and incubated in imaging medium (Hanks balanced salt solution, 20 mM Hepes, 1% FBS, 2 mM l-glutamine, 4.5 g/liter glucose, and 1× amino acids; recipe courtesy of G. Kreitzer, Weill Medical College of Cornell University, New York, NY) at 37°C for 60 min and then processed for time lapse imaging.
DP-GFP fluorescence time-lapse recordings were obtained at consistent time intervals of 1–1.5 min with rapid z stack acquisition (12–15; 0.4–0.5-μm stacks) using an Application Solution Multidimensional Workstation (ASMDW; Leica), an inverted microscope (DMIRE2; Leica) fitted with a Coolsnap HQ camera (Roper Scientific) and a 63× objective (HCX PL APO, glycerine, NA 1.3) with a piezo element. Cells were subjected to imaging in a 37°C climate chamber. All images were processed using a blind deconvolution synthetic algorithm and z stacks were assembled into multi-image projections using ASMDW software. All movies were compiled using MetaMorph 6.1 imaging software (MDS Analytical Technologies).
Quantitation of fluorescence intensity and immunoblot densitometry
Fluorescence pixel intensity at cell borders was determined by multiplying the mean pixel intensity by the area. Background intensity was subtracted from border intensity. Calculations were performed using MetaMorph 6.1 software. Densitometric analyses were performed using Officejet 5610 scan software (Hewlett-Packard) and analyzed using Image J (National Institutes of Health).
Statistical analysis
Error bars represent SEM. Statistical analysis was performed using two-tailed t test.
Online supplemental material
All videos show DP-GFP dynamics during cell–cell contact-initiated junction assembly. Videos 1–3 show PKP2 siRNA- (1 and 3) and control siRNA-transfected cells (2). Video 4 shows DP dynamics during PKC inhibition. Video 5 shows DPS2849G dynamics. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200712133/DC1.
Supplementary Material
Acknowledgments
We thank V. Bennett, V. Cryns, M. Hatzfeld, G. Nolan, J. Stanley, A. Toker, and F. van Roy for generously providing reagents. We thank A. Kowalczyk, S. Brady-Kalnay, and C. Gottardi for critical reading of the manuscript and L. Godsel for critical reading of the manuscript, expert guidance, and support. We thank H. Miao and S. McMillan for technical assistance.
This work was supported by National Institutes of Health grants R01AR43380 and R01AR41836, with partial support from R01CA122151 (to K.J. Green), 1R01DE016905 (to J. Wahl III), and American Cancer Society grant RSG0424901CCG (to M.F. Denning). A.E. Bass-Zubek was supported in part by grants from National Institutes of Health (T32CA009560) and the American Heart Association (0615631Z).
Note added in proof. While this manuscript was under review, Pieperhoff et al. (Pieperhoff, S., H. Schumacher, and W.W. Franke. 2008. Eur. J. Cell Biol. In press) found that PKP2 knockdown in cultured cardiomyocytes led to the appearance of cytoplasmic DP associated with IFs and reduction of DP at cell–cell borders, resulting in a loss of adhesion junctions.
Abbreviations used in this paper: BIM, bisindolylmaleimide I; DP, desmoplakin; IF, intermediate filaments; IP, immunoprecipitation; MARCKS, myristoylated alanine-rich PKC substrate; PG, plakoglobin; PKP, plakophilin.
