Abstract
Multiple growth factors synergistically stimulate proliferation of primitive hematopoietic progenitor cells. A human myeloid cell line, KPB-M15, constitutively produces a novel hematopoietic cytokine, termed stem cell growth factor (SCGF), possessing species-specific proliferative activities. Here we report the molecular cloning, expression, and characterization of a cDNA encoding human SCGF using a newly developed λSHDM vector that is more efficient for differential and expression cloning. cDNA for SCGF encodes a 29-kDa polypeptide without N-linked glycosylation. SCGF transiently produced by COS-1 cells supports growth of hematopoietic progenitor cells through a short-term liquid culture of bone marrow cells and exhibits promoting activities on erythroid and granulocyte/macrophage progenitor cells in primary semisolid culture with erythropoietin and granulocyte/macrophage colony-stimulating factor, respectively. Expression of SCGF mRNA is restricted to myeloid cells and fibroblasts, suggesting that SCGF is a growth factor functioning within the hematopoietic microenvironment. SCGF could disclose some human-specific mechanisms as yet unidentified from studies on the murine hematopoietic system.
Mature blood cells are supplied through a hematopoietic hierarchy from self-renewable stem cells down to committed progenitors. Primitive hematopoietic progenitor cells are redundantly stimulated by multiple pleiotropic cytokines, including colony-stimulating factors (CSFs) and interleukins (ILs), the synergistic action of which is required for the proliferation of more primitive progenitor cells (1). Stem cell factor (SCF or c-kit ligand) (2–4) and flk-2/flt3 ligand (L) (5) are thought to be key molecules among the cooperative regulators.
Stem cell growth factor (SCGF) was originally detected in mitogen-stimulated leukocyte conditioned medium (CM) (6) and constitutively elaborated by a human myeloid cell line, KPB-M15, derived from chronic myelogenous leukemia in blast crisis (7). It lacks colony-stimulating activity, but has burst-promoting activity (BPA) for human bone marrow (BM) erythroid progenitors (burst-forming unit erythroid), granulocyte/macrophage (GM)-promoting activity (GPA) for GM progenitors [colony-forming unit (CFU)-GM] and a CFU-GM-sustaining or recruiting effect during short-term liquid BM cell culture (ΔGPA).
Human BM cell-dependent assay system is indeed a big hurdle for further characterization of SCGF. However, preliminary studies have shown that human SCGF is strictly species-specific; therefore, screening cDNA samples and fractions at each purification step need to be assessed for BPA not on murine cells but on human BM cells. CD34+ BM cell populations are a likely target for BPA assay but are impractical for repetitive experiments for quite a few samples because sufficient BM cells are not available due to a limited number of donors. KPB-M15 cells secrete a significant level of IL-6, but neither produce IL-1α, IL-1β, IL-3, IL-4, G-CSF or GM-CSF, nor express mRNA for IL-11, leukemia inhibitory factor, SCF, or flk-2/flt3 L, as evidenced by ELISA and reverse transcription–PCR analyses. IL-6 alone never exhibits BPA, and anti-IL-6 antibody has little effect on BPA in KPB-M15-CM, inferring that SCGF activities are not mediated by IL-6 (A.H., A.S., and N.O., unpublished observations). These findings indicate that BPA, GPA, and ΔGPA assay systems are suitable for examining direct activities of SCGF on primitive hematopoietic progenitor cells even in nonseparated BM cells.
MATERIALS AND METHODS
Cell Culture.
KPB-M15, K562, and MOLT-4 cells were cultured in 10% fetal calf serum (FCS)/RPMI 1640 medium, and COS-1 cells in 10% FCS/DMEM. Serum-free CM were recovered from 3- to 4-day cultures of 1 × 106/ml KPB-M15, K562, and MOLT-4 cells.
cDNA Library from KPB-M15 Cells.
