SUMMARY
Mammalian palate development is a multi-step process, involving initial bilateral downward outgrowth of the palatal shelves from the oral side of the maxillary processes, followed by stage-specific palatal shelf elevation to the horizontal position above the developing tongue and subsequent fusion of the bilateral palatal shelves at the midline to form the intact roof of the oral cavity. Whereas mutations in many genes have been associated with cleft palate pathogenesis, the molecular mechanisms regulating palatal shelf growth, patterning, and elevation are not well understood. Genetic studies of the molecular mechanisms controlling palate development in mutant mouse models are often complicated by early embryonic lethality or gross craniofacial malformation. We report here the development of a mouse strain for tissue-specific analysis of gene function in palate development. We inserted an IresCre bicistronic expression cassette into the 3′ untranslated region of the mouse Osr2 gene through gene targeting. We show, upon crossing to the R26R reporter mice, that Cre expression from the Osr2IresCre knockin allele activated beta-galactosidase expression specifically throughout the developing palatal mesenchyme from the onset of palatal shelf outgrowth. In addition, the Osr2IresCre mice display exclusive Cre-mediated recombination in the glomeruli tissues derived from the metanephric mesenchyme and complete absence of Cre activity in other epithelial and mesenchymal tissues in the developing metanephric kidney. These data indicate that the Osr2IresCre knockin mice provide a unique tool for tissue-specific studies of the molecular mechanisms regulating palate and kidney development.
Keywords: cleft palate, glomerulus, kidney, odd-skipped, Osr2, palate development, Cre, loxP
Cleft palate is one of the most common birth defects, affecting approximately one in 1000 infants. The primary reason why cleft palate occurs so frequently is because the palate develops from initially separate primordia through multi-step morphogenetic processes. During mammalian embryogenesis, palatal outgrowths arise from the oral side of the paired maxillary processes flanking the embryonic oral cavity. These outgrowths initially grow vertically along the sides of the developing tongue, forming two palatal shelves. At a precise developmental stage, the bilateral palatal shelves elevate to a horizontal position above the dorsum of the tongue and fuse with each other at the midline, separating the nasal cavity from the oral cavity (Ferguson, 1988). Each of these morphogenetic processes, including growth, elevation, and fusion of the palatal shelves, is regulated by highly coordinated molecular networks of which genetic and/or environmental perturbations could result in cleft palate (Gritli-Linde, 2007, and references therein). Moreover, palate development occurs at the same time when the entire embryonic craniofacial region is undergoing extensive growth and morphogenetic remodeling, of which disruption often cause cleft palate as secondary consequences. For example, cleft palate in infants with the condition referred as the Pierre Robin Syndrome, which is also characterized by small lower jaw and displaced tongue, is believed to result from physical hindrance of palatal shelf elevation by the displaced tongue (Cohen, 1999). Although there have been over 400 inherited syndromes in humans as well as over seventy mutant mouse strains reported to exhibit cleft palate phenotypes, the molecular and cellular mechanisms regulating palate development, particularly palatal growth and patterning, are still not well understood. Whereas targeted mutagenesis in mice have been a powerful approach for characterizing the genetic basis of various disease conditions, many of the mutant mouse strains with cleft palate phenotypes also had gross craniofacial defects that made it difficult to distinguish whether the cleft palate resulted from intrinsic defects in palate development or from a secondary effect of other structural abnormalities (Gendron-Maguire et al., 1993; Rijli et al., 1993; Satokata and Maas, 1994; Martin et al., 1995; Sanford et al., 1997; Peters et al., 1998; Lan et al., 2004; Thyagarajan et al., 2003). Moreover, since palate development occurs relatively late during embryogenesis, the involvement of many genes and molecular pathways in palate growth and patterning has not been systematically investigated because null mutations in important components of major developmental signaling pathways often resulted in embryonic lethality prior to palate development.
