Abstract
Proteins terminating with a CAAX motif, such as the nuclear lamins and the Ras family of proteins, undergo posttranslational modification of a carboxyl-terminal cysteine with an isoprenyl lipid—a process called protein prenylation. After prenylation, the last three residues of CAAX proteins are clipped off by an endoprotease of the endoplasmic reticulum. Rce1 is responsible for the endoproteolytic processing of the Ras proteins and is likely responsible for endoproteolytic processing of the vast majority of CAAX proteins. Prenylation has been shown to be essential for the proper intracellular targeting and function of several CAAX proteins, but the physiologic importance of the endoprotease step has remained less certain. Here, we will review methods that have been used to define the physiologic importance of the endoproteolytic processing step of CAAX protein processing.
Introduction
Eukaryotic proteins that terminate with a “CAAX” sequence typically undergo a series of posttranslational processing reactions, beginning with protein prenylation, which is the attachment of a farnesyl or geranylgeranyl lipid to the thiol group of the cysteine residue (the “C” of the CAAX motif) (Zhang and Casey, 1996). Second, the last three amino acids of the protein (i.e., the –AAX) are removed by a prenylprotein-specific endoprotease of the endoplasmic reticulum (ER) (Ashby, 1998; Boyartchuk et al., 1997; Young et al., 2000). Third, the carboxyl group of the newly exposed isoprenylcysteine is methylated by isoprenylcysteine carboxyl methyltransferase (ICMT) (Dai et al., 1998; Hrycyna et al., 1991; Young et al., 2000). These three processing steps have been studied intensively with yeast a-factor, a farnesylated yeast mating pheromone, and with the yeast and mammalian Ras proteins, small GTP-binding proteins involved in signal transduction. However, large numbers of cellular proteins with diverse biological functions contain a CAAX motif and undergo these modifications.
The enzymes responsible for the prenylation of the CAAX proteins, protein farnesyltransferase and protein geranylgeranyltransferase I, have been characterized extensively in both yeast and higher organisms (Zhang and Casey, 1996). Progress in understanding the importance of the endoprotease step has lagged behind, but during the past few years some progress has been made in understanding the CAAX endoproteases and their functional importance.
Two genes from Saccharomyces cerevisiae, RCE1 and STE24 (AFC1), were identified as playing a role in the endoproteolytic removal of the “-AAX” from Ras2p and the precursor to the yeast mating pheromone a-factor (Boyartchuk et al., 1997). Rce1p (for Ras and a-factor converting enzyme) is essential for cleaving the –AAX from Ras2p. In addition, Rce1p is quite capable of cleaving the –AAX from a-factor. Ste24p is a zinc metalloproteinase that plays dual roles in the endoproteolytic processing of the precursor to a-factor. Along with Rce1p, Ste24p is capable of carrying out the carboxyl-terminal cleavage reaction (i.e., the release of the –AAX from a-factor) (Boyartchuk et al., 1997; Tam et al., 1998). In addition, however, it cleaves seven amino acids from the amino terminus of the protein (Tam et al., 1998). Ste24p plays absolutely no role in the endoproteolytic processing of the yeast Ras proteins (Boyartchuk et al., 1997). In fact, a-factor is the only well-established substrate for Ste24p in yeast (Boyartchuk et al., 1997).
Both yeast Rce1p and Ste24p are endoplasmic reticulum proteins with multiple transmembrane domains (Young et al., 2000). Rce1p is 315-amino acids in length and is predicted to contain multiple transmembrane helices (Boyartchuk et al., 1997). The Rce1p amino acid sequence does not contain sequence motifs characteristic of the well-defined classes of proteases, but there are remote similarities with the type IIb signal peptidase, which cleaves signal sequences from proteins containing nearby lipid modifications (Boyartchuk et al., 1997). Ste24p contains 453 amino acids and also contains multiple transmembrane domains (Boyartchuk et al., 1997). Ste24p contains a HEXXH (H, His; E, Glu) motif between amino acids 297–301, a sequence shared by many zinc-dependent metalloproteases. Mutating either of the conserved histidines in the HEXXH motif within Ste24p blocks a-factor processing (Boyartchuk et al., 1997; Fujimura-Kamada et al., 1997)
In yeast, Rce1p is essential for Ras processing and function (Boyartchuk et al., 1997). Heat shock sensitivity elicited by a mutationally activated Ras2p is suppressed by RCE1 deficiency. However, RCE1 deficiency in yeast almost certainly does not completely block all Ras functions, given that rce1Δ yeast are quite viable (Boyartchuk et al., 1997) and yeast lacking the Ras proteins are not (Kataoka et al., 1984). RCE1 deficiency in yeast mislocalizes Ras2p away from the plasma membrane to the cytosol (Boyartchuk et al., 1997). In wild-type yeast, a green fluorescent protein (GFP)–RAS2 fusion is targeted to the periphery of the cells (i.e., the plasma membrane). In contrast, the GFP–RAS2 fusion in rce1α yeast is located inside the cell (i.e., either in the cytosol or associated with internal membrane compartments) (Boyartchuk et al., 1997).
The identification of RCE1 and STE24 in yeast made it possible to identify the human and mouse orthologues, RCE1 and ZMPSTE24 (Boyartchuk et al., 1997; Fujimura-Kamada et al., 1997; Kumagai et al., 1999; Leung et al., 2001; Young et al., 2000). Human RCE1 is absolutely required for normal processing of the Ras proteins; the RCE1 gene and is expressed ubiquitously, with the highest levels of expression in placenta, heart, and skeletal muscle (Kim et al., 1999; Otto et al., 1999). Human RCE1 is 329 amino acids in length, 27.6% identical to Rce1p in S. cerevisiae, and contains multiple transmembrane helices (Young et al., 2000). The human orthologue for yeast STE24, ZMPSTE24, is also expressed ubiquitously (Bergo et al., 2002b; Leung et al., 2001). It is 475 amino acids in length, with 36% identity to the yeast protein (Young et al., 2000). Like yeast Ste24p, human ZMPSTE24 contains the zinc metalloprotease motif HEXXH and multiple transmembrane helices (Young et al., 2000). Human and mouse ZMPSTE24 faithfully carries out the processing of yeast a-factor (Leung et al., 2001); however, no a-factor orthologues have been identified in mammals. During the past few years, several laboratories have shown that Zmpste24 is critical for the endoproteolytic processing of prelamin A, a farnesylated CAAX protein that is the precursor to lamin A, a structural protein of the nuclear envelope (Bergo et al., 2002b; Pendás et al., 2002). Akin to Ste24p-processing of a-factor, Zmpste24 is almost certainly capable of cleaving the –AAX from prelamin A (Bergo et al., 2002b; Corrigan et al., 2004). In addition, it cleaves prelamin A a second time, 15 residues upstream from the farnesylcysteine, releasing mature lamin A (Corrigan et al., 2004; Dalton and Sinensky, 1995; Kilic et al., 1997). Remarkably, prelamin A and yeast a-factor are apparently cleaved twice by Zmpste24 in a highly specific fashion, despite very little amino acid similarity surrounding the four different sessile bonds.