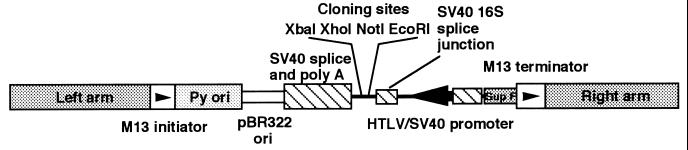
Total RNA was extracted from KPB-M15 cells by the acid guanidinium thiocyanate-phenol-chloroform method (8). Poly(A)+ RNA was isolated with Oligotex-dT30 (Takara Shuzo, Kyoto). cDNA was synthesized from KPB-M15 mRNA (5 μg) using Superscript RTase (GIBCO/BRL) and ZAP cDNA synthesis kit (Stratagene), and size fractionated by Bio-Gel A50m (Bio-Rad). cDNA ≥500 bp was ligated into the EcoRI–XhoI cloning site of λSHDM vector and phage-packaged using a Gigapack-Gold packaging extracts kit (Stratagene) to provide a λSHDM(KPB) phage cDNA library. SURE Escherichia coli was infected with λSHDM(KPB) phage, and seeded onto Luria–Bertani agar plates (1.67 × 106 plaque-forming units). λSHDM vector was constructed by insertion of SHDM plasmid into λ phage (see Fig. 1), which was produced from CDM8 (9) via CDMflit, as described (A.S., A.H., and S.S., unpublished work).
Figure 1.
Structure of λSHDM vector for differential and expression cloning. HTLV, human T-lymphotropic virus; SV40, simain virus 40.
Differential Cloning of SCGF cDNA by Subtractive Probe.
λSHDM(KPB) phage in SURE E. coli plaques was transferred to duplicate nitrocellulose filters (10–12). The phage DNA fixed on each filter was hybridized with 32P-labeled (+) and (−) probes (13). The (+) probe was KPB-M15 cDNA subtracted with MOLT-4 mRNA; single-stranded cDNA, synthesized from KPB-M15 mRNA (14), was annealed with an excess of MOLT-4 mRNA at 65°C for ≥40 hr, and fractions flowing through hydroxyapatite were collected (15). MOLT-4 single-stranded cDNA was used as the (−) probe. The 32P-labeled single-stranded cDNA probes were prepared using the Multiprime DNA labeling system (Amersham). The KPB-M15-specific differential phage library was made of clones both positive for the (+) probe and negative for the (−) probe. The selected phage clones were coinfected with helper phage R408 into XL1-Blue E. coli, replicating to circularize the region between M13 initiator and terminator of λSHDM vector to develop pSHDM cDNA-carrying phagemid. The phagemid was infected into MC1061/P3/PCJ E. coli (at 37°C for 15 min), established by transformation of MC1061/P3 E. coli with CJ236 E. coli-derived PCJ105. Plasmid DNA was prepared from ampicillin/tetracyclin-resistant colonies by alkaline lysis procedure (16, 17), and digested with EcoRI and XhoI.
Transient Gene Expression in COS-1 Cells.
Plasmid cDNA was purified by equilibrium centrifugation in CsCl-ethidium bromide gradients. COS-1 cells (106) were transfected at 37°C for 5 hr with 0.3–3 μg/ml plasmid cDNA in 100 μg/ml DEAE-dextran/100 μM chloroquine/DMEM. They were incubated at room temperature for 2 min in 10% dimethyl sulfoxide/10% FCS/DMEM and then cultured overnight in 10% FCS/DMEM. Another cycle of transfection was followed by 2- to 3-day cultures in serum-free DMEM. Double transfection enhanced gene expression to twice the level by single procedure.
Northern Blot Analysis.
Total RNA (20 μg) from each human cell line was electrophoresed on a 2.2 M formaldehyde/1% agarose gel and transferred to a nitrocellulose filter (18). The RNA blots and the filter fixed with poly(A)+ RNA from various human tissues (Human Multiple Tissue Northern Blots; CLONTECH) were hybridized with fluorescein-labeled pSHDM(116-10C) cDNA fragments covering the 5′ noncoding and most of the coding region (from EcoRI to the first XhoI site in Fig. 2A), and analyzed with the Gene Images detection system (Amersham).
