Abstract
Background
Highly fecund mouse strains provide an ideal model to understand the factors affecting maternal performance. The QSi5 inbred strain of mice was selected for high fecundity and low inter-litter interval, and is very successful at weaning large numbers of offspring when compared to other inbred strains.
Results
Post-natal pup weight gain was used to estimate mammary gland output and to compare the performance of QSi5 mice to CBA mice. Cumulative litter weights and individual pup weight gain was significantly higher throughout the first eight days of lactation in QSi5 mice compared to CBA mice. Morphometric analysis of mammary glands during pregnancy in QSi5 mice revealed a 150 percent greater ductal side branching compared to CBA mice (P < 0.001). Ontology and pathway classification of transcript profiles from the two strains identified an enrichment of genes involved in a number of pathways, including the MAPK, tight junction, insulin signalling and Wnt signalling. Eleven of these genes, including six genes from the MAPK signalling pathway, were identified as associated with postnatal growth. Further, positive mediators of Wnt signalling, including Wnt4, Csnk2a1 and Smad4, were over-represented in the QSi5 strain profile, while negative regulators, including Dkkl1, Ppp2r1a and Nlk, were under-represented. These findings are consistent with the role of Wnt and MAPK signalling pathway in ductal morphogenesis and lobuloalveolar development suggesting enhanced activity in QSi5 mice. A similar pattern of phenotype concordance was seen amongst 12 genes from the tight junction pathway, but a pattern did not emerge from the insulin signalling genes. Amongst a group of differentially expressed imprinted genes, two maternal imprinted genes that suppress growth induced via the IGF signalling pathway, Grb10 and Igf2r, were under-represented in QSi5 mice. Whereas Peg3 and Plagl1, both paternally imprinted genes that enhance neonatal growth, were over-represented in QSi5 mice.
Conclusion
We propose that the combined action of at least three major signalling pathways involved in mammary gland development and milk secretion, namely Wnt, MAPK and tight junction pathways, contribute to the superior maternal performance phenotype in QSi5 mice. Additionally, favourable expression patterns of the imprinted genes Peg3, Plagl1, Grb10 and Igf2r may also contribute.
Background
The capacity of highly fecund mouse strains to successfully reproduce is accompanied by a concomitant increase in maternal performance at sustainable litter sizes [1]. The increase in lactation demanded by larger litters can be met by a combination of physiological strategies that allow for enhanced mammary development, increased milk output and improved milk nutritional quality. The mechanisms that underpin these strategies are complex and are under the control of multiple regulatory pathways that may each contribute at various levels. An effective way to analyse these mechanisms is to identify key genes or regulatory pathways that influence lactation phenotype. Recently, functional genomic approaches have been successfully employed to identify genes that have altered expression during mammary gland development and initiation of secretion. As a result numerous gene candidates have been implicated during different stages of the lactation cycle with some mapped to distinct signalling pathways [2-4].
A complementary approach involves the comparison of mouse strains that have divergent phenotypes, in particular strains that are representative of the high and low extremes of the variation that exists in the mouse population. The QSi5 inbred strain of mice was established from an outbred Quackenbush-Swiss strain by full-sib inbreeding and selection on the basis of increased litter size and shortened inter-litter interval [5]. The strain has an average litter size greater than 13 pups, and females commonly nurse up to 18 pups with greater than 90% survival to weaning. Along with an increased body weight (BW), these traits are indicative of enhanced lactation capacity [6]. Indeed lactation performance, assessed by a weigh-suckle-weigh method, was 3-fold greater in QSi5 mice than in the CBA strain [1].
In this study, we utilised the divergent phenotypes of QSi5 and CBA/CaH (hereafter referred as CBA) mice to identify genes and associated pathways that correspond to enhanced mammary gland capacity. We identified a significant enrichment among the differentially expressed genes in the Wnt and MAPK signalling pathways, both of which are implicated as effectors of ductal side-branching during mammary gland development [2,7-9], and a finding that is consistent with anatomical differences found in the two strains. We also identified favourable expression patterns of genes in the tight junction pathway, and four imprinted genes that influence maternal performance in mice. We propose that the action of these signalling pathways and the effects of gene imprinting contribute to the superior maternal performance in the QSi5 strain.