Establishing that Human RCE1 is a CAAX Endoprotease
The laboratory of Patrick Casey (Duke University) expressed human RCE1 protein Sf9 insect cells and performed biochemical assays to show that it was a CAAX endoprotease (Kim et al., 1999; Otto et al., 1999). Extracts from the Sf9 cells were mixed with recombinant isoprenylated CAAX proteins. In this assay, recombinant RCE1 cleaves the –AAX from the isoprenylated proteins, rendering the proteins susceptible to methylation with recombinant Ste14p (the yeast isoprenylcysteine carboxyl methyltransferase) and [methyl-3H]-S-adenosylmethionine. With this “coupled endoproteolysis/methylation assay,” Casey’s laboratory demonstrated that human RCE1 is capable of processing farnesylated K-Ras, farnesylated H-Ras, farnesylated N-Ras, the farnesylated heterotrimeric G protein Gα1 subunit, geranylgeranylated K-Ras, and geranylgeranyl-Rap1B. Both farnesylated and geranylgeranylated K-Ras exhibited a Km value of approximately 0.5 μM and similar Kcat values. Prenylated CAAX peptides, but not nonisoprenylated peptides, were able to compete for the processing of isoprenylated K-Ras (Otto et al., 1999). Also, a previously identified inhibitor of endoproteolytic processing, RPI, was shown to be an effective inhibitor of RCE1, with an IC50 of ~5 nM (Otto et al., 1999).
Rce1 Knockout Mice
The laboratory of Stephen Young (Univ. California, Los Angeles) created Rce1-deficient mice with standard gene-targeting techniques (Kim et al., 1999). Heterozygous knockout mice (Rce1+/−) were normal. However, almost all of the homozygous embryos (Rce1−/−) died, beginning at about embryonic day 15 (E15). Rce1−/− mice were born alive very rarely, and those mice were invariably quite small and died within a few weeks. The precise reason for the death of Rce1−/− mice is not known; the viable Rce1−/− embryos at E15.5 revealed no consistent differences in morphology, organogenesis, stage of development, color, or size.
Analyzing Ras Function in Rce1−/− MEFs
Primary mouse embryonic fibroblasts (MEFS) could be cultured from the Rce1−/− embryos (Kim et al., 1999). To generate fibroblasts, Rce1+/− mice (which can be obtained from the authors of this chapter), are intercrossed, and pregnant females are sacrificed at E13.5. Genotyping of embryos is performed by Southern blot analysis of DNA prepared from fetal membranes (Kim et al., 1999). The embryos are incubated in 5.0 ml of 0.25% trypsin-EDTA (Life Technologies, Gaithersburg, MD) for 8 h at 4°C, followed by a 20-min incubation at 37°C. The embryos are mechanically disrupted by repeated pipetting in 5 ml of DME-FBS medium [Dulbecco’s modified Eagle’s medium supplemented with 10% v/v fetal bovine serum (FBS), l-glutamine, nonessential amino acids, penicillin-streptomycin (all from Life Technologies), and 2-mercaptoethanol (Sigma, 100 μM final concentration)]. The debris is allowed to settle, and the cell suspension, diluted to 25 ml, is plated in T150 tissue culture flasks. The cells are grown in an incubator at 37°C with 7% CO2. (~1 α 107 cells are obtained from each embryo) To immortalize the MEFS, cells are passaged according to a 3T6 protocol (Todaro, 1969), in which cells are harvested and counted every 3 days and replated at a density of 2 α 106 cells per 100-mm dish.
Absence of CAAX Endoprotease Activity in Rce1-deficient Cells
CAAX endoprotease activity in Rce1−/− and Rce1+/+ MEFs can be measured with a coupled endoprotease/methylation assay (Kim et al., 1999; Otto et al., 1999). Substrate proteins (e.g., K-Ras, N-Ras and Rap-1B) containing a CAAX sequence are expressed in Escherichia coli and then prenylated in vitro with either recombinant protein farnesyltransferase or protein geranylgeranyltransferase (Otto et al., 1999). To prepare membranes from Rce1+/+ and Rce1−/− embryonic fibroblasts, cells were lysed by sonication. After centrifugation at 500 α g for 10 min (to remove debris and cell nuclei), the membrane fraction is prepared by spinning at 200,000 α g for 1.5 h. The membranes are then resuspended in 50 mM Tris-HCl at protein concentrations of 10–25 mg/ml. To measure the prenylprotein–specific CAAX endoprotease activity, the prenylated protein substrates and membrane fractions are mixed in 50 μl of a 100 mM Hepes buffer (pH 7.5) containing 5 mM MgCl2. After a 30-min incubation at 37°C, the proteolysis reactions are stopped and the methyltransferase reaction is initiated by adding a cocktail (20 μl) containing 5 mM NaHPO4, 87.5 mM EDTA, several protease inhibitors (300 μM N-tosyl-l-phenylalanine chloromethylketone, 1 mM phenylmethylsulfonyl fluoride (PMSF), 10 mM 1,10-phenanthroline), 20 μg of Sf9 cell membranes expressing yeast STE14, and 17.5 μM [3H] AdoMet (1.5 Ci/mmol). After a 20-min incubation at 37°C, the methylation reaction is terminated by adding 0.5 ml of 4% sodium dodecyl sulfate (SDS). Bovine brain cytosol (50 μg protein) is added as a carrier, and the mixture is incubated for 20 min at room temperature, after which the proteins were precipitated with 0.5 ml of 30% trichloroacetic acid. Precipitated proteins are collected on glass fiber or nitrocellulose filters and the extent of proteolysis is determined by quantifying 3H incorporation into the protein by scintillation counting. With this assay, Rce1−/− membranes were shown to lack CAAX processing activity for farnesylated heterotrimeric Gα1 subunit, farnesylated H-Ras, farnesylated N-Ras, geranylgeranylated K-Ras, and geranylgeranyl-Rap1B (Kim et al., 1999; Otto et al., 1999).
Documenting an Accumulation of Uncleaved Rce1 Substrates in Rce1-deficient Cells
Rce1−/− MEFs accumulate uncleaved Rce1 substrates, and this accumulation of Rce1 substrates can be detected with a coupled endoproteolysis/methylation assay (Leung et al., 2001). Whole-cell extracts (150 μg) from Rce1+/+ and Rce1−/− MEFs are incubated with 10 μM S-adenosyl-l-[methyl-14C] methionine (55 Ci/mol, Amersham Pharmacia), Sf9 cells expressing high levels of Rce1 (100 μg), and Sf9 cells expressing high levels of yeast Ste14p (100 μg). The reaction is incubated for 2 h at 37°C. The reaction is stopped by adding 50 μl of 1.0 M NaOH containing 0.1% SDS. The reaction mixture (90 μl) can then be spotted onto a pleated 2 α 8-cm filter paper wedged in the neck of a 20-ml scintillation vial containing 5 ml of scintillation fluid (ScintiSafe Econo 1, Fisher). The vials are capped and incubated at room temperature for 5 h to allow the [14C] methanol (formed by base-hydrolysis of methyl esters) to diffuse into the scintillation fluid. The amount of base-releasable methanol is far higher in the Rce1−/− fibroblasts then in the Rce1+/+, reflecting the accumulation of uncleaved Rce1 substrates (substrates that can be readily cleaved and methylated in the setting of high levels of Rce1 and Ste14p) (Fig. 1). Importantly, there was no accumulation of “methylatable” protein substrates in fibroblasts lacking Zmpste24 (Leung et al., 2001) (Fig. 1).