References
- Chen, X., S. Bonne, M. Hatzfeld, F. van Roy, and K.J. Green. 2002. Protein binding and functional characterization of plakophilin 2. Evidence for its diverse roles in desmosomes and beta-catenin signaling. J. Biol. Chem. 277:10512–10522. [DOI] [PubMed] [Google Scholar]
- Denning, M.F. 2004. Epidermal keratinocytes: regulation of multiple cell phenotypes by multiple protein kinase C isoforms. Int. J. Biochem. Cell Biol. 36:1141–1146. [DOI] [PubMed] [Google Scholar]
- Fontao, L., B. Favre, S. Riou, D. Geerts, F. Jaunin, J.-H. Saurat, K.J. Green, A. Sonnenberg, and L. Borradori. 2003. Interaction of the bullous pemphigoid antigen 1 (BP230) and desmoplakin with intermediate filaments is mediated by distinct sequences within their COOH terminus. Mol. Biol. Cell. 14:1978–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke, W.W., C.M. Borrmann, C. Grund, and S. Pieperhoff. 2006. The area composita of adhering junctions connecting heart muscle cells of vertebrates. I. Molecular definition in intercalated disks of cardiomyocytes by immunoelectron micrsocopy of desmosomal proteins. Eur. J. Cell Biol. 85:69–82. [DOI] [PubMed] [Google Scholar]
- Garrod, D.R., M.Y. Berika, W.F. Bardsley, D. Holmes, and L. Tabernero. 2005. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J. Cell Sci. 118:5743–5754. [DOI] [PubMed] [Google Scholar]
- Gerull, B., A. Heuser, T. Wichter, M. Paul, C.T. Basson, D.A. McDermott, B.B. Lerman, S.M. Markowitz, P.T. Ellinor, C.A. MacRae, et al. 2004. Mutations in the desmosomal protein plakophilin-2 are common in arrhythmogenic right ventricular cardiomyopathy. Nat. Genet. 36:1162–1164. [DOI] [PubMed] [Google Scholar]
- Getsios, S., E.V. Amargo, R.L. Dusek, K. Ishii, L. Sheu, L.M. Godsel, and K.J. Green. 2004. Coordinated expression of desmoglein 1 and desmocollin 1 regulates intercellular adhesion. Differentiation. 72:419–433. [DOI] [PubMed] [Google Scholar]
- Godsel, L.M., S. Getsios, A.C. Huen, and K.J. Green. 2004. The molecular composition and function of desmosomes. In Handbook of Experimental Pharmacology: Cell Adhesion. J.N. Behrens and W.J. Nelson, editors. Springer-Verlag, New York. 137–193. [DOI] [PubMed]
- Godsel, L.M., S.N. Hsieh, E.V. Amargo, A.E. Bass, L.T. Pascoe-McGillicuddy, A.C. Huen, M.E. Thorne, C.A. Gaudry, J.K. Park, K. Myung, et al. 2005. Desmoplakin assembly dynamics in 4D: multiple phases differentially regulated by intermediate filaments and actin. J. Cell Biol. 171:1045–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann, K.S., C. Grund, J. Huelsken, M. Behrend, B. Erdmann, W.W. Franke, and W. Birchmeier. 2004. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J. Cell Biol. 167:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzfeld, M. 2005. The p120 family of cell adhesion molecules. Eur. J. Cell Biol. 84:205–214. [DOI] [PubMed] [Google Scholar]
- Hatzfeld, M. 2007. Plakophilins: Multifunctional proteins or just regulators of desmosomal adhesions? Biochim. Biophys. Acta. 1773:69–77. [DOI] [PubMed] [Google Scholar]
- Jansen, A.P., N.E. Dreckschmidt, E.G. Verwiebe, D.L. Wheeler, T.D. Oberley, and A.K. Verma. 2001. Relation of the induction of epidermal ornithine decarboxylase and hyperplasia to the different skin tumor-promotion susceptibilities of protein kinase C alpha, -delta and -epsilon transgenic mice. Int. J. Cancer. 93:635–643. [DOI] [PubMed] [Google Scholar]
- Kimura, T., A. Merritt, and D. Garrod. 2006. Calcium-independent desmosomes of keratinocytes are hyper-adhesive. J. Invest. Dermatol. 127:775–781. [DOI] [PubMed] [Google Scholar]
- Kinsella, T.M., and G.P. Nolan. 1996. Episomal vectors rapidly and stably produce high-titer recombinant retrovirus. Hum. Gene Ther. 7:1405–1413. [DOI] [PubMed] [Google Scholar]