The development of tissue-specific gene inactivation strategies offers great potential to systematically dissect the roles of different molecular pathways in specific developmental processes. The currently most efficient tissue-specific gene inactivation strategy uses the Cre/loxP system (Lewandoski, 2001). Cre is a bacteriophage P1 recombinase that specifically recognizes and effectively excises any DNA sequence flanked by two directly repeated 34-bp specific DNA sequences named loxP sites (Sauer and Henderson, 1988). This bacteriophage P1 DNA recombination system has been adapted with great success for conditional gene inactivation in mice by introducing two loxP sites into noncoding regions of specific genes through gene targeting (Jiang and Gridley, 1997; Lewandoski, 2001). Upon crossing to transgenic mice with the Cre transgene under the control of a tissue-specific promoter, the gene containing loxP sites (“floxed” allele) is specifically inactivated only in the tissues where Cre is expressed. This experimental strategy has greatly advanced genetic analyses of cell lineages and gene function in organogenesis and postnatal development. Whereas mice carrying “floxed” alleles of many signaling pathway genes potentially involved in palate development have been generated in recent years (e.g., Liu et al., 2005), however, the investigation of molecular mechanisms of palate development has been hindered by the lack of a palate-specific Cre-expressing mouse strain.
We previously reported that expression of the Odd-skipped related-2 (Osr2) gene is specifically activated in the palatal mesenchyme at the onset of palatal outgrowth and persists strongly in the downward growing palatal shelves (Lan et al., 2001; 2004). We reasoned that expressing Cre from the Osr2 locus would provide a valuable mouse strain for fine dissection of the molecular network regulating palate development. Since Osr2 gene function is required for palatal shelf growth (Lan et al., 2004), we chose to generate a knockin mouse strain carrying an internal ribosomal entry sequence (Jackson et al., 1990; Jang and Wimmer, 1990) followed by the Cre cDNA (IresCre) targeted into the 3′-untranslated region (UTR) of the Osr2 gene so that Cre would be co-expressed with but would not disrupt the function of the Osr2 gene.
We successfully targeted the IresCre cassette together with an FRT-flanked neo expression cassette into an XbaI restriction enzyme recognition site in the 3′-UTR of the Osr2 gene through homologous recombination in mouse embryonic stem cells (see MATERIALS AND METHODS, and FIG. 1). Mice heterozygous for the targeted Osr2IresCre allele were intercrossed to generate Osr2IresCre/IresCre homozygous mice, which did not show any obvious defects in the B6/129Sv mixed genetic background. Since targeted disruption of Osr2 resulted in cleft palate and neonatal lethality of homozygotes (Lan et al., 2004), these data indicate that insertion of the IresCre and FRT-flanked neo expression cassettes in the 3′-UTR did not significantly affect Osr2 gene function.
FIG. 1.

Targeted insertion of the IresCre bicistronic expression cassette into the 3′ UTR of the mouse Osr2 gene. (A) The Osr2 gene consists of four exons spanning approximately 8 kb of genomic DNA. Boxes indicate exons, with the protein-coding region marked in black. The positions of the translation start (ATG) and stop (TAG) codons are also indicated. Restriction sites are: B, BamHI; E, EcoRI; H, HindIII; P, PstI; X, XbaI. The targeting vector used the 2.6 kb PstI-XbaI fragment containing sequences from Exon 2 to the 3′ UTR region in Exon 4 as the 5′ arm and the 3.3 kb XbaI-HindIII fragment 3′ to the Osr2 coding region as the 3′ arm. The IresCre cassette and a FRT-flanked neo expression cassette were inserted in between the arms and a diptheria toxin A (DTA) expression cassette was cloned 3′ to the 3′ arm for negative selection against random integration. Correct targeting results in insertion of the IresCre cassette and the neo cassette at the XbaI site in the 3′ UTR. (B) Southern hybridization analysis of selected embryonic stem cell DNA samples to confirm correct targeting. Genomic DNA samples were digested with BamHI, separated by electrophoresis through a 1% agarose gel, transferred onto a GeneScreen Plus nylon membrane (PerkinElmer), and hybridized with random prime-labeled probes made from the 600 bp HindIII-EcoRI fragment isolated from the Intron 1 region 5′ to the targeted region. The 14 kb BamHI fragment corresponding to the wild-type allele was detected in all ES DNA samples, whereas the 7.2 kb fragment was only detectable in the DNA samples from correctly targeted ES clones. (C) PCR analysis of tail DNA samples from a litter of newborn F2 progeny. The fragments amplified from wild-type and Osr2IresCre alleles are 490 bp and 706 bp, respectively. Homozygous mutants were born at the expected Mendelian frequency and lived normally. +/+, wild-type; +/−, heterozygote; −/−, homozygote.