Fig 1.
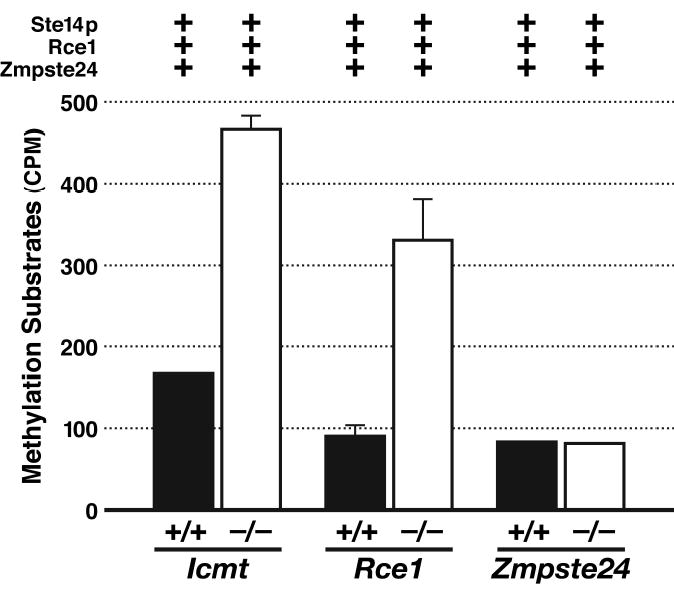
An accumulation of “methylatable” substrates in Rce1 −/− cells. Methylation of whole-cell extracts from primary fibroblasts derived from Zmpste24+/+, Zmpste24 −/−, Icmt+/+, Icmt −/−, Rce1+/+, and Rce1 −/− embryos. The cell extracts were incubated with S-adenosyl-l-[methyl-14C]methionine, membranes containing high levels of Ste14p, yeast membranes containing high levels of mouse Rce1, and yeast membranes containing high levels of mouse Zmpste24. The relative level of methylatable substrates in wild-type and knockout cells was assessed with a base hydrolysis/methanol diffusion assay. For the Zmpste24+/+ and Zmpste24 −/− cells, we observed identical results when membranes expressing Rce1 were left out of the reaction mixture. Reproduced, with permission from The Journal of Biological Chemistry (Leung et al., 2001).
Altered Electrophoretic Migration of Ras proteins in Rce1−/− Fibroblasts
The electrophoretic migration of the Ras proteins in Rce1−/− cells is abnormally slow, reflecting an absence of endoproteolytic processing (Kim et al., 1999). To study the electrophoretic migration of Ras proteins, it is first necessary to prepare lysates of MEFs (Kim et al., 1999). Confluent cultures of MEFS are harvested from a 100-mm plate in 0.5 ml of RIPA lysis buffer [50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 0.5 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml of aprotinin] and disrupted by sonication for 10 s in a Branson model 400 sonifier (duty cycle constant, output control 0.3; Danbury, CT). The samples are then heated to 95°C for 5 min and loaded onto an SDS/10–20% gradient polyacrylamide gel. After electrophoretic transfer of the proteins to a nitrocellulose membrane, western blotting is performed with the “pan-Ras” monoclonal antibody Ab-4 (i.e., binds to N-, K-, and H-Ras; Oncogene Science, Uniondale, NY) or the K-Ras-specific monoclonal antibody Ab-1 (Oncogene Science), followed by an incubation with horseradish peroxidase–conjugated sheep anti-mouse IgG (Amersham). The binding of the secondary antibodies is detected by enhanced chemiluminescence (Amersham). The electrophoretic mobility of the Ras proteins from Rce1−/− MEFs (or Rce1−/− embryos) is distinctly abnormal, even on a minigel. In fact, Ras proteins with a normal electrophoretic mobility are undetectable in Rce1−/− cells (Fig. 2). When equal amounts of Rce1−/− and Rce1+/+ MEF lysates are mixed, one observes a doublet Ras band, consisting of the processed Ras proteins in Rce1+/+ MEFs and the unprocessed Ras proteins in Rce1−/− MEFs (Kim et al., 1999).
Fig 2.
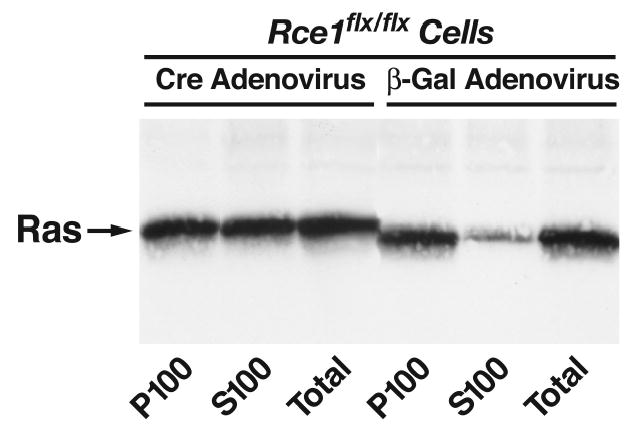
Intracellular localization of Ras proteins in immortalized fibroblasts. Rce1 flx/flx cells express normal levels of Rce1, whereas Rce1 α/α fibroblasts are completely deficient in Rce1 expression. Cre adenovirus fully converted the Rce1flx/flx fibroblasts into Rce1 α/α fibroblasts. Fibroblasts were fractionated into cytosolic (S100) and membrane (P100) fractions; Ras proteins were immunoprecipitated and analyzed on western blots of sodium dodecyl sulfate-polyacrylamide gels with antibody Ab-4. Note higher molecular weight of the Ras proteins in Rce1 α/α fibroblasts. Reproduced, with permission from Molecular and Cellular Biology (Bergo et al., 2002a).