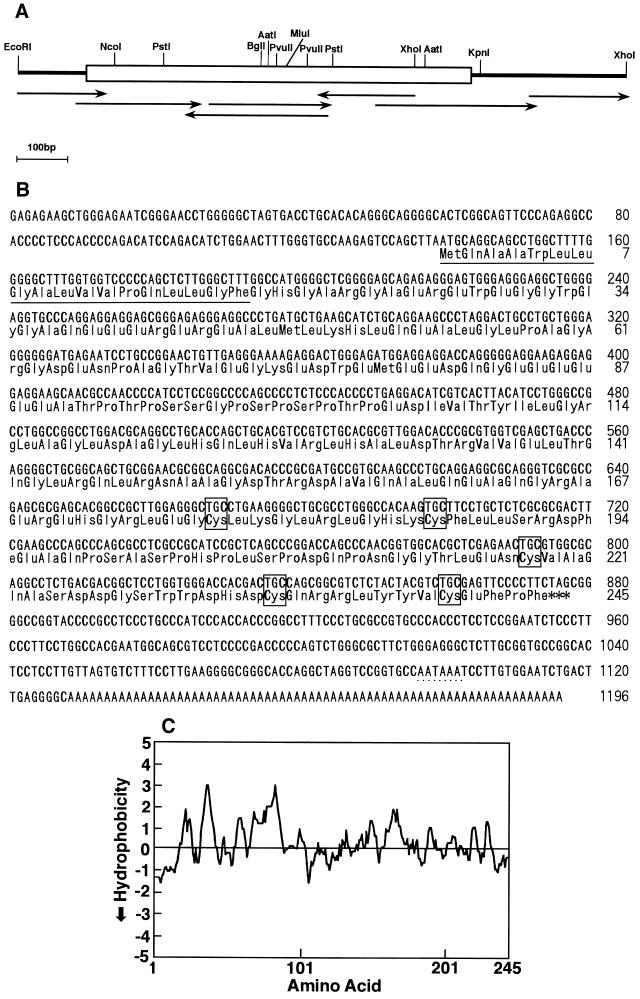
Figure 2.
(A) Restriction endonuclease map and sequencing strategy for pSHDM(116-10C) cDNA. Lines with arrowheads indicate direction and extent of sequencing. A predicted open reading frame is boxed. (B) Nucleotide sequence of SCGF cDNA and deduced amino acid sequence. pSHDM(116-10C) cDNA was sequenced using sets of oligonucleotide sequencing primers and dideoxy terminator cycle sequencing AmpliTaq on an Applied Biosystems model 373A sequencer. Numbering is from the 5′ end of the DNA sequence and from initiation methionine of the amino acid sequence. The putative signal peptide sequence is underlined. Cysteine residues are boxed. The stop codon is indicated by asterisks. Poly(A) signal is dot-underlined in the 3′-untranslated region. (C) Hydrophobicity of SCGF amino acid sequence. Each plot was calculated by the method of Hopp and Wood (22) using the dnasis Mac program (Hitachi, Tokyo).
Assay for Hematopoietic Activities.
Human BM cells were aspirated from the sternum of healthy volunteers after obtaining informed consent. Semisolid culture (1 ml) for BPA and GPA assay contained 5 × 104 nonadherent BM cells at a density of <1.077 g/ml in 2.5–20% samples/5 × 10−5 M 2-mercaptoethanol/20% FCS/Iscove’s modified Dulbecco’s medium/0.3% Bacto agar with 1 unit/ml recombinant human (rh) erythropoietin (Espo; Kirin, Tokyo) and 5 ng/ml rhGM-CSF (Genzyme), respectively (6). The preparations were incubated at 37°C under 5% CO2/100% H2O in air for 14 and 10 days, and then erythroid bursts and GM colonies were enumerated. For ΔGPA assay, an initial 7-day liquid culture of 5 × 105/ml BM cells in 2.5–20% samples/10% FCS/Iscove’s modified Dulbecco’s medium was followed by a secondary semisolid 10-day CFU-GM culture of the harvested cells with 10 ng/ml rhGM-CSF.
