Abstract
Fus2p is a pheromone-induced protein associated with the amphiphysin homologue Rvs161p, which is required for cell fusion during mating in Saccharomyces cerevisiae. We constructed a functional Fus2p–green fluorescent protein (GFP), which exhibits highly dynamic localization patterns in pheromone-responding cells (shmoos): diffuse nuclear, mobile cytoplasmic dots and stable cortical patches concentrated at the shmoo tip. In mitotic cells, Fus2p-GFP is nuclear but becomes cytoplasmic as cells form shmoos, dependent on the Fus3p protein kinase and high levels of pheromone signaling. The rapid cytoplasmic movement of Fus2p-GFP dots requires Rvs161p and polymerized actin and is aberrant in mutants with compromised actin organization, which suggests that the Fus2p dots are transported along actin cables, possibly in association with vesicles. Maintenance of Fus2p-GFP patches at the shmoo tip cortex is jointly dependent on actin and a membrane protein, Fus1p, which suggests that Fus1p is an anchor for Fus2p. In zygotes, Fus2p-GFP forms a dilating ring at the cell junction, returning to the nucleus at the completion of cell fusion.
Introduction
Haploid cells of the yeast Saccharomyces cerevisiae conjugate to form diploid cells capable of entering meiosis. The two haploid mating types (MATa and MATα) detect each others' peptide pheromones (a-factor or α-factor) via G protein–coupled cell surface receptors, which activate an intracellular mitogen-activated protein (MAP)-kinase cascade (Sprague et al., 1983; Dohlman and Thorner, 2001). The downstream MAP-kinase Fus3p regulates at least three different pathways required for efficient mating, including transcriptional regulation, cell cycle arrest, and polarized growth. Polarized growth is directed toward the highest pheromone concentration (Segall, 1993; Barkai et al., 1998; Madden and Snyder, 1998). After mating partners make contact, they adhere stably, forming a prezygote (Lipke and Kurjan, 1992). Cell fusion entails removal of the intervening cell wall (Trueheart et al., 1987; Marsh and Rose, 1997; Gammie et al., 1998) and subsequent fusion of the plasma membranes (Heiman and Walter, 2000; Jin et al., 2004) to produce a zygote with a common cytoplasm. As zygotes mature, cell wall remnants are removed, expanding the diameter of the zone of cell fusion (ZCF) and forming a smooth intracellular margin (Gammie et al., 1998). After cell fusion, nuclei are pulled together by microtubules (Meluh and Rose, 1990; Molk et al., 2006) and the nuclear envelopes fuse to form a diploid nucleus (Kurihara et al., 1994; Beh et al., 1997; Melloy et al., 2007). The newly formed diploid cells then resume mitotic cell growth (Marsh and Rose, 1997).
Cell fusion remains a poorly understood process. Several genes are known to play a role in cell fusion (White and Rose, 2001), which fall into five broad classes: (1) adhesion (e.g., FIG2; Zhang et al., 2002), (2) actin organization and cell polarization (e.g., BNI1, PEA2,and SPA2; Chenevert et al., 1994; Valtz and Herskowitz, 1996; Evangelista et al., 1997; Sheu et al., 1998; Matheos et al., 2004), (3) membrane fusion (e.g., PRM1; Heiman and Walter, 2000; Jin et al., 2004), (4) negative regulation (e.g., PKC1; Philips and Herskowitz, 1997), and (5) cell wall degradation at the ZCF (e.g., FUS2, RVS161, and LRG1; Elion et al., 1995; Brizzio et al., 1998; Gammie et al., 1998; Fitch et al., 2004). Several of these genes also affect the formation of a pheromone-induced low-affinity calcium channel (Muller et al., 2003). The precise mechanism of cell fusion remains unclear and little is known about how these processes are coordinated.
Based on genetic criteria, Fus1p and Fus2p are thought to act in separate pathways; double mutants show a significantly stronger defect than single mutants (Trueheart et al., 1987; Elion et al., 1995; Brizzio et al., 1998). Electron microscopy suggested that Fus1p and Fus2p act at different stages of cell fusion (before and after vesicle clustering at the ZCF, respectively; Gammie et al., 1998). Other data suggest that the two proteins interact or have partially overlapping functions; overexpression of either protein partially suppresses deletion of the other gene (Trueheart et al., 1987; Gammie et al., 1998) and a two-hybrid study identified Fus1p and Fus2p as interacting proteins (Nelson et al., 2004).
Rvs161p and Fus2p function together at the same stage of cell fusion (Brizzio et al., 1998; Gammie et al., 1998). The defect of fus2 rvs161 double mutants is not more severe than the single mutants (Brizzio et al., 1998), and rvs161 mutant cell fusion defects are identical to fus2 mutants (Gammie et al., 1998). Overexpression of Fus2p partially suppresses the requirement for Rvs161p but not vice versa, indicating that Rvs161p likely assists Fus2p (Gammie et al., 1998). Fus2p and Rvs161p have been shown to interact physically (Brizzio et al., 1998) and in two-hybrid studies (Bon et al., 2000; Talarek et al., 2005; Germann et al., 2005), and Fus2p stability depends on Rvs161p (Brizzio et al., 1998).
Rvs161p consists of only a BAR domain, a conserved three-helix bundle shared with amphiphysins (Peter et al., 2004). BAR domains dimerize to form crescent-shaped membrane-binding domains that preferentially bind to curved membranes (Peter et al., 2004). Rvs161p interacts with a second BAR domain protein, Rvs167p, to form an obligate heterodimer that binds lipids (Friesen et al., 2006). Although both proteins are required for endocytosis (Munn et al., 1995), only mutations in RVS161 cause a strong defect in cell fusion (Brizzio et al., 1998; Friesen et al., 2006). Alleles of RVS161 have been identified that specifically effect either endocytosis or cell fusion, indicating that the two functions are independent (Brizzio et al., 1998). How Rvs161p could mediate different functions has remained unclear.
Rvs161p localizes to the tip of the shmoo and the ZCF (Brizzio et al., 1998). A nonfunctional Fus2p-lacZ hybrid was found in a cytoplasmic aggregate in shmoos, although some hybrid protein localized near the ZCF (Elion et al., 1995). To reconcile this apparent conflict, we constructed a fully functional Fus2p-GFP hybrid using a novel internal tagging approach. Like Rvs161p, functional Fus2p-GFP localized to the tip of the shmoo and to the ZCF. However, live-cell observations revealed that Fus2p localization is both highly dynamic and subject to complex regulation. Remarkably, in zygotes, Fus2p-GFP localized to an expanding ring at the cell fusion junction.
Results
Localization of Fus2p-GFP in wild-type cells
FUS2 encodes a 677-residue, 79-kD protein (Fig. 1 A), including a region (residues 116–327) that shares a homology with the Dbl family of guanine nucleotide exchange factors (GEFs) for Rho-like GTPases (Rho-GEF, P = 1.9 × 10−35, analyzed by SMART; Letunic et al., 2006). A short motif (PTRRKYS) resembling an NLS is present within the Rho-GEF domain (residues 313–319). A region near the C terminus (538–574) is predicted to form a coiled coil (probability 0.8, analyzed by COILS; Lupas et al., 1991). Bordering on the coiled-coil region (570–582) is a motif (FQNLQNQMKRELP) that is partially homologous to a highly conserved motif in Rvs161p. Homologues of FUS2 are found within closely related yeast species as well as more distantly related hemiascomycetes (e.g., Candida glabrata, Debaryomyces hansenii, Ashbya gossypii, and Kluyveromyces lactis; Dujon et al., 2004).
Figure 1.
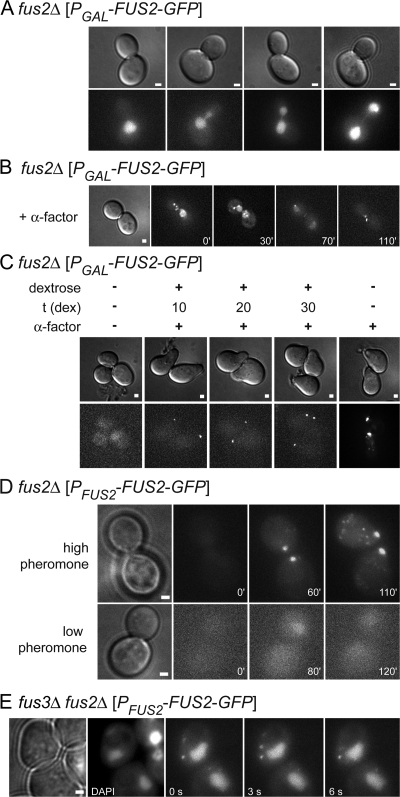
Fus2p-GFP is functional and localizes to the shmoo tip. (A) FUS2 encodes a 79-kD protein with a Dbl Rho-GEF domain (DBH). A putative NLS is present at amino acid residues 313–319. A putative coiled-coil domain is present near the C terminus. (B) Plate mating assays show complementation of the fus2Δ by FLAG and GFP-tagged FUS2. Patches of MY9182, MY9183, MY9184, JM269, and JM362 were mated to JY429 and replica printed to selective media (SC-leu-trp) to select for diploids. (C) Quantitative mating assays. Ratios of diploids formed/total cells (±SEM) are reported for matings of MY9182, MY9183, MY9184, and JM269 to JY429. Experiments were preformed in triplicate. (D) Fus2p-GFP transport in live cells. MY9184 was treated with α-factor for 1.5 h. Images were acquired every 1.5 s. (E) Fus2p-GFP localization during shmoo formation. MY9184 was treated with α-factor at 0 min. Images were acquired every 10 min. The localization to the shmoo tip (arrowhead), rapid movement through the cell (filled arrow), faint nuclear localization (open arrow), and unstable cortical localization (asterisk) are indicated in D and E. Bars, 1 μm.