Methylation of Ras Proteins in Rce1−/− Fibroblasts
The absence of Ras endoproteolysis means that the Ras proteins cannot be methylated. The absence of a methyl ester in the Ras proteins from Rce1−/− MEFs can be documented by a metabolic labeling assay (Clarke et al., 1988) (Kim et al., 1999). 100-mm dishes containing approximately 2.5 α 106 Rce1−/− or Rce1+/+ MEFs are incubated in the presence of 1 mCi of l-[methyl-3H]methionine (80Ci/mmol, Amersham-Pharmacia, Chicago, IL) and [35S] cysteine (>1000 Ci/mmol, Amersham-Pharmacia) in methionine- and cysteine-free DMEM/10% FBS in a 7% CO2 incubator at 37°C for 24 h. The medium is removed, and the cells are washed with 5 ml of ice-cold PBS, scraped from the dish into 1 ml of ice-cold PBS, and centrifuged at 500 α g. Cell pellets are resuspended in 1.0 ml of RIPA lysis buffer and disrupted by sonification, as described above. Ras proteins are then immunoprecipitated with antibody Y13-259, and the immunoprecipitate is size-fractionated on an SDS/10–20% polyacrylamide gel. The gel is fixed in isopropanol, water, and acetic acid (25:65:10, v/v), soaked for 30 min in Amplify (Amersham-Pharmacia), and dried. The gel is imaged with a phosphorimager, and the region of the gel corresponding to the Ras band is excised. The amount of [3H] methanol released from the gel fragment by base hydrolysis of the farnesylcysteine methyl esters is quantified by scintillation counting. The gel slice is placed in a 1.5-ml capless microcentrifuge tube and mixed with 200 μl of 2 M NaOH; the tube is lowered into 20-ml scintillation vial containing 6 ml of Safety-Solve II counting fluor (RPI) fluid. The scintillation vial is then capped and incubated at 55°C. The [3H]methanol released from the farnesylcysteine methyl esters diffuses into the fluor within the capped vial, while the 35S-labeled protein remains in the gel slice. After 24 h, the microcentrifuge tubes are removed, and the radioactivity in the fluor is counted. As judged by the [3H]methanol release, the Ras proteins from Rce1+/+ cells contain a 3H-labeled farnesylcysteine methyl ester, whereas Ras proteins from the Rce1−/− cells do not (Kim et al., 1999).
Subcellular Fractionation and Localization of Ras Proteins in Rce1−/− MEFs
To determine the degree of membrane association of Ras proteins in Rce1−/− MEFs, subcellular fractionation studies can be performed (Bergo et al., 2002b; Bergo et al., 2000; Kim et al., 1999). 100-mm dishes containing confluent MEFs are washed with ice-cold PBS, and cells are collected in 1.0 ml of PBS and centrifuged at 500 α g for 10 min. Cell pellets are incubated with 1225 μl of hypotonic buffer (10 mM Tris-HCl, pH 7.5, 1.0 mM MgCl2, 0.5 mM PMSF, 10 μg/ml leupeptin, and 10 μg/ml of aprotinin, and 1μM DDT) and placed on ice for 10 min. Cells are disrupted with an ice-cold dounce tissue homogenizer, after which 225 μl of 1 M NaCl is added. A total of 450 μl of this solution (“total lysate”) is transferred to a microfuge tube and set aside. The remaining 1000 μl is transferred to a polycarbonate ultracentrifuge tube and spun at 100,000 α g for 30 min at 4°C. The supernatant fluid (S100, representing the cytosolic fraction) is transferred to a new microcentrifuge tube; the pellet (P100, representing the membrane fraction) is resuspended in 850 μl of hypotonic buffer and 150 μl of 1 M NaCl. Next, 50 μl of 10αRIPA buffer is added to the “total lysate” sample, and 110 μl is added to the S100 and P100 fractions. After incubation on ice for 10 min, the lysates are clarified by centrifugation at 25,000 α g for 30 min at 4°C. Supernatants are transferred to new microfuge tubes, and immunoprecipitation, gel fractionation, and western blot detection of the Ras proteins are performed as described earlier. A substantial difference in the intracellular distribution of the Ras proteins is apparent by immunoblot analysis of P100 and S100 fractions of Rce1−/− and Rce1+/+ fibroblasts (Kim et al., 1999). Virtually all of the Ras proteins in Rce1+/+ cells are located in the P100 fraction, whereas about one-half of the Ras proteins in Rce1−/− cells is located in the S100 fraction (Fig. 2).
To further assess the association of the Ras proteins with membranes of Rce1−/− and Rce1+/+ embryonic fibroblasts, enhanced GFP–Ras fusion protein constructs can be expressed in MEFs and then visualized by fluorescence microscopy (Bergo et al., 2002a; Kim et al., 1999). Rce1−/− and Rce1+/+ MEFs are transfected with 2.0 μg of GFP–Ras fusion plasmids with SuperFect Reagent (Qiagen, Valencia, CA). Cells are fixed in 4% formalin 24–28 h after transfection, mounted, and viewed by confocal microscopy. In Rce1+/+ cells, the fluorescence is localized almost entirely to the plasma membrane. In contrast, the vast majority of the fluorescence in the Rce1−/− cells is cytosolic or associated with internal membranes. However, very small amounts of fluorescence can be observed along the plasma membrane in the Rce1−/− cells.
The importance of Rce1 for the localization of GFP-Ras fusion proteins is dependent on the fact that the Ras proteins are farnesylated. When mutations are introduced into the CAAX motif so as to create GFP-Ras fusions that are geranylgeranylated, the impact of Rce1 deficiency on intracellular localization is no longer detectable (Michaelson et al., 2005). Thus, geranylgeranylated GFP–N-Ras and geranylgeranylated GFP–K-Ras are largely localized to the plasma membrane in Rce1−/− MEFs and not mislocalized to the cytosol (i.e., a pattern indistinguishable from that in Rce1+/+ cells). Changing the isoprenyl chain length of Rac1 and RhoB (proteins that are normally geranylgeranylated) had the opposite effect. The intracellular localization of geranylgeranylated GFP–Rac1 and geranylgeranylated GFP–RhoB is unaffected by Rce1 deficiency. However, the intracellular localization of farnesylated GFP–Rac1 and farnesylated GFP–RhoB is sensitive to Rce1 deficiency (Michaelson et al., 2005).
Defining the Impact of Rce1 Deficiency on MAP Kinase Activation in Rce1−/− Fibroblasts
We suspected that absent endoproteolysis of the Ras proteins might result in impaired growth factor–mediated activation of the Ras effector Erk1/2. Serum-stimulated activation of Erk1/2 was assessed by seeding 1 α 105 Rce1+/+ and Rce1−/− cells on 60-mm dishes followed by overnight serum starvation (Bergo et al., 2004). The next morning, medium containing 10% serum was added to the cells. Cells were harvested at various time points after serum stimulation and total cell lysates were analyzed by immunoblotting with an antibody recognizing phosphorylated Erk1/2 (phospho-p44/42 MAP kinase E10 monoclonal, Cell Signaling Technology, Beverly, MA), and total Erk1/2 (p44/42 MAP kinase polyclonal, Cell Signaling Technology). Contrary to our expectation, we observed no effect of Rce1 deficiency on the activation of Erk1/2 (i.e., activation was no different in Rce1−/− and Rce1+/+ cells). We were concerned that intrinsic genetic differences between independent lines of immortalized Rce1−/− and Rce1+/+ fibroblasts might conceivably have prevented us from observing subtle differences in Erk1/2 activation. Accordingly, we repeated the Erk1/2 activation experiments in Rce1flx/flx cells (which have normal levels of Rce1 expression) and the Rce1-deficient Rce1α/α cells that were derived from them by Cre expression (i.e. two fibroblast cell lines that were identical except for Rce1 expression). Once again, however, we observed no differences in Rce1-expressing and Rce1-deficient cells in Erk1/2 activation, either in response to serum or epidermal growth factor (Bergo et al., 2004).