Purification of SCGF from KPB-M15-CM.
Serum-free KPB-M15-CM (1,500 ml) was diluted with H2O (9,750 ml) and applied to a DEAE-Sephacel column (5 cm × 6.1 cm; diameter × height) equilibrated with 0.01% Tween 80/20 mM Hepes-NaOH (pH 6.0) (all buffers used contained 0.01% Tween 80). Fractions eluted with a linear gradient of 0.1–0.25 M NaCl were next applied to Cu2+ chelating-Sepharose column (1.6 cm × 4 cm; diameter × height) equilibrated with 20 mM Hepes-NaOH (pH 7.4). Fractions eluted with a linear gradient of 0.05–0.1 M glycine were reconcentrated through a mini-DEAE-Sephacel column (1.6 cm × 0.5 cm; diameter × height). The condensed fractions were applied to Sephacryl S-200HR column (1.6 cm × 64 cm; diameter × height) equilibrated with 20 mM Hepes-NaOH (pH 7.0).
RESULTS AND DISCUSSION
We developed a new λ phage cloning vector, λSHDM (A.S., A.H., and S.S., unpublished work) more effective for differential and expression cloning (19, 20). It was characterized as being intrinsically convertible without any enzymatic manipulation; from a phage vector where cDNA could hybridize with differential probes, to a plasmid vector pSHDM where cDNA could be expressed in mammalian cells. The λSHDM vector possessed human T-lymphotropic virus type I long terminal repeat (R/U5)/simian virus 40 (SV40) early promoter (21), SV40 16S splice junction/SV40 early splice, and poly(A) signal segment for expressing foreign genes efficiently in mammalian cells, pBR322 origin of replication that could be replicated in E. coli, and Sup F, a selection marker in p3 vector-harboring E. coli (9) located between M13 initiator and terminator flanked by λ phage-derived left and right arms (Fig. 1). cDNA inserted into the EcoRI–XhoI site could be oriented so as to be subject to upper human T-lymphotropic virus type I long terminal repeat (R/U5)/SV40 early promoter.
About 120,000 cDNA clones were prepared from KPB-M15 cells as the starting cohort. These were sorted to ≈14,400 based on hybridization with KPB-M15 cDNA subtracted with MOLT-4 mRNA, but not with cDNA from SCGF-infertile MOLT-4 cells. The clones were distributed individually, 48 to a pool, to make up a total of 300 pools. For 137 pools at the first screening, mixed plasmid cDNA from each pool was transfected into COS-1 cells, and CM was concentrated 7- to 8-fold through DEAE-Sephacel (pH 6). A total of 192 clones from 4 pools were selected as positive for BPA and directly stepped up to a secondary trial, where samples from single plasmid cDNA were screened as untreated. The sample from pSHDM(116-10C) cDNA presented the highest BPA at the second screening. Double-stranded pSHDM(116-10C) cDNA was sequenced according to the strategy outlined in Fig. 2A. It was demonstrated to have an open reading frame encoding a novel polypeptide.