To identify the localization of functional Fus2p, we attempted to epitope tag both the N and C termini of the protein. In both cases, the resulting proteins were only partially functional (7 and 29% of wild-type levels of mating, respectively). Immunofluorescent microscopy showed two different patterns of localization in shmoos; the N-terminally tagged protein localized to the shmoo-tip and the C-terminally tagged protein localized to the nucleus (unpublished data). However, both proteins localized to the ZCF in prezygotes. Both termini are strongly conserved in other yeasts, which suggests that these regions are required for normal function. To identify functional sites to tag Fus2p, we compared the FUS2 sequence of closely related yeasts (Cliften et al., 2003; Kellis et al., 2003) to identify regions that were especially nonconserved and therefore likely to reside in nonessential surface loops. Two regions were chosen for internal epitope tagging (residues 104–109 and 410–419). In both cases, the FLAG-tagged proteins were fully functional (100% of wild-type mating efficiency) and localized to the shmoo tip and ZCF (unpublished data).
To observe Fus2p localization in live cells, we used the upstream internal site to insert GFP. Using a sensitive replica-plate mating assay to a fus1 fus2 partner, the Fus2p-GFP–tagged protein supported mating as well as either the Fus2p-FLAG construct or untagged wild-type Fus2p (Fig. 1 B). In quantitative matings with a fus1 fus2 partner, the Fus2p-FLAG construct supported mating as well as wild-type Fus2p, and Fus2p-GFP retained at least 90% of wild-type function (Fig. 1 C).
To observe the dynamic behavior of Fus2p-GFP, cells were first treated with α-factor to induce expression and allow cells to progress into the mating pathway. In cells that had arrested their cell cycle and formed shmoos, Fus2p-GFP exhibited three general types of localization (Fig. 1 D): diffuse in the nucleus, cytoplasmic dots, and cortical patches. Many of the cytoplasmic dots moved rapidly from the cell body to the shmoo tip, following roughly linear tracks at a mean velocity of 0.38 ± 0.19 μm/s (n = 135, maximal velocity = 0.99 μm/s). A few dots moved away from the shmoo tip and others exhibited random short-range movements. Cortical patches at or near the shmoo tip were very stable, showing no movement and no decrease in intensity over the course of the observation. Patches appearing at other cortical sites were not stable and disappeared over time.
To determine the order at which each localization pattern appeared, mitotic cells were treated with pheromone and observed by time-lapse microscopy at 10-min intervals (Fig. 1 E). In cells that had not completed mitosis, faint Fus2p-GFP fluorescence was observed in the nucleus (20–60 min). After cells completed mitosis, Fus2p-GFP was observed at the nascent shmoo tip (80 min). As the shmoo became more pronounced, Fus2p-GFP fluorescence at the tip increased, and cytoplasmic dots were observed (100 min). The later appearance of dots may reflect ectopic localization as cortical sites become saturated or the early low level of expression may have precluded detection. Regardless, these results show that Fus2p-GFP initially localizes to the nucleus and localizes to cytoplasmic sites after cells exit mitosis and enter the mating pathway.
Fus2p nuclear localization is regulated by the mating response
FUS2 expression is pheromone regulated; in vegetative cells, FUS2 mRNA is virtually undetectable, increasing >32-fold after pheromone treatment (Roberts et al., 2000). To eliminate expression effects on localization studies, FUS2-GFP was placed under the control of the inducible GAL1 promoter. In vegetative cells in which PGAL-FUS2-GFP was expressed by induction with galactose, Fus2p-GFP accumulated in the nucleus at all stages of the cell cycle (Fig. 2 A). When cells expressing PGAL-FUS2-GFP were treated with α-factor, Fus2p-GFP stayed in the nucleus until the completion of mitosis and subsequently localized to the shmoo tip (Fig. 2 B). Thus, the export of Fus2p from the nucleus depends on signals from the pheromone response pathway, possibly in coordination with cell cycle control.
Figure 2.
Fus2p localization is regulated by the mating response. (A) Mitotically expressed Fus2p-GFP localizes to the nucleus at all stages of mitotic growth. MY9201 containing PGAL-FUS2-GFP was grown in media containing galactose (top, DIC; bottom, GFP). (B) α-factor treatment causes Fus2p-GFP to exit the nucleus. MY9201 was grown in galactose-containing media and induced with α-factor. As the cell completed mitosis, nuclear Fus2p exited the nucleus and localized to the shmoo tip in pheromone-responding cells (left, DIC at 0 min; remaining panels, GFP). (C) Preexisting nuclear Fus2p-GFP exits the nucleus. MY9201 grown in galactose was transferred to media containing dextrose for the indicated times (10, 20, or 30 min) to shut off expression before treatment with pheromone for 1.5 h. Fus2p-GFP was nuclear in mitotic cells but localized to the shmoo tip in shmoos (top, DIC; bottom, GFP). (D) Fus2p-GFP nuclear exit is dependent on high levels of pheromone. MY9184 was treated with 6 μM α-factor (top) or 0.6 μM α-factor (bottom) and imaged at various times (left, transmitted or DIC at 0 min; right, GFP). (E) Nuclear exit requires the Fus3p MAP-kinase. MY9211 was treated with α-factor for 1.5 h. Transmitted light (first panel), DAPI of the nucleus (second panel), and GFP (remaining panels) are shown. Bars, 1 μm.
The apparent relocalization of Fus2p-GFP to the cortex could be caused by efflux from the nucleus or new synthesis of cytoplasmic protein coupled with turnover of nuclear protein. To distinguish between these two possibilities, vegetative PGAL-FUS2-GFP was induced with galactose and transcription was then repressed by the addition of glucose. The mRNA was allowed to decay (10, 20, and 30 min), after which α-factor was added to induce the mating pathway for 1.5 h (Fig. 2 C). For all periods of glucose repression, Fus2p-GFP localized to the shmoo tip and no Fus2p-GFP localized to the nucleus. The level of fluorescence at the shmoo tip was similar for all time points, indicating that residual FUS2-GFP mRNA did not make a substantial contribution to the signal. In cells where expression was maintained by continued presence of galactose, Fus2p-GFP stayed nuclear in the absence of pheromone. When pheromone was added to these cells, Fus2p-GFP localized to the shmoo tip and cytoplasmic dots. The signal at the shmoo tip was significantly stronger than when glucose was present, as expected for continued synthesis after pheromone addition. The results strongly suggest that nuclear Fus2p-GFP exits the nucleus and localizes to the shmoo tip in response to pheromone signaling.
Yeast cells exhibit a complex dose response to pheromone; 10–100-fold higher levels are required for shmoo formation compared with cell cycle arrest (Moore, 1983). To determine whether Fus2p relocalization is simply a consequence of cell cycle arrest, cells containing FUS2-GFP expressed from its normal promoter were treated with either standard (6 μM) or low (0.6 μM) levels of α-factor. At low pheromone levels, cells arrested but did not polarize to form shmoos, and low levels of Fus2p-GFP could be detected in the nucleus, confirming that pheromone induction had occurred. However, the Fus2p-GFP remained in the nucleus and did not localize to the cell cortex during the experiment (Fig. 2 D). In contrast, when cells were treated with high levels of pheromone, cells began to polarize soon after completing mitosis, and Fus2p-GFP localized to the tip of the shmoo and to cytoplasmic dots (Fig. 2 D). Thus, relocalization of Fus2p-GFP from the nucleus to the site of polarized growth requires high levels of pheromone beyond those sufficient for its transcriptional induction and cell cycle arrest.
The pheromone response signal is chiefly transmitted by activation of the MAP kinase Fus3p. The fus3Δ mutants are defective for cell fusion, in part because of a defect in cell polarization (Matheos et al., 2004). The fus3Δ mutants respond transcriptionally to pheromone due to the presence of a second partially redundant MAP kinase, Kss1p. To distinguish the transcriptional response from other aspects of pheromone signaling, we examined a fus3Δ mutant. Fus2p-GFP localized at the shmoo tip in 97% of wild-type cells treated with α-factor; it was retained in the nucleus in only 1% of cells. Fus2p-GFP was well-induced by pheromone in the KSS1+ fus3Δ mutant but localized to the shmoo tip in only 2% of cells (Fig. 2 E). In >85% of cells, Fus2p-GFP was retained in the nucleus. In ∼40% of the fus3Δ cells, a small number of cytoplasmic dots were observed moving in the cytoplasm. These results show that Fus3p is specifically required for relocalization.
Rvs161p is required for Fus2p-GFP localization and movement
Fus2p interacts with Rvs161p during mating and Rvs161p is required for Fus2p's stability and function (Brizzio et al., 1998; Gammie et al., 1998). We therefore examined the localization of Fus2p-GFP in an rvs161Δ mutant. In ∼90% of rvs161Δ shmoos, Fus2p-GFP failed to localize to the cortex or shmoo tip; most cells showed diffuse punctate localization in the cytoplasm (Fig. 3 A). The cytoplasmic dots did not undergo rapid transport; almost all showed short-range movements similar to Brownian motion. In some shmoos, faint nuclear localization was observed as well as a few dots close to the nucleus. In sum, Rvs161p is not required for Fus2p-GFP nuclear export but is required for Fus2p-GFP's rapid cytoplasmic movement and localization to the shmoo tip.