To determine the spatial and temporal patterns of Cre activity in the Osr2IresCre mice, we set up timed mating of Osr2IresCre/IresCre mice with the R26R reporter mice and embryos were harvested at various developmental stages from gestational day (E) 9.5 through E16.5. The R26R reporter mice, which carry a loxP-STOP-loxP-lacZ cassette targeted into the ubiquitously expressed ROSA26 locus (Zambrowwicz et al., 1997; Soriano, 1999), have been commonly used to test Cre-induced deletion of loxP-flanked sequences and to carry out genetic lineage tracing assays. Once Cre deletes the loxP-flanked STOP sequence from the R26R locus, the ROSA26 promoter drives lacZ expression in all progeny cells long after Cre expression is turned off. For example, the Wnt1-Cre:R26R double transgenic mice have been widely used to indelibly mark all cranial neural crest-derived tissues even though Cre is only expressed transiently before neural crest migration in the Wnt1-cre transgenic mice (Danielian et al., 1998; Soriano, 1999; Chai et al., 2000). We previously showed that Osr2 mRNA expression during embryogenesis was first activated specifically in the mesonephric vesicles at E9.25 (Lan et al., 2001) and that beta-galactosidase activity expressed from an Osr2-lacZ knockin allele (Osr2tm1Jian) was detectable by E9.5 in the same tissues (Lan et al., 2004). X-gal staining of beta-galactosidase activity was not detected in the mesonephric tissues until E10.0 in Osr2IresCre:R26R double heterozygous embryos (FIG. 2A). This delay of about half a day in beta-galactosidase expression in Osr2IresCre:R26R double heterozygous embryos, compared with that in the Osr2-lacZ knockin mouse embryos, most likely reflects the time needed for Cre-mediated activation of transcription of the lacZ mRNA from the ROSA26 locus and subsequent translation and accumulation of active beta-galactosidase enzymes. Similarly, there is also a delay of expression of beta-galactosidase in the craniofacial and limb tissues in Osr2IresCre:R26R double heterozygous embryos, compared with that in the Osr2-lacZ knockin mouse embryos (FIG. 2B–E). By E10.75, beta-galactosidase became robustly expressed in all early Osr2-expressing tissues, including the maxillary-mandibular junction, mesonephros, and specific domains in the limb buds (FIG. 2E, F). X-gal staining of cryostat sections of E11.5 and E12.5 embryos showed that Cre-mediated deletion of the loxP-flanked STOP sequence in the R26R locus occurred highly specifically and efficiently in the palatal mesenchyme (FIG. 3A, B). Since lacZ expression marks the lineages of cells expressing Cre, the highly localized X-gal staining of the palatal mesenchyme at E12.5 (FIG. 3B) indicates that the X-gal stained cells on the oral side of the maxillary processes at E10.5 (FIG. 2F) were nascent palatal mesenchyme cells that had already expressed Cre specifically. X-gal staining of cryostat sections from E12.5 – E15.5 embryos showed Cre-mediated activation of lacZ expression in all palatal mesenchymal cells throughout the anterior-posterior axis of the palatal shelves (FIG. 3B–D), indicating that the Osr2IresCre mouse strain provides a valuable new tool for genetic dissection of molecular pathways regulating palatal mesenchyme growth and patterning.
FIG. 2.