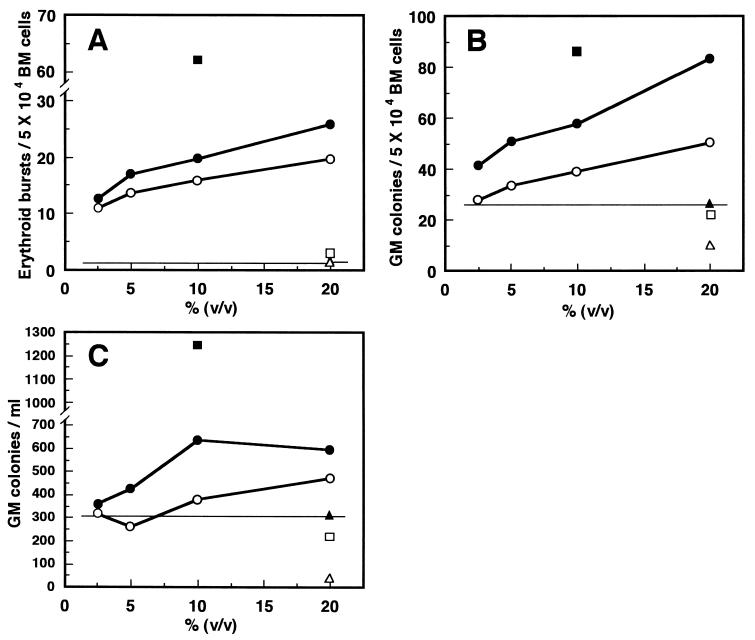
The sample from pSHDM(116-10C) cDNA also exhibited distinct dose-dependent GPA and ΔGPA (Fig. 3). Its BPA and GPA were 2- to 4-fold higher than that of original KPB-M15-CM. In the presence of erythropoietin or GM-CSF, the sample from pSHDM(116-10C) cDNA formed colonies identical in morphology and size with those grown with KPB-M15-CM. The findings therefore confirmed that pSHDM(116-10C) cDNA encoded SCGF as a factor possessing BPA, GPA, and ΔGPA in KPB-M15-CM. SCGF exhibited less BPA than SCF, as evidenced by the number and size of erythroid bursts, and moderate GPA, indicating that the target cell population for SCGF differed from that for SCF. SCGF resembles flk-2/flt3 L in that it has no definite colony-stimulating activity on hematopoietic progenitor cells, while the latter is devoid of erythroid lineage-specific BPA (5). SCGF alone could not support colony formation—i.e., SCGF did not stimulate colony growth in culture containing only SCGF except in 20% FCS/Iscove’s modified Dulbecco’s medium.
Figure 3.
Hematopoietic activities of SCGF on human BM cells. CM samples were reevaluated at various concentrations for BPA (A), GPA (B), and ΔGPA (C), as was CM from COS-1 cells transfected with pSHDM(116-10C) cDNA (•), mock-transfected COS-1 cells (▴), COS-1 cells transfected with pSHDM (soluble rhSCF) cDNA (▪), KPB-M15 cells (○), K562 cells (▵), and MOLT-4 cells (□). In C, the number of preexisting CFU-GM before initial liquid culture was 293 per ml. Data are given as means of three dishes.
Human SCGF cDNA had 1,196 nucleotides containing a long open reading frame of 738 nucleotides that encoded a 245-amino acid polypeptide (Fig. 2B). No homology with the database in the EMBL, GenBank and Swiss-Prot was found for the SCGF cDNA and amino acid sequence. About 20 amino acids, starting from initiation methionine, were hydrophobic (Fig. 2C), corresponding to a putative signal peptide. Unlike in most hematopoietic growth factors with the exception of G-CSF (23) and IL-11 (24), there existed no consensus sequence of potential NH2-glycosylation sites (Asn-Xnn-Thr/Ser). In fact, SCGF bound neither to Con A-Sepharose nor to WGA-agarose (data not shown). The minimal molecular mass of SCGF was calculated to be 29,038 Da. Five cysteine residues were located close to the carboxyl terminus of the polypeptide. Noteworthy was a glutamic acid-rich sequence (13.2%), particularly six successive glutamic acids (Glu-84–Glu-89). A motif sequence search showed that acidic poly-Glu was relatively ubiquitous in parathymosin, bone morphogenetic protein, Na+/H+ exchanger β, CD20, L-/N-myc, HOX-A2/A7/B7, and so forth. A Pro/Ser/Thr-rich PT box (positions 91–104) was found in the carboxyl- and amino-terminal domain of thrombopoietin (25) and GM-CSF (26), respectively. Their functional significance is not clear.