Impact of Endoproteolysis on the Binding of K-Ras to Microtubules
The laboratory of Pat Casey identified a specific and prenylation-dependent interaction between tubulin/microtubules and K-Ras, but not H-Ras or several other small GTPases (Thissen et al., 1997). In a follow-up study, they found that the interaction between K-Ras and microtubules is highly dependent both on the polylysine domain with K-Ras as well as endoproteolysis and methylation at the carboxyl terminus (Chen et al., 2000). Partially processed K-Ras that was farnesylated (but retained the –AAX residues) bound microtubules. However, surprisingly, endoproteolytic removal of the –AAX from K-Ras abolished all binding. Even more surprising, the binding of K-Ras to microtubules was restored by methylation of the C-terminal prenylcysteine. Consistent with these results, localization of the green fluorescent protein–K-Ras fusion was paclitaxel-sensitive in cells lacking Rce1, while no paclitaxel effect was observed in cells lacking the methyltransferase Icmt. These studies show that the polylysine domain is critical for the interaction between K-Ras and tubulin/microtubules and provide the first evidence for a functional consequence of CAAX-processing steps (Chen et al., 2000).
Defining the Impact of Rce1 Deficiency on Cell Growth and on Transformation by Activated Forms of Ras
To study the role of Rce1 on cell growth and Ras-transformation, we examined Rce1+/+ and Rce1−/− MEFs. We also constructed a conditional knockout allele in which the entire Rce1 coding sequence was “floxed” (flanked by loxP sites). Mice homozygous for the conditional allele (Rce1flx/flx) were viable, healthy, and fertile (Bergo et al., 2002a). Rce1 flx/flx MEFs were produced, and it was possible to completely delete the Rce1 gene from these cells by treating the cells with Cre adenovirus (thereby creating Rce1α/α MEFs). Treating Rce1 flx/flx MEFs with very small amounts of Cre adenovirus resulted in mixed cultures of Rce1flx/flx and Rce1α/α MEFs (Bergo et al., 2002a).
To define the effect of the Rce1 excision on cell growth, we examined mixed cultures of Rce1flx/flx and Rce1α/α cells by transfecting them with large amonts of a lacZ adenovirus and small amounts of Cre adenovirus (so as to delete Rce1 from only ~50% of the cells in the culture). The cells were then allowed to grow for multiple passages, and the Rce1 genotype was checked by Southern blotting at frequent intervals (Bergo et al., 2002a). If the Rce1 excision had no effect on cell growth (i.e., if there were no differences in the growth of Rce1flx/flx and Rce1α/α cells), one would expect that the Southern blots would reveal, over multiple passages, stability in the relative intensities of the 5.0-kb Rce1flx band and the 6.5-kb Rce1α band. This was not the case. In each of multiple experiments, the ratio of the 5.0-kb Rce1flx to the 6.5-kb Rce1α band increased steadily, indicating a competitive growth advantage of the Rce1flx/flx cells over Rce1α/α cells (Fig. 3). Control experiments established that the competitive growth advantage of Rce1flx/flx cells over Rce1α/α cells was due to the excision of the Rce1 gene rather than to the loss of the neo (a drug selection marker in the Rce1flx allele) (Bergo et al., 2002a).
Fig 3.
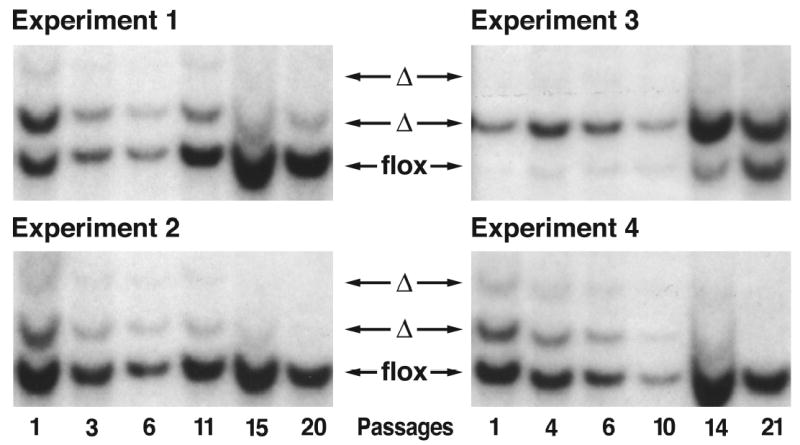
Southern blot assessment of the ratio of Rce1flx and Rce1α alleles during the growth of mixed cultures of Rce1flx/flx and Rce1α/α fibroblasts. Mixed cultures of Rce1 flx/flx and Rce1α/α cells were obtained by infecting immortalized Rce1flx/flx fibroblasts with lacZ adenovirus and Cre adenovirus. Southern blots showing the ratio of Rce1α and Rce1flx alleles at different passages in four independent experiments. These experiments show that the Rce1flx/flx cells, which express Rce1, grow more rapidly than the Rce1α/α fibroblasts, which lack Rce1 expression. Reproduced, with permission from Molecular and Cellular Biology (Bergo et al., 2002a).
Effects of an Rce1 Excision on the Growth of Ras-transfected Cells in Soft Agar and in Nude Mice
In mammalian cells, the expression of activated Ras proteins allow fibroblasts the ability to grow in soft agar (anchorage-independent growth). To test whether a Rce1 deletion would influence this phenotype, Rce1flx/flx fibroblasts were first infected with a retrovirus coding for a mutationally activated K-Ras (Bergo et al., 2002b). After selection for retroviral infection with puromycin (4 μg/ml), the overexpression of K-Ras was documented by western blotting. The Ras-transfected Rce1flx/flx fibroblasts were incubated with a lacZ adenovirus or a Cre adenovirus (to produce Ras-transfected Rce1α/α cells) (Bergo et al., 2002a). Equal numbers (3000 cells/well) of Ras-transfected Rce1flx/flx or Rce1α/α cells were mixed with medium containing 0.35% agarose and poured onto wells of 12-well plates that contained a 0.7% agarose base. The plates were incubated in a humidified culture incubator at 37°C, 7% CO2, for 14–21 d. Colonies were stained with MTT (3-[4,5-dimethylthiazol-2-yl] 2, 5-diphenyltetrazolium bromide; thiazolyl blue, Sigma, 1 mg/ml in phosphate-buffered saline). Each well was photographed with a digital camera. The images were imported into Adobe PhotoShop, and colony numbers were determined with an image-processing tool kit, volume 3.0 (http://members.aol.com/ImagProcTK/). The deletion of Rce1 resulted in a ~30% reduction in colonies in each of four independent experiments (P = 0.029; P = 0.09; P = 0.005; P = 0.008) (Bergo et al., 2002b) (Fig. 4). A ~75% reduction in transformed colonies (P < 0.001) was observed when using a second pair of Rce1flx/flx and Rce1α/α cells (not shown). In parallel, we generated H-Ras–transformed Rce1flx/flx and Rce1α/αcells, and found that deletion of Rce1 resulted in a 65% reduction in colonies (P = 0.002). We also obtained K-Ras–transformed Rce1−/− and Rce1+/+ cells by transfecting primary embryonic fibroblasts with E1A and K-Ras. We observed a ~50% reduction in numbers of colonies from Rce1−/− cells, compared with Rce1+/+ cells in three independent experiments (P = 0.003, P < 0.001, P < 0.001) (Bergo et al., 2002a).