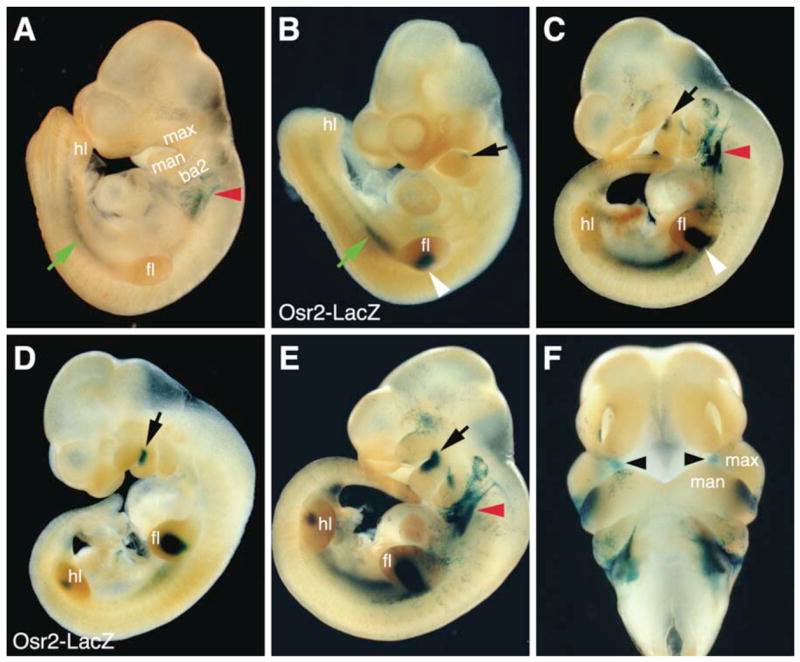
Whole mount X-gal staining detection of beta-galactosidase expression in Osr2IresCre:R26R double heterozygous embryos (A,C,E,F) in comparison with that in Osr2tm1Jian (Osr2-lacZ) heterozygous embryos. (A) At E10.0, beta-galactosidase expression (blue color) was detected in the mesonephric tissues (green arrow) in Osr2IresCre:R26R double heterozygous embryos. There was also beta-galactosidase expression in the region immediately caudal to the second branchial arches (red arrowhead), which was not observed in Osr2-lacZ embryos (B). (B) At E10.0, beta-galactosidase activity was detected in the mesonephros (green arrow), in the forelimb buds (white arrowhead), and was beginning to be expressed in the rostro-proximal mandibular mesenchyme (black arrow), in the Osr2-lacZ heterozygous embryos. (C) By E10.5, beta-galactosidase activity was detected in the forelimb buds (white arrowhead) and in the rostro-proximal mandibular region (black arrow) in Osr2IresCre:R26R double heterozygous embryos. (D) At the same developmental stage (E10.5) as the embryo in C, Osr2-lacZ heterozygous embryos exhibit more intense X-gal staining in the rostro-proximal mandibular region (black arrow) as well as expression in the hindlimb buds. (E, F) By E10.75 to E11.0, the Osr2IresCre:R26R double heterozygous embryos exhibit robust beta-galactosidase expression in the rostroproximal region of the mandibular processes (black arrow) and in the nascent palatal mesenchyme (black arrowheads) next to the oral side of the maxillary processes. ba2, second branchial arch; fl, forelimb bud; hl, hindlimb bud; man, mandibular process; max, maxillary process.
FIG. 3.