Results
Maternal Performance
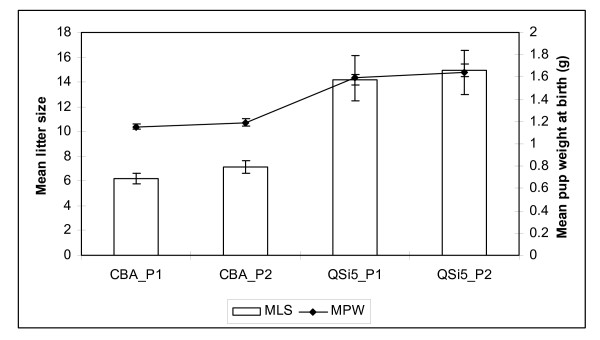
We compared a highly fecund QSi5 inbred strain of mice to a non-selected CBA strain of mice raised on a similar genetic background for reproductive performance. The number of pups born alive (NBA) in the first and second parities respectively for QSi5 mice was 14.2 and 14.9 pups/litter, whereas for the CBA strain it was 6.2 and 7.2 pups/litter (Figure 1). Thus significant differences in fecundity exist between QSi5 and CBA strains of mice both in first and second parities (P < 0.001).
Figure 1.
Comparison of litter size and pup weight at birth in the two inbred strains. oth number of pups born alive and mean pup weight at birth at the first two parities in CBA (n = 20) and QSi5 (n = 20) were significantly higher in the QSi5 strain of mice (p < 0.0001). Note: all error bars are + SEM.
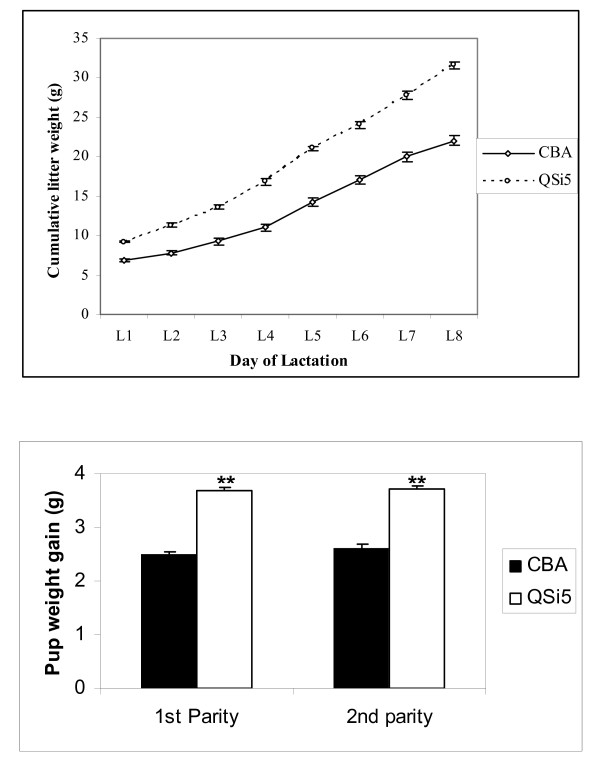
To determine whether the increased maternal performance of QSi5 mice was independent of litter size we adjusted litter sizes on day of parturition for both the QSi5 and CBA strains, and calculated cumulative weights for a standardized six pup litter. On this basis, significant strain-specific differences in maternal performance (P < 0.001) were observed throughout the first eight days of lactation based on the cumulative litter weight (Figure 2a). Additionally, maternal performance assessed by individual pup growth rate was 150% greater (P < 0.001) in QSi5 mice when compared to CBA mice (Figure 2b). Further, there was no significant difference in pup growth rate between first and second parities within each strains. These results confirm that the superior maternal performance of the QSi5 mice was independent of the large litter size phenotype, and that significant differences in maternal performance exist between the QSi5 and CBA inbred strains of mice.
Figure 2.
Assessment of maternal performance in two inbred mice strains. (a) Cumulative litter weights in two strains – Cumulative litter weights measured using 6 pups/dam during the first 8 days of lactation in CBA (n = 16) and QSi5 (n = 20) strains; Weights are reported as Mean ± SEM. Lactation performance was significantly greater in QSi5 dams compared to CBA. (b) Individual pup weight gain at 8 days – Mean pup weight gain at the first 2 parities in CBA (n = 16) and QSi5 (n = 20) strains of mice. Pup weight gain was significantly higher in QSi5 strain relative to CBA strain of mice in both parities.