Figure 3.
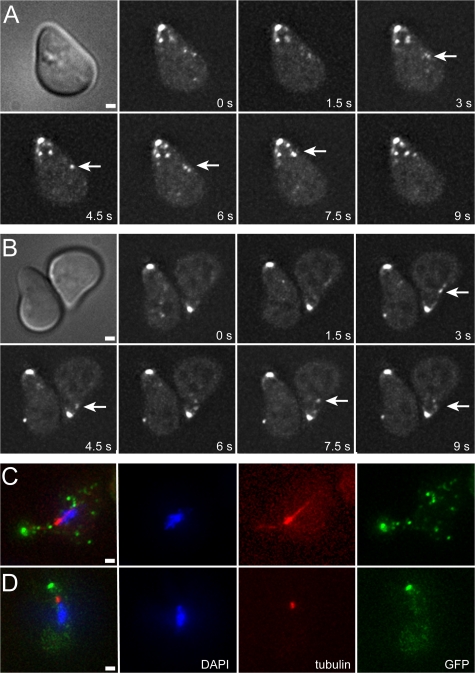
Rvs161p is required for Fus2p movement and localization. (A) Fus2p-GFP is not transported or localized in an rvs161Δ mutant. MY9214 was induced with pheromone for 1.5 h. (B) Fus2p-GFP localization is dependent on Rvs161p's cell fusion function. MY9231 (rvs161Δ), MY9233 (cell fusion defective allele rvs161-P203Q), and MY9239 (endocytosis-defective allele rvs161-R113K) were treated with α-factor for 1.5 h. Bars, 1 μm. (C) Fus2p interacts with Rvs161p but not Rvs167p. Wild-type cells expressing Rvs167p-HA, Fus2p-FLAG, or both epitope-tagged proteins were treated with α-factor, and proteins were immunoprecipitated with anti-HA or anti-FLAG antibodies. Fus2p-FLAG and Rvs167p-HA could immunoprecipitate Rvs161p but the two tagged proteins did not coimmunoprecipitate with each other. (D) Rvs161p association with Fus2p is dependent on pheromone induction. Fus2p-GFP was expressed from the PGAL1 promoter (MY9201) by induction with galactose, mock treated, or treated with α-factor for 1.5 h. Protein extracts were prepared and Fus2p-GFP was immunoprecipitated.
Rvs161p has two independent functions, endocytosis and cell fusion, and function-specific mutations have been isolated (end and fus). The fus mutations are specifically defective for binding Fus2p (Brizzio et al., 1998). To differentiate between defects in endocytosis or cell fusion, Fus2p-GFP was transformed into end and fus mutants. In an end-rvs161 mutant, Fus2p-GFP localized normally at the shmoo tip. In a fus-rvs161 mutant, Fus2p-GFP exhibited abnormal localization similar to the rvs161Δ null mutant, neither localizing to the shmoo tip nor moving rapidly (Fig. 3 B). Therefore, Fus2p-GFP localization is dependent on the interaction with Rvs161p, and the mutant defect is not due to an indirect effect on endocytosis.
Rvs161p and Rvs167p form an obligate heterodimer during vegetative growth, and this complex is present after pheromone stimulation (Friesen et al., 2006). To determine whether Rvs161p and Rvs167p are both associated with Fus2p in a ternary complex during mating, we performed coimmunoprecipitation (coIP) experiments in which all three proteins were detected (Fig. 3 C). In α-factor–treated cells, IP of either functional Fus2p-FLAG or Rvs167p-HA pulled down Rvs161p as expected (Brizzio et al., 1998; Friesen et al., 2006). However, in cells expressing both tagged proteins, IP of Rvs167p-HA was unable to pull down Fus2p-FLAG, and IP of Fus2p-FLAG was unable to pull down Rvs167-HA. We conclude that Fus2p and Rvs161p form a heterodimer during mating, separate from the Rvs161p/Rvs167p heterodimer, reflecting the different functions of Rvs161p in mating and endocytosis. When Fus2p-GFP was constitutively expressed under the control of the galactose promoter, formation of the Rvs161p–Fus2p complex was dependent on the pheromone response (Fig. 3 D), which is consistent with mitotic Fus2p being retained in the nucleus.
Fus2p transport and localization are not dependent on microtubules
The rapid linear movement of Fus2p-GFP dots suggested that they are actively transported along cytoskeletal filaments. Shmoos contain two different cytoskeletal elements along which movement might occur: (1) microtubules that extend from the nuclear envelope to the tip of the shmoo and (2) actin cables that form a network that emanates from the shmoo tip into the cytoplasm. To determine whether microtubules play a role in Fus2p localization, shmoos were treated with nocodazole to depolymerize microtubules (Fig. 4, A and B). In both mock-treated and nocodazole-treated cells, Fus2p-GFP localized normally to the shmoo tip and Fus2p-GFP dots moved rapidly through the cell (Fig. 4, arrows). Immunofluorescent staining for tubulin confirmed that the microtubules were depolymerized (Fig. 4, C and D). Thus, microtubules do not play a significant role in Fus2p localization or dynamics.
Figure 4.
Fus2p movement and localization are independent of microtubules. MY9184 expressing Fus2-GFP expressed from its own promoter was treated with α-factor for 1.5 h and either mock treated (A and C) or treated with nocodazole (B and D) for 10 min. Live cell image stacks were acquired (A and B). Arrows indicate directed movement of Fus2p-GFP puncta. Alternatively, cells were fixed and stained with anti-tubulin antibody (C and D) to demonstrate microtubule depolymerization. Bars, 1 μm.
Fus2p localization is aberrant in polarity mutants
To determine if Fus2p-GFP localization is dependent on the actin cytoskeleton, we first examined mutants with defects in cell fusion due to defective actin organization: bni1, pea2, and spa2. BNI1 encodes one of two formins required for actin cable nucleation (Evangelista et al., 1997; Pruyne et al., 2002; Sagot et al., 2002). Bni1p is a substrate for the Fus3p MAP kinase and bni1 mutants fail to form polarized shmoos (Evangelista et al., 1997; Matheos et al., 2004). PEA2 and SPA2 mutants form peanut-shaped shmoos with broad shmoo tips due to disorganized actin cytoskeletons (Gehrung and Snyder, 1990; Chenevert et al., 1994; Valtz and Herskowitz, 1996). Bni1p, Pea2p, and Spa2p are part of the polarisome complex, which helps orient the cytoskeleton toward the bud site in mitotic cells and the shmoo tip during mating (Sheu et al., 1998).
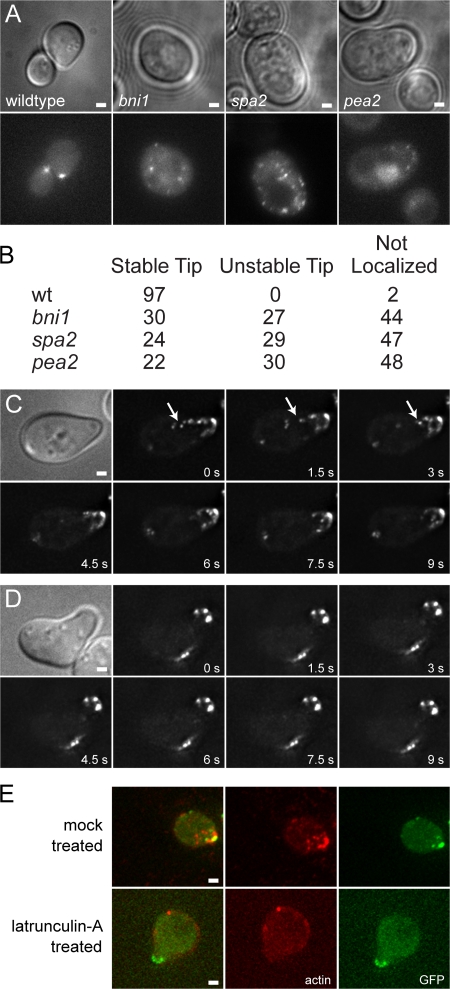
Fus2p-GFP localized to the shmoo tip cortex in 97% of wild-type shmoos (Fig. 5, A and B). In bni1, pea2, and spa2 mutants, stable localization to defined regions of the cortex was observed in only 20–30% of cells. In most mutant cells, the Fus2p-GFP dots were randomly distributed. In cells where some dots were seen at the cortex, localization was unstable, disappearing after 10–20 min. We infer that the polarisome complex and/or proteins dependent on the polarisome are required for localization and maintenance of Fus2p-GFP at the cortex.
Figure 5.
Fus2p localization and transport is dependent on actin organization. (A) Fus2p-GFP localization is disrupted in cell polarity mutants. Strains (MY9184 MY9208, MY9213, and MY9216) were treated with α-factor for 1.5 h (top, DIC or transmitted; bottom, GFP). (B) Quantitation of Fus2p-GFP localization in polarity mutants. See text for details. Numbers indicate the percentage of cells with indicated patterns of localization (50 < n < 100). (C–E) Actin depolymerization disrupts Fus2p-GFP movement but not maintenance. MY9184 was treated with α-factor for 1.5 h and either mock-treated (C) or treated with lat-A (D) for 10 min. Fus2p-GFP moved rapidly in mock-treated cells (arrows) and was stably localized to the shmoo tip. Lat-A–treated cells showed no movement of Fus2p-GFP dots, but Fus2p-GFP stayed localized at the shmoo tip. (E) Cells in C and D were fixed was stained with Texas red–phalloidin to demonstrate actin depolymerization. Bars, 1 μm.