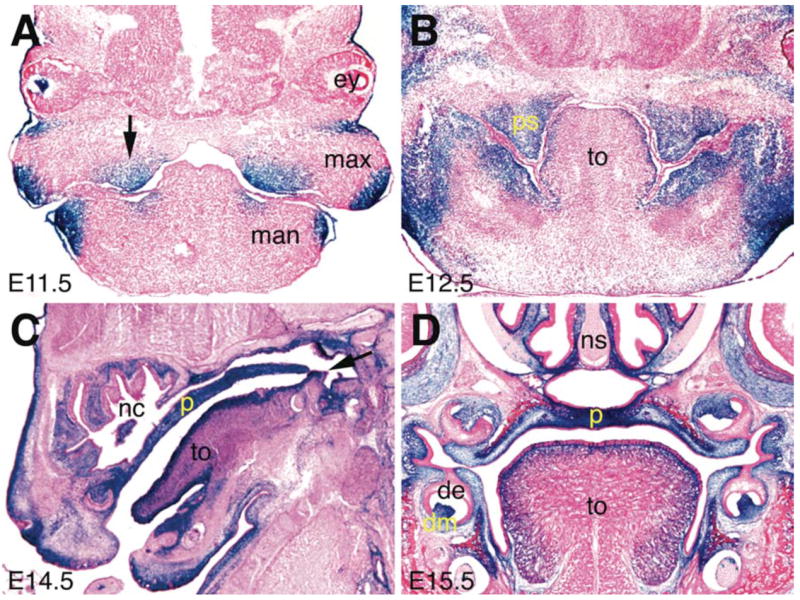
Detection of Cre-mediated activation of LacZ expression during palate development in the Osr2IresCre:R26R double heterozygous embryos. (A) At E11.5, palatal outgrowths arise from the oral side of the maxillary processes. Beta-galactosidase activity was detected highly specifically in the nascent palatal mesenchyme (black arrow). (B) Frontal section of an E12.5 Osr2IresCre:R26R double heterozygous embryo showing tissue-specific X-gal staining in the palatal mesenchyme, and parts of the mandibular mesenchyme. (C) Sagittal section of an E14.5 Osr2IresCre:R26R double heterozygous embryo showing X-gal staining throughout the anteroposterior axis of the secondary palate. (D) Frontal section of an E15.5 Osr2IresCre:R26R double heterozygous embryo showing intense X-gal staining in the palatal mesenchyme and in the molar dental mesenchyme. de, dental epithelium; dm, dental mesenchyme; ey, eye; man, mandible; max, maxillary process; nc, nasal cavity; p, palate; ps, palatal shelf; to, tongue.
Osr2 mRNA is also expressed during kidney development (Lan et al., 2001). To examine the utility of the Osr2-IresCre mice for tissue-specific genetic analysis of kidney development, we carried out X-gal staining of cryostat sections through the developing kidneys in Osr2IresCre:R26R embryos. At E12.5, specific and intense X-gal staining was detected throughout the mesonephric mesenchyme-derived epithelial tubules (FIG. 4A). In contrast, the Wolffian duct epithelium completely lacked beta-galactosidase activity (FIG. 4A). X-gal staining in the metanephric kidney was observed exclusively in the metanephric mesenchyme-derived epithelial tissues (FIG. 4B–D). Neither the epithelial structures derived from branching morphogenesis of the ureteric bud (reviewed in Dressler, 2006), including the collecting duct and the distal tubules, nor the kidney stroma cells showed any LacZ activity. These data indicate that Cre expression in the Osr2IresCre mice is specifically activated following epithelial transformation of the metanephric mesenchyme. Thus, the Osr2IresCre mice provide a unique tool for genetic dissection of the molecular pathways regulating cell differentiation during kidney development.
FIG. 4.

Detection of Cre-mediated activation of beta-galactosidase expression during kidney development in the Osr2IresCre:R26R double heterozygous embryos. (A) A transverse section through the mesonephros and the rostral cortex of the developing metanephric kidney of an E12.5 Osr2IresCre:R26R double heterozygous embryo showing intense X-gal staining in the mesonephric tubules. The Woffian duct epithelium is devoid of beta-galactosidase activity whereas the mesenchyme immediately surrounding the Woffian duct shows X-gal staining. (B) A transverse section through the middle of the developing metanephric kidney of an E12.5 Osr2IresCre:R26R double heterozygous embryo showing highly specific and restricted X-gal staining in the metanephric mesenchyme derived epithelial S-shaped bodies. (C) High magnification view of a region of the section shown in B containing one well-recognizable S-shaped body structure. The X-gal staining is restricted to the developing glomerulus capsule epithelia and is completely absent from the distal tubule connecting to the S-shaped body. The collecting duct epithelia and the kidney stromal mesenchyme are completely devoid of beta-galactosidase activity. (D) A transverse section through the developing metanephric kidney of an E14.5 Osr2IresCre:R26R double heterozygous embryo showing intense X-gal staining in the maturing glomeruli (black arrows). A subset of the epithelial tubules, most-likely the metanephric mesenchyme derived proximal tubules of the nephrons, were also intensely stained. The collecting duct epithelia and the kidney stromal mesenchyme remain negative for beta-galactosidase activity. cd, collecting duct; dt, distal tubule; gl, glomerulus; gr, genital ridge; ki, kidney; mt, mesonephric tubule; pt, proximal tubule; sb, S-shaped body; wd, Wolffian duct.