Northern blot analysis was carried out to elucidate the types of cells expressing SCGF mRNA. Hybridization of total or poly(A)+ RNA from miscellaneous cells with pSHDM(116-10C) cDNA fragments demonstrated that a single 1.8-kb mRNA was expressed in KPB-M15, HL-60, THP-1, U-937, HEL, NHF, and CCD-8Lu cells, but not in K562, MOLT-4, U266Bl, IM-9, HeLa S3, A431, Bowes, and 293 cells (Fig. 4A); in particular, no transcript was identified in the 20 μg/lane poly(A)+ RNA from K562 and MOLT-4 cells (data not shown). G-CSF (27), IL-11 (24), and thrombopoietin (28) are all hematopoietic growth factors, for which mRNA has been detected in myeloid cells and fibroblasts, but not in lymphocytes. This implies that SCGF is a growth factor functioning within the hematopoietic microenvironment. Low-level expression of SCGF mRNA species of 1.6 kb was widespread throughout the tissues examined (Fig. 4B); an additional 2.6-kb transcript was noted in the heart.
Figure 4.
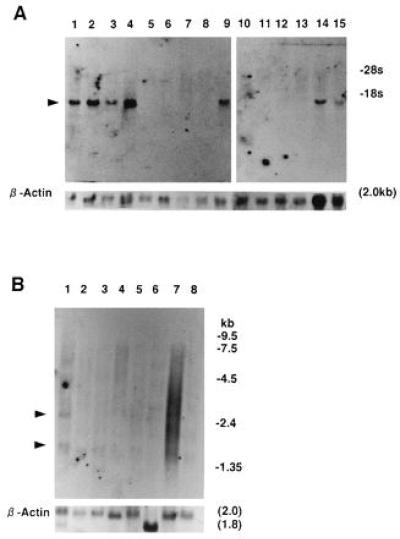
Northern blot hybridization. (A) pSHDM(116-10C) cDNA fragment was hybridized with total RNA (20 μg) from human cell lines. Lanes: 1, HL-60 (myeloid); 2, THP-1 (monocyte); 3, U-937 (monocyte); 4 KPB-M15 (myeloid); 5, U266Bl (B cell); 6, IM-9 (B cell); 7, MOLT-4 (T cell); 8, K562 (erythroid); 9, HEL (erythroid); 10, HeLa S3 (cervical carcinoma epithelial-like); 11, A431 (epidermoid carcinoma epithelial-like); 12, Bowes (melanoma cell); 13, 293 (adenovirus type 5-transformed embryonic kidney cell); 14, NHF (fibroblast); and 15, CCD-8Lu (fibroblast). (B) pSHDM(116-10C) cDNA fragment was hybridized with poly(A)+ RNA (2 μg) from human tissues. Lanes: 1, heart; 2, brain; 3, placenta; 4, lung; 5, liver; 6, skeletal muscle; 7, kidney; and 8, pancreas. The RNA blots were rehybridized with β-actin cDNA.
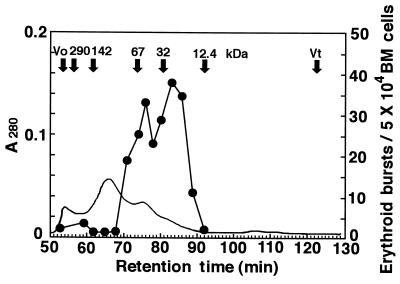
SCGF was partially purified from KPB-M15-CM by sequential DEAE-Sephacel (pH 6) and Cu2+ chelating-Sepharose column chromatography, and applied to Sephacryl S-200HR gel filtration. Two elution peaks of 29 and 58 kDa were obtained (Fig. 5); the former roughly corresponded to the molecular mass predicted from the SCGF amino acid sequence, and the latter was most likely a homodimer of the former, because the elution peak was not bimodal in DEAE-Sephacel and Cu2+ chelating-Sepharose column chromatography (data not shown). It is presently unclear, however, whether KPB-M15-CM contains another SCGF-like factor.