Morphometric analysis
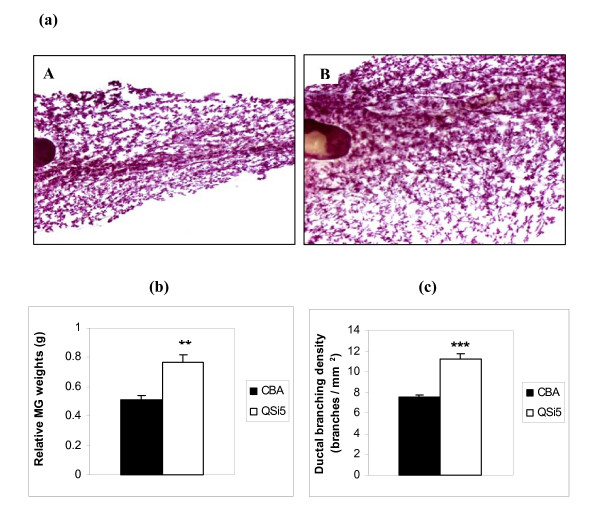
To ascertain whether the strain-specific differences in maternal performance can be attributed to anatomical differences in the mammary gland during development, morphometric analysis of mammary whole mounts from mice at pregnancy day 12 (Figure 3a). When adjusted for body weight, a significant difference in relative mammary gland weight existed between the two strains (P < 0.001), calculated as 150% higher in the QSi5 strain relative to the CBA strain of mice (Figure 3b). Quantitation of ductal density in mammary gland whole mounts using image analysis software (NIH-ImageJ) revealed a significant difference in the proportion of ductal branching between the two strains of mice (P < 0.001) (Figure 3c). Ductal branching was approximately 150% higher in the QSi5 strain relative to the CBA strain. Similarly, relative quantitation of the terminal end buds (TEB) as an estimate of alveolar epithelial density showed a 140% increase in QSi5 mice compared to the CBA strain of mice (P < 0.0001). Thus, the QSi5 mice had both a greater mammary tissue mass and enhanced ductal side-branching; both consistent with a superior lactation phenotype.
Figure 3.
Histological analysis of mammary glands from pregnant mice. (a) Whole mounts of the inguinal mammary gland from day 12 pregnant mice A. CBA strain and B. QSi5 strain. (b) Comparison of relative mammary gland weights of the two different strains (n = 5 each). (c) Quantitation of ductal branching Both relative mammary gland weights (p < 0.001) and density of ductal branching (p < 0.001) were significantly higher in the QSi5 strain compared to CBA strain of mice.
Mammary gene expression profiling
Gene expression profiling of mammary gland tissue is an effective approach to identifying pathways that are active during mammary gland development and lactation. To identify the genes associated with the enhanced maternal performance phenotype of the QSi5 strain, gene expression profiles of mammary gland tissue from five animals each of lactating QSi5 and CBA mice were compared. Lactation day 9 was chosen for quantitative measurement because neonatal growth is a better correlate of lactation performance during the plateau phase of milk production in mice, that is at 6–10 days post-partum [6]. Further, measurements on day 9 were comparable to other lactation cycle studies performed in mice [10].
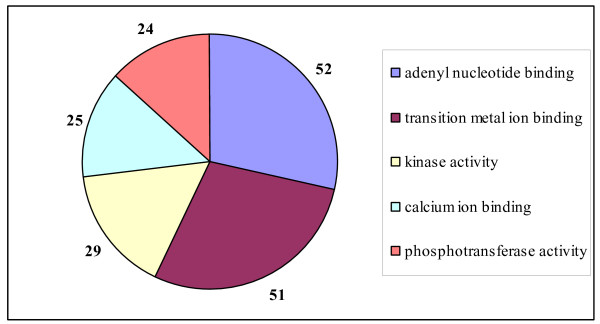
We identified 740 probesets (640 genes) and 331 probesets (282 genes) using a permutation based FDR of 8.2%, and BH FDR of 5% respectively, as being differentially expressed between the two strains of mice. Gene ontology classification of the differentially expressed genes based on molecular function identified the dominant categories, including calcium ion-binding and kinase activity, to be significantly over-represented among the differentially expressed genes (Figure 4). The genes identified in the calcium ion-binding category, included some well-characterised genes associated with lactation, such as lactalbumin (Lalba), Egf, phospholipase C, epsilon 1 (Plce1), annexins in addition to Phospholipid scramblase 1 (Plscr1), phospholipase A2, group XIIA (Pla2g12a) and nucleobindins (Nucb1, Nucb2). Similarly, a number of known kinases associated with mammary gland development, including Nemo like kinase (Nlk), casein kinase (Csnk1a1), ribosomal protein S6 kinase, polypeptide 1 (Rps6kb1), and proviral integration site 1 & 2 kinases (Pim1/2). Importantly, positive mediators of mammary gland development such as Lalba, Egf, Csnk1a1, Rps6kb1, Pim1 all had higher levels of expression in the QSi5 mice relative to the CBA mice.