Whereas most Osr2IresCre:R26R embryos exhibited tissue-specific beta-galactosidase expression patterns consistent with specifc Cre expression from the Osr2 locus, a subset of the embryos showed ectopic X-gal staining, particularly in the central nervous system (FIG. 5). Similar ectopic Cre activity was observed from mice derived from two independent ES clones. We crossed the Osr2IresCre mice to the Flipper mice, which express the FLP DNA recombinase from the ROSA26 locus and efficiently deletes FRT-flanked sequences (Farley et al., 2000), and generated Osr2IresCre mice that lacked the neo expression cassette. However, deleting the neo cassette did not eliminate ectopic Cre activity in the small subset of progeny embryos. Since we have never detected ectopic lacZ expression in the Osr2-lacZ knockin mice, it is likely that the Ires sequence enabled efficient translation of the Cre mRNA from transcriptional noise in a subset of Osr2IresCre mouse embryos. Whereas the small subset of embryos displaying ectopic Cre activity should not prevent using this mouse strain for tissue-specific genetic analysis of palate and kidney development, interpretation of phenotypes observed in mice with Osr2-IresCre-mediated gene inactivation needs to distinguish tissue-specific functional requirements from possible secondary effects of the ectopic Cre activity.
Fig. 5.

Ectopic Cre activity in a subset of Osr2IresCre:R26R double heterozygous embryos. (A–C) Three E12.5 Osr2IresCre:R26R double heterozygous embryos representing the three different X-gal staining patterns. Most embryos analyzed displayed tissue-specific X-gal staining consistent with specific Cre expression from the Osr2 locus, as shown in A. Approximately 20% of the double heterozygous embryos displayed some ectopic X-gal staining, as shown in B. Approximately 10% of the double heterozygous embryos displayed almost ubiquitous X-gal staining, as shown in C.
MATERIALS AND METHODS
Generation of mice carrying the Osr2IresCre allele
A 9.4 kb HindIII genomic DNA fragment containing all three coding exons of the Osr2 gene was subcloned from a 129/Sv mouse BAC clone as described previously (Lan et al., 2004). To target the IresCre cassette into the Osr2 locus, the targeting vector was constructed by inserting a DNA fragment containing the IresCre cassette and a FRT-flanked neo cassette (Macatee et al., 2003) into a unique XbaI site in the middle of the 3′ UTR of the Osr2 gene. The completed targeting vector contained a 2.6 kb 5′ homology arm and a 3.3 kb 3′ homology arm flanking the IresCre-neo cassettes (FIG. 1A). A PGK-DTA expression cassette was subcloned downstream to the 3′ homology arm for negative selection. The targeting vector was linearized and electroporated into CJ7 ES cells as previously described (Swiatek and Gridley, 1993). G418 resistant ES colonies were screened by Southern hybridization for homologous recombination (FIG. 1B). Two independently targeted ES cell clones were injected into blastocysts from C57BL/6J mice and the resultant chimeras bred with C57BL/6J females. F1 mice were genotyped by Southern hybridization analysis of tail DNA. Mice and embryos from subsequent generations were genotyped by PCR. PCR with Primer 1 (5′-GAT ACG GGT AAG ACA GAA ACT G-3′) and Primer 2 (5′-CTA CAA GGA TCT AGC ACA TGC TG-3′) amplifies a product of 490 bp from the wild-type Osr2 allele. PCR with Primer 3 (5′-GTC CCA TTT ACT GAC CGT ACA CC-3′) and Primer 4 (5′-GTT ATT CGG ATC ATC AGC TAC ACC-3′) amplifies a product of 706 bp from the Cre sequence in the Osr2IresCre allele.
Other mouse strains
The R26R and Flipper mice were purchased from the Jackson Laboratory (Bar Harbor, ME). The Osr2-lacZ knockin (Osr2tm1Jian) mice have been described previously (Lan et al., 2004).