Latrunculin disrupts Fus2p-GFP movement but not maintenance
To directly examine the requirement for actin, we used latrunculin-A (lat-A), which depolymerizes actin within a few minutes after addition. Because polarization requires an intact actin cytoskeleton, cells were first treated with α-factor to form shmoos and then treated with lat-A. In mock-treated cells, 94% of shmoos showed rapid movement of Fus2p-GFP dots (Fig. 5 C). After lat-A treatment, rapid movement was not seen in any cell (Fig. 5 D). In 11% of cells, we observed short-range movements, similar to Brownian motion. We conclude that rapid movement of the Fus2p-GFP dots is caused by actin-dependent transport.
In lat-A–treated cells, Fus2p-GFP localized to a broad region or several dots at the shmoo tip. To confirm that actin was depolymerized, cells were fixed and stained with Texas red–conjugated phalloidin. In control cells, cortical actin patches were concentrated at the shmoo tip (Fig. 5 E, top) and Fus2p-GFP colocalized with a subset of the patches. In lat-A–treated cells, phalloidin staining was diffuse or nonexistent (Fig. 5 E, bottom). Nevertheless, Fus2p-GFP remained at the shmoo tip, showing that actin is not uniquely required for the maintenance of cortical localization.
Cortical localization is dependent on both actin and Fus1p
We hypothesized that Fus2p-GFP remained localized to the shmoo tip in the absence of actin because it is anchored at the cortex by other proteins. The defects of the polarisome mutants implied that the putative cortical anchor might also be polarized. One candidate for a cortical anchor is Fus1p, a pheromone-regulated O-glycosylated integral membrane protein (Trueheart and Fink, 1989). Fus1p localizes to a broad region at the shmoo tip (Trueheart et al., 1987; Nelson et al., 2004).
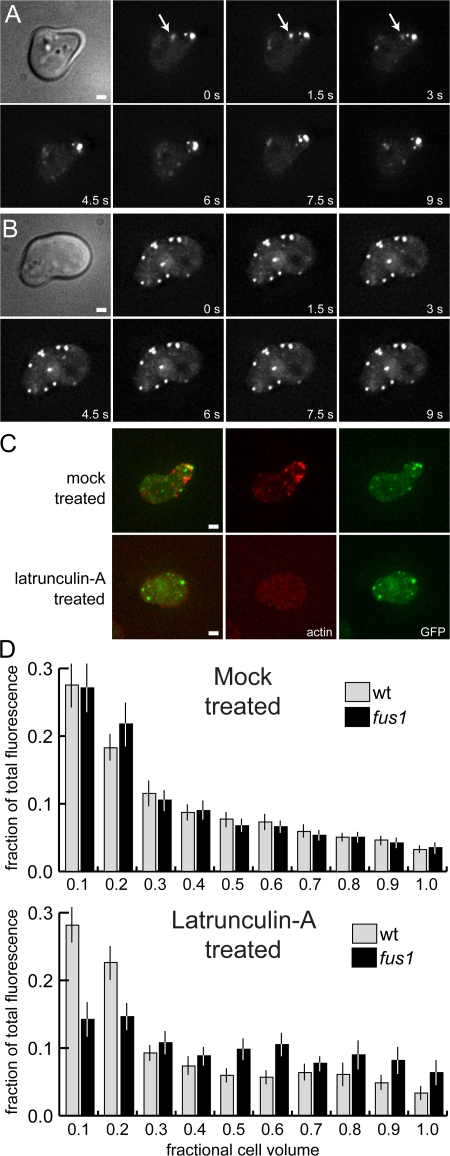
Deletion of FUS1 had surprisingly little effect on Fus2p-GFP localization. Cytoplasmic dots of Fus2p-GFP moved with wild-type velocity and concentrated at the shmoo tip (Fig. 6 A). However, actin-dependent transport might concentrate Fus2p-GFP at the shmoo tip and obscure an anchoring defect. Accordingly, we blocked actin-dependent processes in the fus1Δ mutant with lat-A. Strikingly, within 10 min after lat-A treatment, Fus2p-GFP became randomly distributed throughout the fus1Δ shmoos (Fig. 6 B). Texas red–phalloidin staining of the fixed cells confirmed that actin localization was normal in untreated fus1Δ cells and that lat-A treatment caused complete depolymerization (Fig. 6 C).
Figure 6.
Cortical localization is dependent on both actin and Fus1p. (A and B) Fus2p-GFP localization is disrupted by lat-A in a fus1 mutant. MY9217 was treated with α-factor for 1.5 h and either mock treated or treated with lat-A as in Fig. 5. (A) The fus1 mutation had no effect on the transport or localization of Fus2p-GFP. Arrows indicate rapid movement of Fus2p-GFP dots. (B) Treatment with lat-A (10 min) leads to a rapid loss of Fus2p-GFP localization at the shmoo tip. (C) Texas red–phalloidin staining of fixed cells after lat-A treatment demonstrating actin depolymerization. Bars, 1 μm. (D) Distribution of GFP fluorescence in wild-type and fus1 mutant shmoos. The fraction of total fluorescence was measured for every 1/10 of the cell, starting at the shmoo tip. 50 cells were measured for each strain and condition. Bars represent the mean values; error bars indicate 95% confidence intervals.
To quantify the effects of fus1Δ and lat-A on Fus2p-GFP localization, cell images were divided into 10 equal-width strips, orthogonal to the length of the cell. GFP fluorescence was measured in each strip and expressed as the fraction of total cell fluorescence (Fig. 6 D). In wild-type shmoos, almost half of the total fluorescence was found within the first 2/10 of the cell; the first 1/10 contained significantly more than the second 1/10 (28 vs. 18%). Remaining sections each contained 5–10% of the total in a decreasing gradient from the tip. The GFP fluorescence in untreated fus1Δ and lat-A–treated wild-type shmoos was very similar, albeit slightly less concentrated at the tip (fluorescence in the second 1/10 increased to 22 and 23%, respectively). In contrast, lat-A treatment of fus1Δ shmoos greatly reduced fluorescence at the tip (14%), and GFP became evenly distributed throughout all cell sections. Therefore, Fus1p and actin are jointly required for localization of Fus2p-GFP at the shmoo tip.
Localization of Fus2p in prezygotes and zygotes: an expanding ring during cell fusion
Localization at the shmoo tip positions Fus2p-GFP at the presumptive ZCF. In wild-type zygotes, Fus2p-GFP initially localized to a small patch at the ZCF (0.73 ± 0.17 μm, n = 11) that is considerably narrower than the overall width of the ZCF (Fig. 7 A, top). As cell fusion progressed, Fus2p-GFP expanded at a mean rate of 0.065 ± 0.02 μm/min along the ZCF to a diameter comparable to the junction between the cells (1.87 ± 0.31 μm, n = 20). Coincident with the expansion, Fus2p-GFP intensity at the ZCF progressively diminished, and increased localization was observed in the nucleus (Fig. 7 A, 30′–50′). In wild-type zygotes, 24 ± 5 min (n = 10) elapsed from the initiation of expansion to the final disappearance of Fus2p-GFP at the ZCF.
Figure 7.
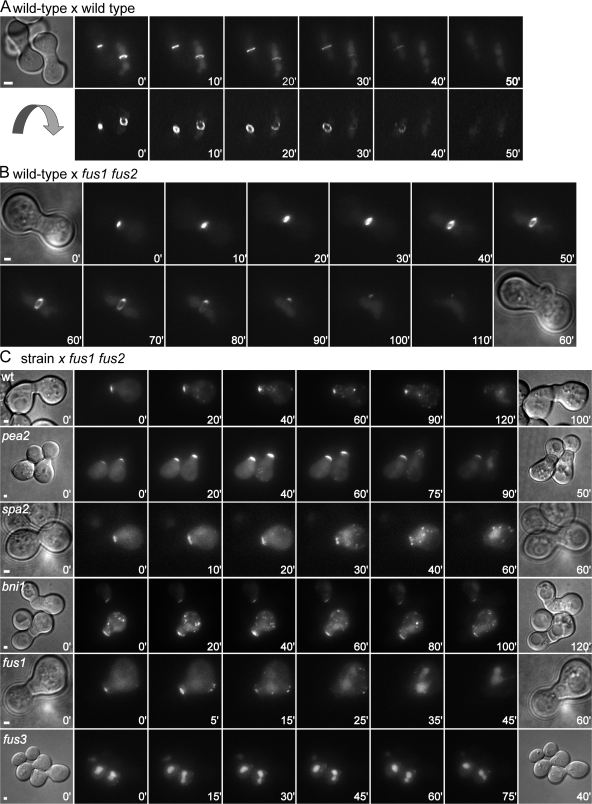
Fus2p-GFP localizes of to an expanding ring at the ZCF. (A) Mating of wild type (MY9184) expressing Fus2p-GFP to wild type (MY8093) for 1 h. Flattened projections of deconvolved images are shown (top). To reveal the ring, deconvolved images were rotated ∼150° around the x axis. Fus2p-GFP localizes first to a restricted patch at the ZCF (left zygote, 0 min), dilates into a ring (10–40 min), and then accumulates in the nucleus (50 min). (B) Mating of wild type (MY9184) expressing Fus2p-GFP to a fus1 fus2 mutant (JY429). Deconvolved images were rotated slightly around the y axis to show the ring. (C) Indicated strains expressing Fus2p-GFP (MY9184, MY9213, MY9216, MY9208, MY9217, and 9211) were mated to a fus1 fus2 mutant (JY429). Bars, 1 μm.