Figure 4.
Significant GO categories classified by DAVID. Pie chart showing the significantly enriched molecular function categories (Level 5) based on gene ontological classification of the differentially expressed genes (p < 0.01) between the two strains of mice using DAVID.
Gene ontology analysis of the differentially expressed genes based on the biological process category did not reveal any significant enrichment of carbohydrate and lipid metabolism. Subsequently, analysis of 37 genes corresponding to enzymes known to be involved in these metabolic processes [10] identified Agpat1 as the only gene to be differentially expressed.
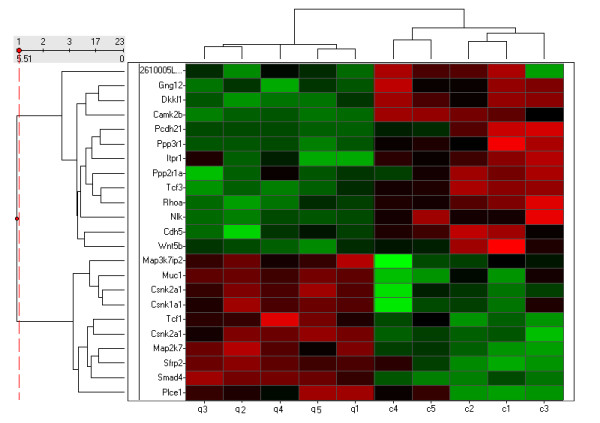
Pathway classification of the differentially expressed genes using the Onto-Express pathway analysis tool identified 16 genes in the MAPK signalling pathway, 12 genes in the tight junction signalling pathway and 10 genes each from the Wnt signalling pathway, cell adhesion molecules and insulin signalling pathways, as enriched among the differentially expressed genes (Table 1 and Additional file 1). Similar pathway analysis results were obtained when a group of 282 differentially expressed genes selected below a 5% FDR cut-off were analysed, and additionally included adherens, junction and TGF-beta signalling pathways. We also examined whether the differentially expressed genes in these pathways were associated with any of the mammary gland associated phenotypes found in the mouse genome informatics (MGI) database. Ten of the 48 genes in these pathways were associated with postnatal growth retardation or postnatal lethality, which is substantially influenced by maternal/lactation performance. This included seven genes, namely Egf, Cdc42, Nlk, Jund1, Kras, Casp3 and Map3k12, from the MAPK signalling pathway, a pathway that plays a critical role in mammary gland development via the EGFR response. Additionally, two genes that are associated with abnormal mammary gland development when deleted, Egf and Neo1, were both over-expressed in the QSi5 mice. Besides the ten unique Wnt signalling pathway genes identified in the above pathway analysis, five other genes closely associated with the canonical Wnt/β-catenin/TCF pathway, namely Dkkl1, Muc1, Rps6kb1, Sdc 2 and Sdc 4, were also differentially expressed between the two mouse strains. The Wnt pathway plays a significant role in initiation of mammary gland morphogenesis. Wnts are secreted glycoproteins which stabilize β-catenin by preventing its degradation by the PP2A-GSK3β-Axin complex, resulting in translocation of β-catenin to the nucleus where it activates downstream target genes. Further, all positive mediators of the canonical Wnt-β-catenin signalling pathway in this group, such as Csnk1a1, Csnk2a1, Smad 4, were over-represented, while negative regulators, including Ppp2r1a, Cdh5 and Nlk, were under-represented in the QSi5 mice relative to CBA mice (Figure 5).