Fig 4.
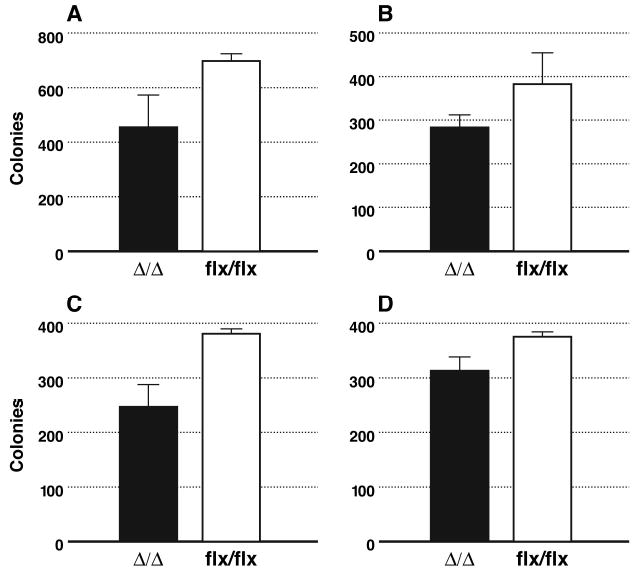
Comparison of the ability of K-Ras–transformed immortalized Rce1α/α and Rce1flx/flx fibroblasts to form colonies in soft agar. Rce1flx/flx fibroblasts were infected with a K-Ras retrovirus, then treated with either lacZ adenovirus or Cre adenovirus. In Rce1flx/flx cells treated with Cre adenovirus, the Rce1 was fully deleted, generating Rce1α/α fibroblasts. In each experiment, the excision of Rce1 resulted in fewer colonies in soft agar (P = 0.029, P = 0.005; P = 0.09; P = 0.008). Reproduced, with permission from Molecular and Cellular Biology (Bergo et al., 2002a).
The reduced ability of K-Ras–transfected Rce1α/α cells to grow in soft agar was associated with a reduced capacity to contribute to the growth of tumors in nude mice (Bergo et al., 2002a). Nude mice were injected with a mixed culture of K-Ras-transfected Rce1flx/flx and Rce1α/α cells. The Rce1flx/flx cells manifested an increased capacity to contribute to the formation of tumors relative to the Rce1α/α cells. As judged by Southern blots, the ratio of Rce1α to Rce1flx alleles in the injected cells was 1.99. The ratio increased to 11.8 ± 2.9 (mean ± SD) in the DNA prepared from established tumors, reflecting more rapid growth of Rce1flx/flx cells relative to Rce1α/α cells.
A Rce1flx/flx skin carcinoma cell line harboring a mutationally activated H-Ras was produced by treating the skin of Rce1flx/flx mice with mutagens (Bergo et al., 2002a). We treated the Rce1flx/flx cells with 108 pfu of Cre adenovirus, successfully deleting Rce1 from 90% of the cells. However, we found that the small fraction of Rce1flx/flx cells overgrew the culture within a few days in culture, strongly suggesting that Rce1flx/flx skin carcinoma cells grow more rapidly than the Rce1α/α cells.
Minimal Effects of Rce1 Deficiency on the Hematopoietic System in Mice
Initially, we considered the possibility that Rce1−/− embryos might die from defective hematopoiesis. However, this did not appear to be the case. Lethally irradiated mice were successfully rescued with hematopoietic stem cells from the livers of Rce1−/− embryos (Kim et al., 1999). Both the red and white cell counts remained stable for more than 6 months of follow-up, suggesting that there was no significant impact of Rce1 deficiency on hematopoiesis. ERK activities were measured in bone marrow collected 3 to 6 months after adoptive transfer. In multiple experiments, Rce1−/− and Rce1+/+ bone marrow demonstrated equivalent basal and GM-CSF-stimulated ERK activities.
We considered the possibility that the potential of Rce1−/− stem cells might have a relatively reduced capacity to repopulate the bone marrow when compared directly with stem cells from Rce1+/+ bone marrow. However, this was not the case (Aiyagari et al., 2003). Wild-type or Rce1−/− fetal liver cells were injected into irradiated hosts with the same reference population of BoyJ competitor cells to directly compare their potential to repopulate the bone marrow. The Rce1−/− and Rce1+/+ hematopoietic cells exhibited equivalent bone-marrow repopulating potential over a dose range that produced 10% to 70% donor cell chimerism. These data strongly suggest that inactivation of Rce1 does not impair the proliferative capacity of hematopoietic cells.
Analyzing the Functional Importance of Rce1 in the Liver and Heart
To inactivate Rce1 in the liver, Rce1flx/flx mice were given an intravenous injection of a Cre adenovirus (2 α 1011 plaque-forming units). After 3 days, livers were removed, and the levels of CAAX endoprotease activity in the tissues were determined, and the electrophoretic mobilities of the Ras proteins were analyzed (Bergo et al., 2004). Western blot analysis of the Ras proteins in the liver revealed that approximately one-half of the Ras proteins had retarded electrophoretic mobility, indicating a loss of endoproteolytic processing in a large fraction of the liver cells. Despite the loss of Rce1 in the liver, the mice remained quite healthy. Mice lacking Rce1 in the liver were also generated by breeding Rce1flx/flx mice carrying the interferon-inducible Mx1-Cre transgene, and then inducing Cre expression with intraperitoneal injections of polyinosinic-polycytidylic ribonucleic acid (pI-pC, Sigma, St. Louis MO). As judged by Southern blots, the excision of Rce1 in the livers of the pI-pC-treated mice was complete, and Rce1 activity levels were dramatically reduced. Again, despite the absence of Rce1 in the liver, the pI-pC-treated Rce1flx/flxMx1-Cre+/+ mice gained weight and exhibited normal vitality over 2–3 months of observation. During this time, transaminase levels remained normal, and the histological appearance of the liver on hematoxylin and eosin-stained sections was indistinguishable from that of wild-type mice (Bergo et al., 2004).
Mice lacking Rce1 in the heart were created by breeding Rce1flx/α mice carrying a Cre transgene driven by the α-myosin heavy chain (αMyhc) promoter (Bergo et al., 2004). The cardiac myocytes in these mice would be expected to lack Rce1 expression in cardiac myocytes. Indeed, quantitative PCR studies revealed lower Rce1 mRNA levels in the hearts of Rce1flx/ααMyhc-Cre+/o mice than in the hearts of Rce1flx/α controls (P < 0.01) (Bergo et al., 2004). Similarly, using the coupled endoprotease/methylation assay, we observed lower CAAX endoprotease activity levels in the hearts of Rce1flx/ααMyhc-Cre+/o mice than in the hearts of Rce1flx/α control mice (P < 0.01). As expected, we observed retarded electrophoretic mobility of the Ras proteins in hearts of Rce1flx/ααMyhc-Cre+/o mice, and we were able to document, with the coupled endoprotease/methylation assay, a significant accumulation of Rce1 protein substrates in the hearts of Rce1flx/ααMyhc-Cre+/o mice.