X-gal staining
X-gal staining of whole mount embryos and cryostat sections for beta-galactosidase detection was performed as described previously (Hogan et al., 1994). Cryostat sections were counterstained with nuclear fast red following X-gal staining.
Acknowledgments
We thank Anne Moon and Phil Soriano for the IresCre-frt-neo-frt and PGK-DTA plasmids, respectively. We thank Tom Gridley for the CJ7 ES cells. This work was supported by a NIH/NIDCR grant (R01DE013681) to RJ.
Contract grant sponsor: National Institutes of Health (NIH)/National Institute of Dental and Craniofacial Research (NIDCR). Contract grant number: DE013681.
LITERATURE CITED
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Cohen MM., Jr Robin sequences and complexes: causal heterogeneity and pathogenetic variability. Am J Med Genet. 1999;84:311–315. [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, McMahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- Dressler GR. The cellular basis of kidney development. Annu Rev Cell Dev Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (Flipper) mice. Genesis. 2000;28:106–10. [PubMed] [Google Scholar]
- Ferguson MWJ. Palate Development. Development [suppl] 1988;103:41–60. doi: 10.1242/dev.103.Supplement.41. [DOI] [PubMed] [Google Scholar]
- Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- Gritli-Linde A. Molecular control of secondary palate development. Dev Biol. 2007;301:309–326. doi: 10.1016/j.ydbio.2006.07.042. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1994. [Google Scholar]
- Jackson RJ, Howell MT, Kaminski A. The novel mechanism of initiation of picornavirus RNA translation. Trends Biochem Sci. 1990;15:477–483. doi: 10.1016/0968-0004(90)90302-r. [DOI] [PubMed] [Google Scholar]
- Jang SK, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- Jiang R, Gridley T. Gene targeting: Things go better with Cre. Curr Biol. 1997;7:R321–323. doi: 10.1016/s0960-9822(06)00149-7. [DOI] [PubMed] [Google Scholar]
- Lan Y, Kingsley PD, Cho E-S, Jiang R. Osr2, a new mouse gene related to Drosophila odd-skipped, exhibits dynamic expression patterns during craniofacial, limb, and kidney development. Mech Dev. 2001;107:175–179. doi: 10.1016/s0925-4773(01)00457-9. [DOI] [PubMed] [Google Scholar]
- Lan Y, Ovitt CE, Cho ES, Maltby KM, Wang Q, Jiang R. Odd-skipped related 2 (Osr2) encodes a key intrinsic regulator of secondary palate growth and morphogenesis. Development. 2004;131:3207–3216. doi: 10.1242/dev.01175. [DOI] [PubMed] [Google Scholar]
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin JF, Bradley A, Olson EN. The paired-like homeo box gene Mhox is required for early events of skeletogenesis in multiple lineages. Genes Dev. 1995;9:1237–1249. doi: 10.1101/gad.9.10.1237. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubuser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes Dev. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFβ knockout phenotypes. Development. 1997;124:2659–2670. doi: 10.1242/dev.124.13.2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–356. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- Sauer B, Henderson N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc Natl Acad Sci USA. 1988;85:5166–5170. doi: 10.1073/pnas.85.14.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Swiatek P, Gridley T. Perinatal lethality and defects in hindbrain development in mice homozygous for a targeted mutation of the zinc finger gene Krox20. Genes Dev. 1993;7:2071–2084. doi: 10.1101/gad.7.11.2071. [DOI] [PubMed] [Google Scholar]
- Thyagarajan T, Totey S, Danton MJS, Kulkarni AB. Genetically altered mouse models: the good, the bad, and the ugly. Crit Rev Oral Biol Med. 2003;14:154–174. doi: 10.1177/154411130301400302. [DOI] [PubMed] [Google Scholar]
- Zambrowicz BP, Imamoto A, Fiering S, Herzenberg LA, Kerr WG, Soriano P. Disruption of overlapping transcripts in the ROSA beta geo 26 gene trap strain leads to widespread expression of beta-galactosidase in mouse embryos and hematopoietic cells. Proc Natl Acad Sci USA. 1997;94:3789–3794. doi: 10.1073/pnas.94.8.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]