Table 1.
Enriched biological pathways among differentially expressed genes. Table showing the most enriched biological pathways (p < 0.1) based on pathway classification of differentially expressed genes (p < 0.01) between the two strains of mice using Onto-Express Pathway analysis tool. Rank, impact factor, p-values and the number of input genes associated with significant pathways are listed.
| Rank | Pathway Name | Impact Factor | # Input Genes in Pathway | p-value |
| 2 | Tight junction | 5.772 | 12 | 0.00 |
| 7 | Insulin signaling pathway | 3.197 | 10 | 0.04 |
| 9 | MAPK signaling pathway | 3.022 | 16 | 0.05 |
| 10 | Cell adhesion molecules (CAMs) | 3.002 | 10 | 0.05 |
| 14 | Wnt signaling pathway | 2.371 | 10 | 0.09 |
Figure 5.
Hierarchical clustering and heat map of differentially expressed Wnt signalling genes. Clustering was performed on all ten transcript profiled samples (QSi5 – 1st 5 vertical columns and CBA – last 5 vertical columns) for ten genes up-regulated in QSi5 (bottom ten horizontal rows) and 15 down-regulated genes (top 15 horizontal rows) relative to the CBA strain of mice. Expression levels are colour coded with shades of red, green and black corresponding to an increase, decrease and no change in gene expression, respectively.
Amongst the 12 differentially expressed genes in the significantly enriched tight junction signalling pathway, seven and five genes were over- and under-represented respectively, in the QSi5 mice relative to CBA mice. Genes that play an important role in tight junction stability, including Tjp2, Csnk2a1, Pard6g (Par6a), Llgl1 (Mgl1), were all over-represented in the high lactation performance QSi5 mice, whereas genes associated with tight junction permeability, namely Ppp2r1a (PP2A) and Rhoa, were over-represented in the low performance CBA mice. Thus the decreased tight junction permeability of the QSi5 mice may contribute to enhanced milk secretion. Also, amongst seven strain-specific differentially expressed genes in the TGF-β pathway,Tgfb2 and Rhoa, both negative regulators of mammary gland development, were over-represented in the low performance CBA mice relative to the QSi5 mice. However, despite over-representation in the gene ontology analysis, when genes belonging to the insulin pathway were examined there was insufficient evidence to conclude that a pattern of expression between the two strains reflected lactation performance.
Imprinted genes play a crucial role in facilitating transfer of nutrients from the mother to the offspring. Analysis of the differentially expressed genes identified four imprinted genes (Additional file 2). The paternally expressed genes, Peg3 and Plagl1, that are both growth promoting, were over-represented in the QSi5 strain relative to the CBA strain of mice. Conversely, two maternally expressed genes that are pup-growth limiting, Grb10 and Igf2r, were over-represented in the CBA mice.
The expression patterns of 17 milk protein genes previously identified as being differentially expressed in the mammary gland during the lactation cycle were also examined [10]. Five of these genes, namely Xdh, Muc1, Lalba, Egf, and Csn1s2a, were differentially expressed between the two strains and all were over-represented in the high lactation performance QSi5 mice. One other pathway known to be associated with mammary development and lactation, the prolactin receptor pathway was also examined. Key prolactin receptor-Stat5 signalling pathway encoding genes, such as Sta5a, Stat5b, Prl, Prlr, Jak1, Jak2, or Socs1, were not differentially expressed between the two strains. However, amongst the 25 Stat5 target genes identified in mammary epithelial cells [11], three genes, Socs2, Pim1 and Grb10, were differentially expressed. Of these Grb10 was over-represented in the CBA mice whereas the other two genes were over-represented in QSi5 mice.
Thus, differential expression of components of the tight junction, Wnt and MAPK signalling pathways together with favourable genomic imprinting may contribute to the increased maternal performance phenotype of the QSi5 strain of mice.