Figure 5.
Sephacryl S-200HR gel filtration of SCGF, partially purified from KPB-M15-CM through DEAE-Sephacel (pH 6) and Cu2+ chelating-Sepharose column chromatography. Each fraction was monitored by BPA (•). A solid line indicates A280 nm, and Mr markers are given as arrows.
The effects of rhSCGF on surface phenotype-sorted hematopoietic progenitor cells are currently being investigated. Murine SCGF, if available in the future, could be a useful tool for elucidating the mechanisms of primitive hematopoiesis in conjunction with SCF, flk-2/flt3 L, IL-1, IL-3, IL-6, IL-11, G-CSF, and GM-CSF. The λSHDM vector devised in the present study could be applied to the search for other novel genes by expression cloning. Clinically, in vivo administration of SCGF should ameliorate myelosuppression after irradiation or chemotherapy, or hematopoietic insufficiency including aplastic anemia, myelodysplastic syndrome, and leukemia. Elucidating the mechanisms of SCGF production should help to establish the pathogenesis of hematological disorders.
Acknowledgments
We thank Jun Fujita for providing SCF cDNA.
ABBREVIATIONS
- SCGF
stem cell growth factor
- BPA
burst-promoting activity
- GPA
granulocyte/macrophage-promoting activity
- BM
bone marrow
- CM
conditioned medium
- CSF
colony-stimulating factor
- IL
interleukin
- GM
granulocyte/macrophage
- SCF
stem cell factor
- FCS
fetal calf serum
- rh
recombinant human
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. D86586).
References
- 1.Metcalf D. Blood. 1993;82:3515–3523. [PubMed] [Google Scholar]
- 2.Zsebo K M, Wypych J, McNiece I K, Lu H S, Smith K A, Karkare S B, Sachdev R K, Yuschenkoff V N, Birkett N C, Williams L R, Satyagal V N, Tung W, Bosselman R A, Mendiaz E A, Langley K E. Cell. 1990;63:195–201. doi: 10.1016/0092-8674(90)90300-4. [DOI] [PubMed] [Google Scholar]
- 3.Huang E, Nocka K, Beier D R, Chu T-Y, Buck J, Lahm H-W, Wellner D, Leder P, Besmer P. Cell. 1990;63:225–233. doi: 10.1016/0092-8674(90)90303-v. [DOI] [PubMed] [Google Scholar]
- 4.Anderson D M, Lyman S D, Baird A, Wignall J M, Eisenman J, Rauch C, March C J, Boswell H S, Gimpel S D, Cosman D, Williams D E. Cell. 1990;63:235–243. doi: 10.1016/0092-8674(90)90304-w. [DOI] [PubMed] [Google Scholar]
- 5.Lyman S D, James L, Bos T V, de Vries P, Brasel K, Gliniak B, Hollingsworth L T, Picha K S, McKenna H J, Splett R R, Fletcher F A, Maraskovsky E, Farrah T, Foxworthe D, Williams D E, Beckmann M P. Cell. 1993;75:1157–1167. doi: 10.1016/0092-8674(93)90325-k. [DOI] [PubMed] [Google Scholar]
- 6.Hiraoka A, Ohkubo T, Fukuda M. Cell Biol Int Rep. 1986;10:347–355. doi: 10.1016/0309-1651(86)90006-8. [DOI] [PubMed] [Google Scholar]
- 7.Hiraoka A, Ohkubo T, Fukuda M. Cancer Res. 1987;47:5025–5030. [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Seed B. Nature (London) 1987;329:840–842. doi: 10.1038/329840a0. [DOI] [PubMed] [Google Scholar]