Because zygotes tend to orient parallel to the microscope slide, details of the expanding ZCF were obscured. To remedy this, images were captured at multiple focal planes during cell fusion and deconvolved image stacks were rotated about the x axis. From these images, it was evident that the initial patch of Fus2p-GFP developed into a ring, which then dilated as cell fusion progressed (Fig. 7 A, bottom).
To facilitate these studies, Fus2p-GFP cells were mated to fus1 fus2 mutants, which impeded cell fusion, making it easier to find and observe prezygotes. Cell fusion succeeded in ∼30% of matings between wild-type and fus1 fus2 cells; a representative zygote is shown in Fig. 7 B. As in fully wild-type matings, Fus2p-GFP initially localized to a small region of the ZCF. Ring expansion began with a significantly larger patch of Fus2p-GFP (0.98 ± 0.33 μm, n = 17, P = 0.014), which expanded into a ring somewhat smaller than in the wild type (1.66 ± 0.42 μm, n = 19) before relocalizing to the nucleus. Ring dilation was significantly slower than in fully wild-type matings, occurring at less than half the rate (0.027 ± 0.009 μm/min, n = 16, P < 0.0001). The mean time between dilation initiation and Fus2p-GFP disappearance from the ZCF was significantly longer (36 ± 8 min, n = 8, P = 0.003). Therefore, in zygotes where functional Fus1p and Fus2p are supplied by only one parent, the overall rate of fusion is limited by their reduced concentration.
Cell fusion did not succeed in ∼70% of wild-type–fus1 fus2 matings. In these prezygotes, Fus2p-GFP persisted at the ZCF for ∼71 ± 17 min (n = 29), forming a patch 1.20 ± 0.21 μm in diameter (n = 32) before relocalizing to the nucleus (Fig. 7 C). During relocalization, Fus2p-GFP was frequently seen in cytoplasmic dots, possibly reflecting intracellular transport. Relocalization may reflect adaptation to pheromone signaling or reinitiation of polarization (Bidlingmaier and Snyder, 2004). In some cases, frustrated prezygotic cells subsequently attempted to conjugate with another neighboring cell (unpublished data). In these cases, Fus2p-GFP relocalized to the ZCF associated with the new partner.
We next examined Fus2p-GFP localization in polarisome mutants (pea2, spa2, and bni1) mated to the fus1 fus2 strain (Fig. 7 C). As expected, very few such matings lead to productive cell fusion (<2%). Surprisingly, in >90% of these prezygotes, Fus2p-GFP did localize to the ZCF, albeit to significantly broader regions than the wild type (pea2, 1.78 ± 0.35 μm; spa2, 1.59 ± 0.13 μm; and bni1, 2.23 ± 0.48 μm; P < 0.0001). Like wild-type–fus1 fus2 prezygotes, at later time points, Fus2p-GFP increasingly localized to cytoplasmic dots and then relocalized to the nucleus (Fig. 7 C). However, Fus2p-GFP localization to the ZCF was significantly less persistent in the mutants than in wild-type cells (pea2, 50 ± 14 min; spa2, 52 ± 22 min; bni1, 48 ± 11 min; P < 0.0002), which suggests that actin organization contributes to the maintenance of Fus2p at the ZCF.
In fus1 mutants mated to fus1 fus2, Fus2p-GFP localized initially to a small patch at the ZCF (1.12 ± 0.29 μm), which is similar to the wild type. However, Fus2p-GFP very rapidly dispersed from the ZCF and relocalized to the nucleus significantly faster than wild-type or polarisome mutants (19 ± 5 min, n = 18, P < 0.0001), which is consistent with a role for Fus1p in retaining Fus2 at ZCF.
Fus2p-GFP was retained in the nucleus when fus3 mutant cells were mated with fus1 fus2 (Fig. 7 C, bottom). Unlike the polarisome mutants, Fus2p was not transiently localized at the ZCF but appeared nuclear at all time points. In ∼30% of prezygotes, a small amount of Fus2p-GFP localized to the ZCF, indicating that the cells were not defective for localization per se. A defect in Fus2p export may contribute to the fus3 mutant cell fusion defect.
Discussion
From these observations, we propose a model for Fus2p during yeast mating (Fig. 8). When initially expressed in mitotic cells exposed to pheromone, Fus2p localizes to the nucleus. Upon cell cycle completion, and dependent on high levels of pheromone signaling, Fus2p exits the nucleus and complexes with the amphiphysin homologue Rvs161p. Cytoplasmic Fus2p–Rvs161p is transported along actin cables and localizes to the shmoo tip cortex. The Fus2p dots may represent secretory vesicles, given the known association of Rvs161p with membranes (Friesen et al., 2006). Fus2p–Rvs161p would be anchored at the cortex, dependent on actin and an integral membrane protein, Fus1p. Based upon the presence of a Rho-GEF domain, we hypothesize that cortical Fus2p regulates a Rho-type G protein. Given the requirement for Fus2p–Rvs161p to remove the intervening cell walls, we speculate that activation of a Rho protein would lead to fusion of secretory vesicles bearing cell wall–degrading enzymes. After cell fusion, Fus2p–Rvs161p would remain associated with remnant cell walls, ultimately returning to the nucleus as cells reenter the mitotic cycle.
Figure 8.
A model for the role of Fus2p in cell fusion. Cytoplasmic Fus2p associates with Rvs161p and attaches to vesicles (ves) as it is transported to the shmoo tip along actin cables by a myosin (myo). Fus2p/Rvs161p would be tethered to the cortex by Fus1p. Fus2p then acts as a Rho-GEF, activating a putative Rho-GTPase to trigger vesicle fusion with the plasma membrane. Cell wall (cw) hydrolytic enzymes, carried by the vesicles, would be released into the extracellular space to degrade the intervening cell wall, allowing plasma membranes (pm) to fuse in subsequent steps.
Given its function in cell fusion, the initial nuclear localization of Fus2p was surprising. However, regulation of Fus2p's localization may be required for proper coordination of mating with respect to mitosis. Fus2p interacts with Rvs161p, which has mitotic functions in the cell unrelated to cell fusion (e.g., actin organization and endocytosis). Premature interaction with Fus2p might sequester Rvs161p and interfere with its mitotic functions. Similarly, we expect that Fus2p's GEF domain regulates one of the six Rho proteins in yeast. The Rho proteins have critical roles in yeast mitosis and inappropriate activation would likely be deleterious. Therefore, nuclear localization may prevent interference with the completion of mitosis. Similarly, Fus2p relocalization to the nucleus after cell fusion would prevent interference with the next cell cycle.
The movement and localization of cytoplasmic Fus2p depended on interaction with the amphiphysin homologue Rvs161p. Most BAR domain proteins contain protein interaction domains and/or G protein regulatory domains (e.g., Rho-GEF, Rho-GAP, or Arf-GAP), which suggests that they often have roles in signaling (Peter et al., 2004). Notably, Rvs161p consists solely of a BAR domain. The mitotic association of Rvs161p with Rvs167p provides an SH3 domain and functions related to endocytosis (Munn et al., 1995; Bon et al., 2000; Talarek et al., 2005; Friesen et al., 2006). The association with Fus2p provides a putative Rho-GEF domain with functions specific to cell fusion. From this perspective, the Fus2p–Rvs161p complex would be similar to a typical BAR domain protein, except that the domains have been split into two proteins to allow greater regulatory flexibility.
The interaction of Fus2p with Rvs161p suggests that Fus2p would be associated with membranes. Indeed, the behavior of Fus2p-GFP closely parallels that of vesicles at the ZCF (Gammie et al., 1998). In wild-type prezygotes, both Fus2p-GFP and the vesicles were initially restricted to a small subregion of the ZCF (0.5 and 0.73 μm in diameter, respectively). Mutations causing dispersal of the vesicles (pea2 and spa2) resulted in broader zones of Fus2p localization. Similarly, in fus1 mutants, vesicles were rare at the ZCF and Fus2p-GFP was significantly less persistent.
Vesicles also cluster around the remnant cell walls that persist after cell fusion (Gammie et al., 1998; Melloy et al., 2007). Consistent with localization to vesicles, Fus2p-GFP localizes to a ring that forms, dilates, and disappears after cell fusion. The ring may represent a site where Fus2p–Rvs161p helps to remove residual cell wall remnants; a feature of fus2 and rvs161 mutants is the failure to remove cell wall remnants even when cell fusion occurs (Brizzio et al., 1998; Gammie et al., 1998).
Fus2p-GFP persisted at the ZCF ring even as it began to accumulate in the nucleus at the end of cell fusion. The regulatory signals that determine cortical versus nuclear localization are unclear. Perhaps Fus2p–Rvs161p binds to the highly curved membrane at the remnant cell wall by virtue of the BAR domain. Removal of remnant cell wall and smoothing of the plasma membrane would then erase the signal for localization. Alternatively, the loss of cortical localization may reflect mating or cell cycle–dependent modification of Fus2p or interacting proteins.