Quantitative RT-PCR
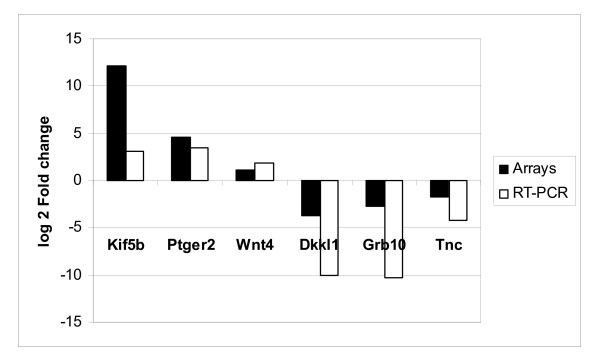
Quantitative RT-PCR (qRT-PCR) was used to evaluate the expression profiles of eight genes, including five genes identified as differentially expressed between the two strains on by microarray. In addition, Wnt4 was selected for validation as it was identified as differentially expressed based on the analysis of a subset of the data, and both c-myc and CyclinD1 (Ccnd1) were examined as potential target genes of Wnt signalling pathways. A significant degree of concordance, approximately 74% (p < 0.05) was observed between the two methods based on the expression patterns for all genes examined (Table 2). Six of the eight genes examined, namely Kif5b, Ptger2, Grb10, Tnc and Dkkl1, were differentially expressed between the two strains. Differential expression was not detected for the remaining two genes, c-myc and Ccnd1, but these genes are tightly regulated and are perhaps transiently expressed during stages of the cell cycle (Figure 6). Thus, the results of the qRT-PCR analysis generally supported those of the microarray data analysis.
Table 2.
Quantitative RT-PCR validation of 8 selected genes.
| Gene Symbol | Microarrays | Real-time PCR | ||
| FC | P-value | FC | P-value | |
| Kif5b | 12.17 | < 0.001 | 3.14 | 0.005 |
| Ptger2 | 4.58 | < 0.001 | 3.42 | 0.01 |
| Dkkl1 | -3.76 | < 0.001 | -10.01 | 0.006 |
| Grb10 | -2.68 | < 0.001 | -10.32 | 0.09 |
| Tnc | -1.76 | 0.001 | -4.16 | 0.06 |
| Wnt4 | 1.13 | 0.229 | 1.82 | 0.047 |
| cyclin D1 | 1.08 | 0.66 | 1.95 | 0.12 |
| cmyc | 0.85 | 0.08 | 1.02 | 0.97 |
Comparison between array experiment and real-time PCR for 8 selected genes is shown. Log-fold change in real-time PCR was calculated from the difference in normalised expression between the CBA and QSi5 strains of mice. P-value was calculated from 2-tailed Student's t-test.
Figure 6.
Comparison of fold change differences in microarrays and qRT-PCR for selected genes. Quantitative RT-PCR validation of the microarray results showing the observed fold change differences in both methods for three up regulated genes in QSi5 strain (Kif5b, Ptger2, Wnt4) and three down regulated genes (Dkkl1, Grb10, Tnc).
Discussion
Highly fecund mice provide an opportunity to investigate the factors that control maternal performance. Comparison of the two phenotypically divergent inbred mouse strains, QSi5 and CBA, identified genes associated with maternal performance. Histological analysis of mammary whole mounts during mid-pregnancy revealed that QSi5 mice have increased ductal branching relative to the CBA strain of mice. Strain-specific differences in mammary gland architecture have been recognised for many years and may be regarded as a correlate of reproductive capacity. The differences may be due to epithelial factors [12,13], stromal factors [14], or hormonal factors [15]. Mammary gland recombination and transplantation experiments using mice from strains with highly branched architecture, the 129 and C57Bl/6J strains, which have a much lower level of side-branching, revealed that stromal factors were a likely source of variation [16], but differential expression of stromal genes has been difficult to demonstrate [17]. In the present study, a comparative analysis of gene expression in QSi5 and CBA mice identified candidate genes in a number of pathways, including tight junction, MAPK and Wnt signalling pathways, as contributors to the superior maternal performance phenotype of the QSi5 strain. Additionally, four imprinted genes that have a role in neonatal growth and mammary development, were identified as candidate genes that may also contribute to the phenotype.