- Lai-Cheong, J.E., K. Arita, and J.A. McGrath. 2007. Genetic diseases of junctions. J. Invest. Dermatol. 127:2713–2725. [DOI] [PubMed] [Google Scholar]
- Lapouge, K., L. Fontao, M.F. Champliaud, F. Jaunin, M.A. Frias, B. Favre, D. Paulin, K.J. Green, and L. Borradori. 2006. New insights into the molecular basis of desmoplakin- and desmin-related cardiomyopathies. J. Cell Sci. 119:4974–4985. [DOI] [PubMed] [Google Scholar]
- Larsson, C. 2006. Protein Kinase C and the regulation of the actin cytoskeleton. Cell. Signal. 18:276–284. [DOI] [PubMed] [Google Scholar]
- Leirdal, M., and M. Sioud. 2002. Gene silencing in mammalian cells by preformed small RNA duplexes. Biochem. Biophys. Res. Commun. 295:744–748. [DOI] [PubMed] [Google Scholar]
- Matsuoka, Y., X. Li, and V. Bennett. 1998. Adducin is an in vivo substrate for protein kinase C: phosphorylation in the MARCKS-related domain inhibits activity in promoting spectrin–actin complexes and occurs in many cells, including dendritic spines of neurons. J. Cell Biol. 142:485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, J.J., E.A. Bornslaeger, K.J. Green, P.M. Steinert, and W. Ip. 1997. Two-hybrid analysis reveals fundamental differences in direct interactions between desmoplakin and cell type-specific intermediate filaments. J. Biol. Chem. 272:21495–21503. [DOI] [PubMed] [Google Scholar]
- Mertens, C., C. Kuhn, and W.W. Franke. 1996. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol. 135:1009–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovitz, I., A. Toker, and A.M. Mercurio. 1999. Protein kinase C–dependent mobilization of the α6β4 integrin from hemidesmosomes and its association with actin-rich cell protrusions drive the chemotactic migration of carcinoma cells. J. Cell Biol. 146:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, A., L. Langbein, M. Rode, S. Pratzel, R. Zimbelmann, and W.W. Franke. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481–499. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., L. Langbein, S. Pratzel, M. Rode, H.R. Rackwitz, and W.W. Franke. 1999. Plakophilin 3–a novel cell-type-specific desmosomal plaque protein. Differentiation. 64:291–306. [DOI] [PubMed] [Google Scholar]
- Sheu, H.M., Y. Kitajima, and H. Yaoita. 1989. Involvement of protein kinase C in translocation of desmoplakins from cytosol to plasma membrane during desmosome formation in human squamous cell carcinoma cells grown in low to normal calcium concentration. Exp. Cell Res. 185:176–190. [DOI] [PubMed] [Google Scholar]
- Stappenbeck, T.S., J.A. Lamb, C.M. Corcoran, and K.J. Green. 1994. Phosphorylation of the desmoplakin COOH terminus negatively regulates its interaction with keratin intermediate filament networks. J. Biol. Chem. 269:29351–29354. [PubMed] [Google Scholar]
- van Hengel, J., L. Gohon, E. Bruyneel, S. Vermeulen, M. Cornelissen, M. Mareel, and F. von Roy. 1997. Protein kinase C activation upregulates intercellular adhesion of α-catenin–negative human colon cancer cell variants via induction of desmosomes. J. Cell Biol. 137:1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis, S., S. Lloyd, I. Wise, G. Ireland, T.P. Fleming, and D. Garrod. 2000. The alpha isoform of protein kinase C is involved in signaling the response of desmosomes to wounding in cultured epithelial cells. Mol. Biol. Cell. 11:1077–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf, A., R. Keil, O. Gotzl, A. Mun, K. Schwarze, M. Lederer, S. Huttelmaier, and M. Hatzfeld. 2006. The armadillo protein p0071 regulates Rho signalling during cytokinesis. Nat. Cell Biol. 8:1432–1440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.