The Rce1flx/ααMyhc-Cre+/o mice and control Rce1flx/flx mice were bred and observed for 10 months. The Rce1flx/ααMyhc-Cre+/o mice appeared healthy at 1 month of age, but started dying by 3–5 months of age (Bergo et al., 2004). By 7 months of age, 50% of the Rce1flx/ααMyhc-Cre+/o mice had died; by 10 months, 70% had died. In contrast, none of the Rce1flx/flx mice or Rce1+/+ αMyhc-Cre+/o mice died during a 10-month follow-up period. Similarly, no premature deaths were observed in Rce1flx/α mice. Several weeks before their deaths, most of the heart-specific Rce1 knockout mice (Rce1+/+ αMyhc-Cre+/o) appeared listless and had ruffled fur. The hearts of Rce1+/+ αMyhc-Cre+/o mice were invariably enlarged (Fig. 5), and echocardiography showed dilated left ventricles (Bergo et al., 2004). Histological sections revealed dilatation of all four chambers of the heart, and organized thrombi were occasionally noted within the left atrium. The left ventricular musculature was thin, and there were increased amounts of collagen between myocytes. Heart tissue from Rce1flx/flx, Rce1+/+αMyhc-Cre+/o, and Rce1flx/α control mice was normal. Thus, in contrast to the situation with the liver and hematopoietic cells, Rce1 deficiency in the heart had significant adverse consequences.
Fig 5.
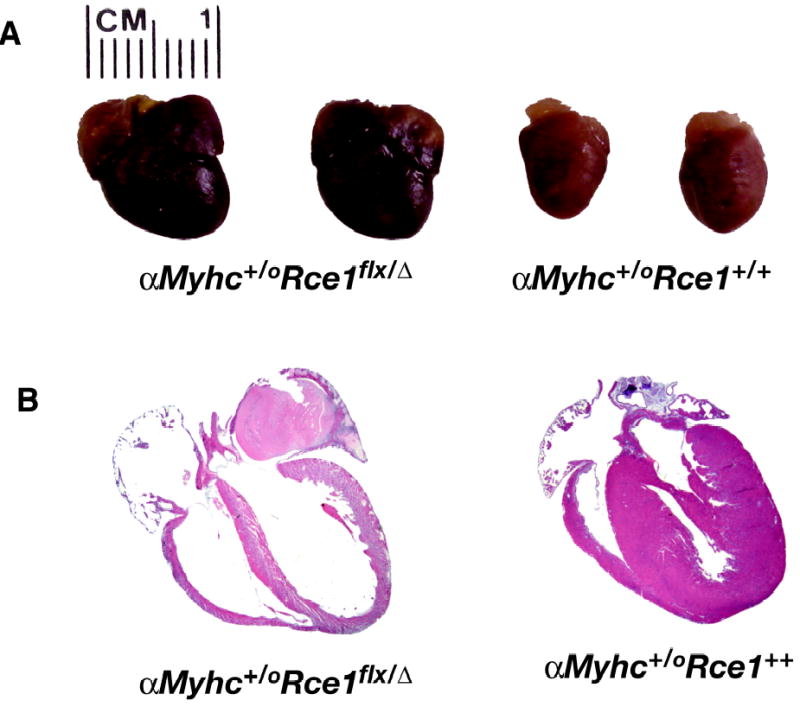
Dilated cardiomyopathy in Rce1 lx/α αMyhc-Cre+/o mice. A, Increased size of hearts from Rce1flx/α αMyhc-Cre+/o mice, compared with Rce1+/+αMyhc-Cre+/o controls. B, H&E-stained sections of an Rce1flx/α αMyhc-Cre+/o heart and an Rce1+/+αMyhc-Cre+/o heart, both from 7-month-old mice. The hearts from Rce1flx/α αMyhc-Cre+/o mice were invariably dilated. Note the organized left atrial thrombus (arrow) in the heart from the Rce1flx/α αMyhc-Cre+/o mouse. Reproduced, with permission from The Journal of Biological Chemistry (Bergo et al., 2004).
It is tempting to speculate that the effects of Rce1 deficiency in the heart are somehow related to the absence of Ras endoproteolytic processing, particularly given that an activated H-Ras transgene in mice causes myocardial hypertrophy (Gottshall et al., 1997) and the expression of a dominant-negative H-Ras causes a dilated cardiomyopathy (Dr. Seigo Izumo, personal communication; see also http://cardiogenomics.med.harvard.edu/groups/proj1/pages/ras_home.html). However, despite the attractiveness of the “Ras hypothesis,” there are clearly reasons for caution. First, we did not observe any effect of Rce1 deficiency in fibroblasts on the activation of Ras effectors by growth factors (Bergo et al., 2004). Second, Rce1 has dozens of protein substrates, and it is conceivable that the elimination of endoproteolysis of some of the non-Ras substrates underlies the pathology in the heart-specific Rce1 knockout mice. For example, the endoproteolytic processing of lamin B1 (a key structural component of the nuclear lamina) does not occur in the absence of Rce1 (Maske et al., 2003). Missense mutations in lamin A/C clearly cause dilated cardiomyopathy (Fatkin et al., 1999), so one could imagine that absent endoproteolytic processing of lamin B1 might affect heart function. One could make similar arguments about the potential involvement of many other CAAX proteins in mediating the cardiomyopathy of Rce1 deficiency.