Localization of Fus2p in shmoos was dependent on both polymerized actin and a pheromone-induced membrane protein, Fus1p. The requirement for actin may arise through multiple pathways. Mutations that affect cell polarity (bni1, pea2, and spa2) cause defects in actin cable assembly and thereby lead to defects in vesicular traffic and Fus1p localization. Accordingly, in these mutants, Fus2p moved randomly throughout the cell and also failed to form stable cortical patches.
Lat-A treatment clearly showed a requirement for polymerized actin in Fus2p transport. Only in the context of a fus1 mutation was actin also seen to be required for Fus2p's localization at the shmoo tip. In the absence of Fus1p, actin-dependent transport may suffice to concentrate Fus2p at the shmoo tip. Alternatively, additional actin-dependent interactions may help stabilize the cortical localization.
Electron microscopy suggested that Fus1p and Fus2p act at different steps in cell fusion (Gammie et al., 1998); fus1Δ mutants show fewer, more dispersed vesicles, and fus2Δ mutants showed wild-type vesicle clustering. These results suggested that Fus1p acts before vesicle clustering and that Fus2p acts later, possibly affecting vesicle fusion. Clearly, the morphology of the fus1 mutants would have obscured evidence for additional downstream functions.
The dependence of Fus2p localization on Fus1p is consistent with genetic data showing both independent and partially overlapping functions. Overexpression of Fus2p might partially suppress the requirement for Fus1p by increasing Fus2p's cortical concentration, independent of anchoring. Overexpression of Fus1p might partially suppress fus2 mutations by influencing the localization or function of an alternate mitotic Rho-GEF protein. The possibility that other Rho-GEF proteins substitute for Fus2p is supported by the fact that high-copy Bem1p, a scaffolding protein that interacts with Cdc24p (a GEF for Cdc42p), also suppresses fus2 (Fitch et al., 2004). Fus2p dependence on Fus1p is also consistent with two-hybrid data for their interaction (Nelson et al., 2004). Collectively, these results support a model in which Fus2p is anchored at the shmoo tip by Fus1p but in which each protein has additional functions.
Remarkably, polarity mutants unable to localize Fus2p-GFP to the shmoo tip were able to localize it to the ZCF in prezygotes, which suggests that there is additional polarizing information in prezygotes. One major difference between shmoos and prezygotes is the way that pheromone is presented. Shmoos are formed by exposure to high levels of isotropic pheromone and polarize toward the preexisting bud site. Mating cells are exposed to a pheromone gradient and polarize toward the mating partner. Thus, conjugating cells receive positional information from the pheromone gradient, which is not available to cells in isotropic pheromone. Consistent with this view, the cell polarity mutants (bni1, spa2, and pea2) were profoundly defective for mating when their pheromone receptors were saturated (default pathway) but showed much fewer severe defects in normal mating conditions, where cells respond to pheromone gradients (chemotropic pathway; Dorer et al., 1997). Hence, Fus2p provides a molecular marker whose localization behavior is acutely sensitive to polarization during chemotropic mating. Note that Fus2p-GFP localization at the ZCF was significantly less stable in these mutants, which is consistent with roles for Bni1p, Pea2p, and Spa2 in the maintenance of Fus2p localization and/or cell polarization.
The pattern of Fus2p-GFP localization in zygotes, forming a ring at a site of localized secretion where cells have formed a tight adhesion, is remarkably similar to metazoan cell junctions, such as the T cell and neuronal synapses. At these sites, adhesion molecules also form rings around sites of directed secretion (Monks et al., 1998; Dustin and Colman, 2002). The similarity may represent a fundamental solution to the problem of how two cells engage in intimate communication.
Materials and methods
General yeast techniques
Yeast media and general techniques used were described previously (Rose et al., 1990). All yeast strains were grown at 30°C. For strains induced with galactose, cultures were grown overnight in media containing 2% galactose, back-diluted to early log phase in 2% galactose, and allowed to grow for 2–4 h before further manipulation. For standard induction with mating pheromones, cells were grown overnight and diluted to early log phase in low-pH media (pH, 3.5) and allowed to grow for 2 h, and then synthetic α-factor (Department of Molecular Biology Syn/Seq Facility, Princeton University) dissolved in methanol was diluted 100-fold to a final concentration of 6 μM for 1.5 h. Low pheromone treatments used pheromone at 0.6 μM. Yeast transformations were performed as described previously (Rose et al., 1990).
Yeast strains and plasmid construction
Yeast strains and plasmids used in this study are listed in Tables I and II. For some experiments, Rho strains were used to facilitate live cell nuclear staining. The Rho character had no obvious effect on the efficiency of mating or the behavior of Fus2p-GFP in otherwise wild-type cells. The fus2∷HIS3 deletion was made by one-step gene replacement. Epitope tagging was performed by in vivo recombination (Ma et al., 1987; Oldenburg et al., 1997). To generate the internally tagged FLAG construct, 476 bp of the 5′ untranslated region (UTR) plus the first 312 bp of FUS2 were amplified with JP21 (5′-TAGGGCGAATTGGGTACCGGGCCCCCCCTCGAGGTCGACGGTATCGATgtcccacctgcttggtgg-3′), a primer that contained a RS416 vector sequence (uppercase letters) and a FUS2 sequence (lowercase letters), and JP52 (5′-CTTCTCATCATCGTCCTTGTAGTCCATgggaatctgagggc-3′), a primer that contained part of the FLAG epitope (uppercase) and FUS2 sequence (lowercase). The C-terminal portion of FUS2 was amplified using JP22 (5′-GCTGGAGCTCCACCGCGGTGGCGGCCGCTCTAGAACTAGTGGATCCCCctgctccagcgcagtagt-3′; RS416 in uppercase, FUS2 in lowercase) and a primer that contained an overlapping part of the FLAG epitope JP51 (5′-AAGGACGATGATGAGAAGCCCCATaattctcctcacgag-3′; FLAG in uppercase, FUS2 lowercase). The two PCR products were transformed along with RS416 and cut with BamHI into MY1817.
Table I. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| JM231 | MATa leu2Δ1 ura3-52 [pRS416] | |
| JM269 | MATa leu2-3,112, ura3-52, his4-34 [pMR3392] | |
| JY429 | MATα fus1Δ1 fus2Δ3 ura3-52 trp1Δ1 | G.R. Fink (Whitehead Institute, Cambridge MA) |
| MY8093 | MATα his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 | Invitrogen |
| MY9182 | MATa fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 [pRS416] | |
| MY9183 | MATa fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 [pMR5480] | |
| MY9184 | MATa fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 [pMR5482] | |
| MY9201 | MATa fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 [pMR5469] | |
| MY9208 | MATa bni1Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9211 | MATa fus3Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9213 | MATa pea2Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9214 | MATa rvs161Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9216 | MATa spa2Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9217 | MATa fus1Δ∷kanMX fus2Δ∷HIS3 his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rho− [pMR5482] | |
| MY9231 | MATa rvs161Δ∷LEU2 fus2Δ∷HIS3 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 [pMR5482] | |
| MY9233 | MATa rvs161-P203Q fus2Δ∷HIS3 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 [pMR5482] | |
| MY9239 | MATa rvs161-A175P fus2Δ∷HIS3 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 [pMR5482] | |
| BY1034 | MATa fus2∷HIS3 RVS167-3xHA∷kanMX ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 | Friesen et al., 2006 |
| MY10025 | MATa fus2∷HIS3 RVS167-3xHA∷kanMX ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 [pMR2414] | From BY1034 |
| MY10026 | MATa fus2∷HIS3 RVS167-3xHA∷kanMX ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 [pMR5480] | From BY1034 |
| BY263 | MATa fus2∷HIS3 ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 | Friesen et al., 2006 |
| MY10027 | MATa fus2∷HIS3 ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 [pMR2414] | From BY263 |
| MY10028 | MATa fus2∷HIS3 ura3-52 lys2-801 ade2-107 his3Δ200 trp1Δ63 leu2-Δ1 [pMR5480] | From BY263 |
All strains were constructed for this study unless otherwise noted.
Table II. Plasmids.
| Strain | Genotype/description | Source |
|---|---|---|
| pRS416 | URA3 CEN3 ARS1 AMPR | Sikorski and Hieter, 1989 |
| pMR2414 | FUS2 URA3 CEN4 ARS1 AMPR | |
| pMR3392 | FUS2 URA3 CEN3 ARS1 AMPR | |
| pMR5469 | GAL-FUS2-GFP104 URA3 CEN | |
| pMR5480 | FUS2-FLAG URA3 CEN3 ARS1 AMPR | |
| pMR5482 | FUS2-GFP104 URA3 CEN3 ARS1 AMPR |
Plasmids were constructed for this study unless otherwise noted.
To generate the internally tagged GFP construct, 476 bp of the 5′ UTR plus the first 312 bp of FUS2 were amplified using MR3373 as a template, with JP21 and a primer that contained part of the N-terminal sequence of GFP (underlined), JP89 (5′-GAAAAGTTCTTCTCCTTTACTCATGGGAATCTGAGGGC-3′). The C-terminal portion of FUS2 was amplified using JP22 and a primer that contained the C-terminal sequence of GFP, JP90 (5′-CATGGCATGGATGAACTATACAAAaattctcctcacgag-3′; GFP in uppercase, FUS2 in lowercase). GFP was amplified using MR3453 as a template and primers PR70 and PR71. The three PCR products were transformed along with RS416, cut with BamHI into MY1817, and ligated by in vivo recombination.