- 10.Hedrick S M, Cohen D I, Nielsen E A, Davis M M. Nature (London) 1984;308:149–153. doi: 10.1038/308149a0. [DOI] [PubMed] [Google Scholar]
- 11.Duguid J R, Dinauer M C. Nucleic Acids Res. 1990;18:2789–2792. doi: 10.1093/nar/18.9.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hara E, Kato T, Nakada S, Sekiya S, Oda K. Nucleic Acids Res. 1991;19:7097–7104. doi: 10.1093/nar/19.25.7097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahl G M, Berger S L. Methods Enzymol. 1987;152:415–423. doi: 10.1016/0076-6879(87)52048-1. [DOI] [PubMed] [Google Scholar]
- 14.Gubler U, Hoffman B J. Gene. 1983;25:263–269. doi: 10.1016/0378-1119(83)90230-5. [DOI] [PubMed] [Google Scholar]
- 15.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 30–33. [Google Scholar]
- 16.Birnboim H C, Doly J. Nucleic Acids Res. 1979;7:1513– 1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ish-Horowicz D, Burke J F. Nucleic Acids Res. 1981;9:2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alwine J C, Kemp D J, Stark G R. Proc Natl Acad Sci USA. 1977;74:5350–5354. doi: 10.1073/pnas.74.12.5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee F, Yokota T, Otsuka T, Gemmell L, Larson N, Luh J, Arai K, Rennick D. Proc Natl Acad Sci USA. 1985;82:4360–4364. doi: 10.1073/pnas.82.13.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang Y-C, Ciarletta A B, Temple P A, Chung M P, Kovacic S, Witek-Giannotti J S, Leary A C, Kriz R, Donahue R E, Wong G G, Clark S C. Cell. 1986;47:3–10. doi: 10.1016/0092-8674(86)90360-0. [DOI] [PubMed] [Google Scholar]
- 21.Takebe Y, Seiki M, Fujisawa J, Hoy P, Yokota K, Arai K, Yoshida M, Arai N. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hopp T P, Wood K R. Proc Natl Acad Sci USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagata S, Tsuchiya M, Asano S, Kaziro Y, Yamazaki T, Yamamoto O, Hirata Y, Kubota N, Oheda M, Nomura H, Ono M. Nature (London) 1986;319:415–418. doi: 10.1038/319415a0. [DOI] [PubMed] [Google Scholar]
- 24.Paul S R, Bennett F, Calvetti J A, Kelleher K, Wood C R, O’Hara R M, Jr, Leary A C, Sibley B, Clark S C, Williams D A, Yang Y-C. Proc Natl Acad Sci USA. 1990;87:7512–7516. doi: 10.1073/pnas.87.19.7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Sauvage F J, Hass P E, Spencer S D, Malloy B E, Gurney A L, Spencer S A, Darbonne W C, Henzel W J, Wong S C, Kuang W-J, Oles K J, Hultgren B, Solberg L A, Jr, Goeddel D V, Eaton D L. Nature (London) 1994;369:533–538. doi: 10.1038/369533a0. [DOI] [PubMed] [Google Scholar]
- 26.Wong G G, Witek J S, Temple P A, Wilkens K M, Leary A C, Luxenberg D P, Jones S S, Brown E L, Kay R M, Orr E C, Shoemaker C, Golde D W, Kaufman R J, Hewick R M, Wang E A, Clark S C. Science. 1985;228:810–815. doi: 10.1126/science.3923623. [DOI] [PubMed] [Google Scholar]
- 27.Demetri G D, Griffin J D. Blood. 1991;78:2791–2808. [PubMed] [Google Scholar]
- 28.Kaushansky K. Blood. 1995;86:419–431. [PubMed] [Google Scholar]