References
- Aiyagari AL, Taylor BR, Aurora V, Young SG, Shannon KM. Hematologic effects of inactivating the Ras processing enzyme Rce1. Blood. 2003;101:2250–2252. doi: 10.1182/blood-2002-07-2250. [DOI] [PubMed] [Google Scholar]
- Ashby MN. CaaX converting enzymes. Curr Opin Lipidol. 1998;9:99–102. doi: 10.1097/00041433-199804000-00004. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Ambroziak P, Gregory C, George A, Otto JC, Kim E, Nagase H, Casey PJ, Balmain A, Young SG. Absence of the CAAX endoprotease Rce1: Effects on cell growth and transformation. Mol Cell Biol. 2002a;22:171–181. doi: 10.1128/MCB.22.1.171-181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Gavino B, Ross J, Schmidt WK, Hong C, Kendall LV, Mohr A, Meta M, Genant H, Jiang Y, et al. Zmpste24 deficiency in mice causes spontaneous bone fractures, muscle weakness, and a prelamin A processing defect. Proc Natl Acad Sci USA. 2002b;99:13049–13054. doi: 10.1073/pnas.192460799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergo MO, Leung GK, Ambroziak P, Otto JC, Casey PJ, Young SG. Targeted inactivation of the isoprenylcysteine carboxyl methyltransferase gene causes mislocalization of K-Ras in mammalian cells. J Biol Chem. 2000;275:17605–17610. doi: 10.1074/jbc.C000079200. [DOI] [PubMed] [Google Scholar]
- Bergo MO, Lieu HD, Gavino BJ, Ambroziak P, Otto JC, Casey PJ, Walker QM, Young SG. On the physiological importance of endoproteolysis of CAAX proteins: heart-specific RCE1 knockout mice develop a lethal cardiomyopathy. J Biol Chem. 2004;279:4729–4736. doi: 10.1074/jbc.M310081200. [DOI] [PubMed] [Google Scholar]
- Boyartchuk VL, Ashby MN, Rine J. Modulation of Ras and a-factor function by carboxyl-terminal proteolysis. Science. 1997;275:1796–1800. doi: 10.1126/science.275.5307.1796. [DOI] [PubMed] [Google Scholar]
- Chen Z, Otto JC, Bergo MO, Young SG, Casey PJ. The C-terminal polylysine region and methylation of K-Ras are critical for the interaction between K-Ras and microtubules. J Biol Chem. 2000;275:41251–41257. doi: 10.1074/jbc.M006687200. [DOI] [PubMed] [Google Scholar]
- Clarke S, Vogel JP, Deschenes RJ, Stock J. Posttranslational modification of the Ha-ras oncogene protein: Evidence for a third class of protein carboxyl methyltransferases. Proc Natl Acad Sci USA. 1988;85:4643–4647. doi: 10.1073/pnas.85.13.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan, D. P., Kuszczak, D., Rusinol, A. E., Thewke, D. P., Hrycyna, C. A., Michaelis, S., and Sinensky, M. S. (2004). Prelamin A endoproteolytic processing in vitro by recombinant Zmpste24. Biochem J epub ahead of print. [DOI] [PMC free article] [PubMed]
- Dai Q, Choy E, Chiu V, Romano J, Slivka SR, Steitz SA, Michaelis S, Philips MR. Mammalian prenylcysteine carboxyl methyltransferase is in the endoplasmic reticulum. J Biol Chem. 1998;273:15030–15034. doi: 10.1074/jbc.273.24.15030. [DOI] [PubMed] [Google Scholar]
- Dalton M, Sinensky M. Expression systems for nuclear lamin proteins: Farnesylation in assembly of nuclear lamina. Methods Enzymol. 1995;250:134–148. doi: 10.1016/0076-6879(95)50068-5. [DOI] [PubMed] [Google Scholar]
- Fatkin D, MacRae C, Sasaki T, Wolff MR, Porcu M, Frenneaux M, Atherton J, Vidaillet HJ, Jr, Spudich S, de Girolami U, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottshall KR, Hunter JJ, Tanaka N, Dalton N, Becker KD, Ross J, Jr, Chien KR. Ras-dependent pathways induce obstructive hypertrophy in echo-selected transgenic mice. Proc Natl Acad Sci USA. 1997;94:4710–4715. doi: 10.1073/pnas.94.9.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrycyna CA, Sapperstein SK, Clarke S, Michaelis S. The Saccharomyces cerevisiae STE14 gene encodes a methyltransferase that mediates C-terminal methylation of a-factor and Ras proteins. EMBO J. 1991;10:1699–1709. doi: 10.1002/j.1460-2075.1991.tb07694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Powers S, McGill C, Fasano O, Strathern J, Broach J, Wigler M. Genetic analysis of yeast RAS1 and RAS2 genes. Cell. 1984;37:437–445. doi: 10.1016/0092-8674(84)90374-x. [DOI] [PubMed] [Google Scholar]
- Kilic F, Dalton MB, Burrell SK, Mayer JP, Patterson SD, Sinensky M. In vitro assay and characterization of the farnesylation-dependent prelamin A endoprotease. J Biol Chem. 1997;272:5298–5304. doi: 10.1074/jbc.272.8.5298. [DOI] [PubMed] [Google Scholar]
- Kim E, Ambroziak P, Otto JC, Taylor B, Ashby M, Shannon K, Casey PJ, Young SG. Disruption of the mouse Rce1 gene results in defective Ras processing and mislocalization of Ras within cells. J Biol Chem. 1999;274:8383–8390. doi: 10.1074/jbc.274.13.8383. [DOI] [PubMed] [Google Scholar]
- Kumagai H, Kawamura Y, Yanagisawa K, Komano H. Identification of a human cDNA encoding a novel protein structurally related to the yeast membrane-associated metalloprotease, Ste24p. Biochim Biophys Acta. 1999;1426:468–474. doi: 10.1016/s0304-4165(98)00170-6. [DOI] [PubMed] [Google Scholar]
- Leung GK, Schmidt WK, Bergo MO, Gavino B, Wong DH, Tam A, Ashby MN, Michaelis S, Young SG. Biochemical studies of Zmpste24-deficient mice. J Biol Chem. 2001;276:29051–29058. doi: 10.1074/jbc.M102908200. [DOI] [PubMed] [Google Scholar]
- Maske CP, Hollinshead MS, Higbee NC, Bergo MO, Young SG, Vaux DJ. A carboxyl-terminal interaction of lamin B1 is dependent on the CAAX endoprotease Rce1 and carboxymethylation. J Cell Biol. 2003;162:1223–1232. doi: 10.1083/jcb.200303113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, Young SG, Philips M. Postprenylation CAAX Processing Is Required for Proper Localization of Ras but Not Rho GTPases. Mol Biol Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto JC, Kim E, Young SG, Casey PJ. Cloning and characterization of a mammalian prenyl protein-specific protease. J Biol Chem. 1999;274:8379–8382. doi: 10.1074/jbc.274.13.8379. [DOI] [PubMed] [Google Scholar]
- Pendás AM, Zhou Z, Cadiñanos J, Freije JMP, Wang J, Hultenby K, Astudillo A, Wernerson A, Rodríguez F, Tryggvason K, Lopéz-Otín C. Defective prelamin A processing and muscular and adipocyte alterations in Zmpste24 metalloproteinase–deficient mice. Nat Genet. 2002;31:94–99. doi: 10.1038/ng871. [DOI] [PubMed] [Google Scholar]
- Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thissen JA, Gross JM, Subramanian K, Meyer T, Casey PJ. Prenylation-dependent association of Ki-Ras with microtubules. Evidence for a role in subcellular trafficking. J Biol Chem. 1997;272:30362–30370. doi: 10.1074/jbc.272.48.30362. [DOI] [PubMed] [Google Scholar]
- Todaro, G. J. (1969). Transformation assay using cell line 3T3, In Fundamental Techniques in Virology, K. Habel, and N. P. Salzman, eds. (New York: Academic Press), pp. 220–228.
- Young, S. G., Ambroziak, P., Kim, E., and Clarke, S. (2000). Postisoprenylation protein processing: CXXX (CaaX) endoproteases and isoprenylcysteine carboxyl methyltransferase, In The Enzymes, F. Tamanoi, and D. S. Sigman, eds. (San Diego: Academic Press), pp. 155–213.
- Zhang FL, Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]