Mating assays
Limited plate matings were performed as described previously (Gammie et al., 1998). In brief, patches of cells were replica-printed to yeast extract/peptone/dextrose (YEPD) plates containing lawns of the opposite mating type. The mating plates were incubated at 30° for 4 h followed by replica printing to the appropriate media to select for diploids.
Quantitative mating assays were performed as described previously (Rose et al., 1990). In brief, overnight cultures were diluted to early log phase and grown 2–4 h. 3 × 106 cells of each mating type were combined and concentrated on 2.5-cm2 nitrocellulose filter discs (Millipore). The filters were placed onto either a YEPD or synthetic complete (SC) plate and the cells were allowed to mate for 4 h at 30°.
Microscopy
For immunofluorescent imaging, exponentially growing cells or cells treated with pheromone were fixed as described previously (Brizzio et al., 1998). In brief, cells were incubated for 1 h in 4% formaldehyde and washed with PBS. Cells were spheroplasted with 25 μg/ml Zymolyase 100,000 T (MP Biomedicals) for 30 min at 30°. Anti-FLAG antibody was used at 1:6,000 dilutions (Sigma-Aldrich) and Alexa-conjugated anti–mouse secondary antibodies were used at 1:500 dilutions (Invitrogen).
For live-cell imaging of GFP-tagged proteins, aliquots of cells were spread onto an agar pad containing the appropriate media with or without α-factor. For time-course experiments observing Fus2p-GFP, cells were grown to early exponential phase, treated with pheromone for various times, and spread on the surface of agar pads containing SC-URA, pH 3.5, + α-factor. All cells were visualized at 23° using a deconvolution microscope (DeltaVision; Applied Precision, LLC) based on a TE200 (Nikon) using a 100× NA 1.4 objective (Nikon), a 50-W Hg burner, and a Cool Snap ER charge-coupled device camera (Roper Scientific). Excitation was attenuated with 50% neutral density filter, 0.2-s exposures, and 2 × 2 binning.
For time-lapse studies after the induction of Fus2p-GFP from t = 0, early logarithmic phase cells were harvested and treated with 6 μM α-factor and immediately spread on agar pads containing SC-URA, pH 3.5, + 6 μM α-factor. Images were acquired using a stack of 10 sections, spaced every 0.5 μm and taken at 10-min intervals. For short time-course experiments designed to follow the rapid movement of the Fus2p-GFP dots, cells were induced with 6 μM α-factor for 1.5 h, harvested, and placed on an agar pad containing SC-URA, pH3.5, + 6 μM α-factor. Images were acquired in a similar way except that the image stack contained only three sections, spaced 0.5 μm apart, and taken at 1.5-s intervals. For experiments observing prezygotes, strains were mated on filters as stated in the Mating assays section for 4 h, lightly sonicated, and placed on an agar pad made from SC media. Images were acquired at 5- or 10-min time intervals.
Images were deconvolved and collapsed using a maximum intensity algorithm (SoftWorx, Applied Precision, LLC). Distance and intensity measurements were made on collapsed images using SoftWorx. Statistical analysis used a Student's t test for single group comparisons. For multiple comparisons, a one-way analysis of variance (ANOVA) with Tukey's honestly significant difference (HSD) test was used for all pairwise comparisons as implemented in KaleidaGraph (Synergy Software). For publication purposes, brightness and contrast of images was enhanced using linear filters in SoftWorx and/or Photoshop CS2 (Adobe).
IPs and Western blotting
IPs were performed as described previously (Brizzio et al., 1998), with modifications. A 100-ml culture grown to early log phase was induced with 6 μM of α-factor for 90 min. Cells were collected, washed once with breaking buffer (50 mM Tris, pH 7.4, 50 mM NaCl, and 0.5% Triton X-100), and resuspended in 1 ml of breaking buffer containing a protease inhibitor cocktail (5 μg/ml chymotrypsin, 5 μg/ml leupeptin, 5 μg/ml aprotinin, 5 μg/ml pepstatin, and 1 mM PMSF). Cells were broken by vortexing with glass beads for 1 min followed by 1 min on ice, repeated five times. The extract was twice cleared by centrifugation at 14,000 rpm in a microcentrifuge for 5 min. Samples were brought to 1 ml and NaCl was adjusted to 150 mM. Fus2p-FLAG was immunoprecipitated by the addition of 30 μl of anti-FLAG M2 affinity gel (Sigma-Aldrich) prepared by washing once with H2O and three times with breaking buffer. Rvs167p-3xHA was immunoprecipitated by the addition of 40 μl monoclonal anti-HA agarose (Sigma-Aldrich) prepared like the anti-FLAG M2 affinity gel. Anti-FLAG and anti-HA IPs were performed for 1 h at 4°C, and the beads were washed five times with lysis buffer.
Fus2p-GFP was immunoprecipitated by the addition of 60 μl of Dynabeads (Invitrogen) washed and conjugated with 40 μl anti-GFP (Sigma-Aldrich). The conjugated mixture was added to extracts, anti-GFP IPs were performed overnight at 4°C, and the beads were washed four times with lysis buffer.
After SDS-PAGE, proteins were transferred to nitrocellulose and detected using anti-FLAG M2 (Roche) at 1:5,000, anti-GFP at 1:250, or a crude anti-Rvs161p polyclonal (Brizzio et al., 1998) at 1:1,000. HRP-conjugated secondary antibodies (GE Healthcare) were used to detect primary antibodies and were detected by chemiluminescence.
Lat-A treatment
Treatment with lat-A (Invitrogen) was performed as described previously (Miller et al., 1999). In brief, pheromone-induced cells (described in the Microscopy section) were concentrated 50-fold and treated with 200 μM lat-A for 10 min; control cells were treated with 2% DMSO. For experiments with live cells, 7 μl of lat-A–treated cells were placed on glass slide with an agar pad containing SC-URA, pH 3.5, + 200 μM lat-A + 6 μM α-factor and imaged immediately by fluorescent microscopy. In experiments with fixed cells, the lat-A–treated cultures were fixed with 4% formaldehyde for 10 min at room temperature and washed with PBS. To stain actin, cells were then resuspended in 50 μl of PBS and treated with 25 μl of Texas red–phalloidin (0.2 U/μl; Invitrogen) for 1 h at room temperature in the dark. The cells were washed in PBS and examined by fluorescence microscopy as described in the Microscopy section for fixed cells.
Nocodazole treatment
Pheromone-induced cells were concentrated 10-fold and treated with 15 μg/ml nocodazole (Sigma-Aldrich), then diluted 100-fold from a 1.5-mg/ml stock in DMSO for 10 min at room temperature; control cells were treated with 1% DMSO. For experiments with live cells, 7 μl of nocodazole-treated cells were placed on a glass slide with an agar pad containing SC-URA, pH 3.5, + 15 μg/ml nocodazole + 6 μM α-factor and examined by fluorescent microscopy. To visualize microtubules by indirect immunofluorescence, cells were prepared and processed for immunofluorescent microscopy as described in the Microscopy section, except that cells were fixed for only 10 min at 23° with 4% formaldehyde. An anti-tubulin antibody, Yol1/34 (Morphosys), was used at 1:10 dilution in 10 mg/ml PBS-BSA. Anti–rat antibody conjugated to Alexa 647 (Invitrogen) was used at a dilution of 1:500. Nuclei were stained with DAPI (Sigma-Aldrich) at 1 μg/ml for 5 min at room temperature.
Acknowledgments
We thank Alison Gammie for the suggestion of tagging FUS2 internally. We thank Soo-Chen Cheng for initial experiments with N- and C-terminal tagged Fus2p. We thank Brenda Andrews for providing strains. We thank Yuling Hua for help on the experiments involving the Rvs161p–Fus2p complex.
This project was supported by a National Institutes of Health grant (R01 GM37739) to M.D. Rose.
Abbreviations used in this paper: GEF, guanine nucleotide exchange factor; IP, immunoprecipitation; lat-A, latrunculin-A; MAP, mitogen-activated protein; ZCF, zone of cell fusion.
References
- Barkai, N., M.D. Rose, and N.S. Wingreen. 1998. Protease helps yeast find mating partners. Nature. 396:422–423. [DOI] [PubMed] [Google Scholar]
- Beh, C.T., V. Brizzio, and M.D. Rose. 1997. KAR5 encodes a novel pheromone-inducible protein required for homotypic nuclear fusion. J. Cell Biol. 139:1063–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidlingmaier, S., and M. Snyder. 2004. Regulation of polarized growth initiation and termination cycles by the polarisome and Cdc42 regulators. J. Cell Biol. 164:207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bon, E., P. Recordon-Navarro, P. Durrens, M. Iwase, E.A. Toh, and M. Aigle. 2000. A network of proteins around Rvs167p and Rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast. 16:1229–1241. [DOI] [PubMed] [Google Scholar]
- Brizzio, V., A.E. Gammie, and M.D. Rose. 1998. Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141:567–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenevert, J., N. Valtz, and I. Herskowitz. 1994. Identification of genes required for normal pheromone-induced cell polarization in Saccharomyces cerevisiae. Genetics. 136:1287–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliften, P., P. Sudarsanam, A. Desikan, L. Fulton, B. Fulton, J. Majors, R. Waterston, B.A. Cohen, and M. Johnston. 2003. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 301:71–76. [DOI] [PubMed] [Google Scholar]
- Dohlman, H.G., and J.W. Thorner. 2001. Regulation of G protein-initiated signal transduction in yeast: paradigms and principles. Annu. Rev. Biochem. 70:703–754. [DOI] [PubMed] [Google Scholar]
- Dorer, R., C. Boone, T. Kimbrough, J. Kim, and L.H. Hartwell. 1997. Genetic analysis of default mating behavior in Saccharomyces cerevisiae. Genetics. 146:39–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon, B., D. Sherman, G. Fischer, P. Durrens, S. Casaregola, I. Lafontaine, J. De Montigny, C. Marck, C. Neuveglise, E. Talla, et al. 2004. Genome evolution in yeasts. Nature. 430:35–44. [DOI] [PubMed] [Google Scholar]
- Dustin, M.L., and D.R. Colman. 2002. Neural and immunological synaptic relations. Science. 298:785–789. [DOI] [PubMed] [Google Scholar]
- Elion, E.A., J. Trueheart, and G.R. Fink. 1995. Fus2 localizes near the site of cell fusion and is required for both cell fusion and nuclear alignment during zygote formation. J. Cell Biol. 130:1283–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista, M., K. Blundell, M.S. Longtine, C.J. Chow, N. Adames, J.R. Pringle, M. Peter, and C. Boone. 1997. Bni1p, a yeast formin linking cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 276:118–122. [DOI] [PubMed] [Google Scholar]
- Fitch, P.G., A.E. Gammie, D.J. Lee, V.B. de Candal, and M.D. Rose. 2004. Lrg1p Is a Rho1 GTPase-activating protein required for efficient cell fusion in yeast. Genetics. 168:733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen, H., C. Humphries, Y. Ho, O. Schub, K. Colwill, and B. Andrews. 2006. Characterization of the yeast amphiphysins Rvs161p and Rvs167p reveals roles for the Rvs heterodimer in vivo. Mol. Biol. Cell. 17:1306–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie, A.E., V. Brizzio, and M.D. Rose. 1998. Distinct morphological phenotypes of cell fusion mutants. Mol. Biol. Cell. 9:1395–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrung, S., and M. Snyder. 1990. The SPA2 gene of Saccharomyces cerevisiae is important for pheromone-induced morphogenesis and efficient mating. J. Cell Biol. 111:1451–1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germann, M., E. Swain, L. Bergman, and J.T. Nickels Jr. 2005. Characterizing the sphingolipid signaling pathway that remediates defects associated with loss of the yeast amphiphysin-like orthologs, Rvs161p and Rvs167p. J. Biol. Chem. 280:4270–4278. [DOI] [PubMed] [Google Scholar]
- Heiman, M.G., and P. Walter. 2000. Prm1p, a pheromone-regulated multispanning membrane protein, facilitates plasma membrane fusion during yeast mating. J. Cell Biol. 151:719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, H., C. Carlile, S. Nolan, and E. Grote. 2004. Prm1 prevents contact-dependent lysis of yeast mating pairs. Eukaryot. Cell. 3:1664–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis, M., N. Patterson, M. Endrizzi, B. Birren, and E.S. Lander. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 423:241–254. [DOI] [PubMed] [Google Scholar]
- Kurihara, L.J., C.T. Beh, M. Latterich, R. Schekman, and M.D. Rose. 1994. Nuclear congression and membrane fusion: two distinct events in the yeast karyogamy pathway. J. Cell Biol. 126:911–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic, I., R.R. Copley, B. Pils, S. Pinkert, J. Schultz, and P. Bork. 2006. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 34:D257–D260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipke, P.N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science. 252:1162–1164. [DOI] [PubMed] [Google Scholar]
- Ma, H., S. Kunes, P.J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene. 58:201–216. [DOI] [PubMed] [Google Scholar]
- Madden, K., and M. Snyder. 1998. Cell polarity and morphogenesis in budding yeast. Annu. Rev. Microbiol. 52:687–744. [DOI] [PubMed] [Google Scholar]
- Marsh, L., and M.D. Rose. 1997. The pathway of cell and nuclear fusion during mating in S. cerevisiae. In The Molecular and Cellular Biology of the Yeast Saccharomyces cerevisiae. J.R. Pringle, J.R. Broach, and E.W. Jones, editors. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 827–888.
- Matheos, D., M. Metodiev, E. Muller, D. Stone, and M.D. Rose. 2004. Pheromone-induced polarization is dependent on the Fus3p MAPK acting through the formin Bni1p. J. Cell Biol. 165:99–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy, P., S. Shen, E. White, J.R. McIntosh, and M.D. Rose. 2007. Nuclear fusion during yeast mating occurs by a three-step pathway. J. Cell Biol. 179:659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and M.D. Rose. 1990. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 60:1029–1041. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., D. Matheos, and M.D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molk, J.N., E.D. Salmon, and K. Bloom. 2006. Nuclear congression is driven by cytoplasmic microtubule plus end interactions in S. cerevisiae. J. Cell Biol. 172:27–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks, C.R., B.A. Freiberg, H. Kupfer, N. Sciaky, and A. Kupfer. 1998. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature. 395:82–86. [DOI] [PubMed] [Google Scholar]
- Moore, S.A. 1983. Comparison of dose–response curves for alpha factor-induced cell division arrest, agglutination, and projection formation of yeast cells. Implication for the mechanism of alpha factor action. J. Biol. Chem. 258:13849–13856. [PubMed] [Google Scholar]
- Muller, E.M., N.A. Mackin, S.E. Erdman, and K.W. Cunningham. 2003. Fig1p facilitates Ca2+ influx and cell fusion during mating of Saccharomyces cerevisiae. J. Biol. Chem. 278:38461–38469. [DOI] [PubMed] [Google Scholar]
- Munn, A.L., B.J. Stevenson, M.I. Geli, and H. Riezman. 1995. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol. Biol. Cell. 6:1721–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B., A.B. Parsons, M. Evangelista, K. Schaefer, K. Kennedy, S. Ritchie, T.L. Petryshen, and C. Boone. 2004. Fus1p interacts with components of the Hog1p mitogen-activated protein kinase and Cdc42p morphogenesis signaling pathways to control cell fusion during yeast mating. Genetics. 166:67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldenburg, K.R., K.T. Vo, S. Michaelis, and C. Paddon. 1997. Recombination-mediated PCR-directed plasmid construction in vivo in yeast. Nucleic Acids Res. 25:451–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter, B.J., H.M. Kent, I.G. Mills, Y. Vallis, P.J. Butler, P.R. Evans, and H.T. McMahon. 2004. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 303:495–499. [DOI] [PubMed] [Google Scholar]
- Philips, J., and I. Herskowitz. 1997. Osmotic balance regulates cell fusion during mating in Saccharomyces cerevisiae. J. Cell Biol. 138:961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science. 297:612–615. [DOI] [PubMed] [Google Scholar]
- Roberts, C.J., B. Nelson, M.J. Marton, R. Stoughton, M.R. Meyer, H.A. Bennett, Y.D. He, H. Dai, W.L. Walker, T.R. Hughes, et al. 2000. Signaling and circuitry of multiple MAPK pathways revealed by a matrix of global gene expression profiles. Science. 287:873–880. [DOI] [PubMed] [Google Scholar]
- Rose, M.D., F.M. Winston, and P. Hieter. 1990. Methods in Yeast Genetics: a Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. 198 pp.
- Sagot, I., A.A. Rodal, J. Moseley, B.L. Goode, and D. Pellman. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4:626–631. [DOI] [PubMed] [Google Scholar]
- Segall, J.E. 1993. Polarization of yeast cells in spatial gradients of alpha mating factor. Proc. Natl. Acad. Sci. USA. 90:8332–8336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu, Y.J., B. Santos, N. Fortin, C. Costigan, and M. Snyder. 1998. Spa2p interacts with cell polarity proteins and signaling components involved in yeast cell morphogenesis. Mol. Cell. Biol. 18:4053–4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague, G.F. Jr., L.C. Blair, and J. Thorner. 1983. Cell interactions and regulation of cell type in the yeast Saccharomyces cerevisiae. Annu. Rev. Microbiol. 37:623–660. [DOI] [PubMed] [Google Scholar]
- Talarek, N., A. Balguerie, M. Aigle, and P. Durrens. 2005. A novel link between a Rab GTPase and Rvs proteins: the yeast amphiphysin homologues. Cell Biochem. Funct. 23:253–266. [DOI] [PubMed] [Google Scholar]
- Trueheart, J., and G.R. Fink. 1989. The yeast cell fusion protein FUS1 is O-glycosylated and spans the plasma membrane. Proc. Natl. Acad. Sci. USA. 86:9916–9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trueheart, J., J.D. Boeke, and G.R. Fink. 1987. Two genes required for cell fusion during yeast conjugation: evidence for a pheromone-induced surface protein. Mol. Cell. Biol. 7:2316–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz, N., and I. Herskowitz. 1996. Pea2 protein of yeast is localized to sites of polarized growth and is required for efficient mating and bipolar budding. J. Cell Biol. 135:725–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, J.M., and M.D. Rose. 2001. Yeast mating: getting close to membrane merger. Curr. Biol. 11:R16–R20. [DOI] [PubMed] [Google Scholar]
- Zhang, M., D. Bennett, and S.E. Erdman. 2002. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell. 1:811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]