Transcript profiling of mammary gland tissue from the two divergent strains during peak lactation revealed that Wnt signalling pathway genes were enriched within the differentially expressed genes. Wnt (wingless) genes are a family of secreted glycoproteins which commonly act through the canonical Wnt/β-catenin/TCF signal transduction pathway both in embryonic and postnatal mammary gland development [9,18]. Wnt genes, especially Wnt4, Wnt5b and Wnt6, are progesterone responsive and are expressed in a temporal pattern during pregnancy and lactation [19]. Wnt4 over-expression in virgin mice results in excessive ductal branching [20], while gene deletion reduces ductal side branching in mammary glands at pregnancy day 12 [21-23]. There is also evidence that different Wnt genes switch on or off at different stages of the lactation cycle [24]. The primary mode of action of the canonical Wnt signalling pathway is the stabilisation of β-catenin [25], which is necessary for normal lobular development of the mammary gland [8]. Csnk1a1 and Csnk2a1, which were over-represented in QSi5 mice gene expression profiles, are positive mediators of the pathway, while a lower level of expression was seen for Ppp2r1a, which has a central role in the β-catenin degradation complex. [26-28]. Thus, stabilization of cytoplasmic β-catenin may provide a mechanism to enhance lobuloalveolar development in the QSi5 strain of mice.
Postnatal growth of pups during the first two weeks is a key indicator of the lactation capacity of the dam. More than ten genes associated with this trait were identified amongst the enriched pathways. This included six genes of the MAPK pathway, namely Egf, Cdc42, Nlk, Jund1, Kras and Map3k12, which is activated during mammary gland development through the Egf receptor. Exogenous administration of Egfr ligands reinitiated ductal outgrowth during puberty in both ovariectomized mice [29] and ER-α knock out mice [30]. A role for the Egfr pathway in ductal development has been demonstrated by the phenotype observed in mice that have a functionally defective pathway [31,32]. There is also evidence that Wnts activate Egfr in murine mammary epithelial cells [33,34]. This is consistent with the findings of the present study, in which a relative enrichment of genes involved in both MAPK and Wnt signalling pathways was observed in QSi5 mice.
Decreased tight junction permeability results in enhanced milk secretion [35]. The significant enrichment of genes from the tight junction signalling pathway amongst those differentially expressed in the present study suggests that the pathway may play a role in the lactation performance differences between the two strains of mice. Positive mediators of the pathway include Tjp2 and Csnk2a,1, are involved inclaudin polymerisation and tight junction formation [36], and were over-represented in the QSi5 mice. Corresponding to this was a relative decreased expression of the negative mediators of this pathway, namely Rhoa and Ppp2r1a. Inhibition of Rhoa is required for tight junction formation in rat mammary epithelial cells [37]. Thus the potential for decreased tight junction permeability in QSi5 mice may contribute to enhanced milk secretion.
Imprinted genes play a critical role both in prenatal and postnatal growth, the latter being mainly influenced by the maternal performance of the dam. According to the "parent-offspring conflict hypothesis" paternally expressed imprinted genes favour the growth of its offspring and facilitates maximum transfer of nutrients from the mother, whereas maternally expressed imprinted genes inhibit growth and favour equal allocation of resources [38]. This hypothesis holds true even for the postnatal growth of offspring prior to weaning for most of the imprinted genes that have been identified in mammals [39]. In the present study, both the paternally-expressed imprinted genes, Peg3 and Plagl1, were over-represented in the superior performing QSi5 strain of mice whereas both the maternally-expressed imprinted genes, Grb10 and Igf2r, were under-represented. Previously it has been reported that pups raised on Peg3 deficient mice have reduced growth rate compared to their wild-type litter mates due to poor maternal care and impaired milk release [40]. Peg3 deficient offspring fostered on wild-type mothers had reduced birth weight and also had inferior growth rates compared to wild-type offspring [41]. Plagl1 deficient mice also produced offspring with reduced birth weight, and postnatal mortality was high, albeit difficult to interpret because of delayed lung maturity [42].
Both the maternally expressed imprinted genes Grb10 and Igf2r inhibit post-natal growth by blocking mitogenic effects of Igf1 and Igf2 [43-45]. Grb10 negatively regulates Igf1 [46] whereas Igf2r over-expression results in decreased cell growth and proliferation due to increased clearance of Igf2 levels as well as reduced phosphorylation of Igf1 receptor [47,48]. Over-expression of Igf1 results in prolonged lactation [49], while deletion of both Igf1 and Igf1r results in reduced ductal side branching and fewer TEB, respectively [50-52]. Igf2 has been shown to mediate prolactin induced alveolar development in the mammary gland and acts upstream of cyclin D1 during mid-pregnancy [53,54]. Hence, a relatively lower level of Grb10 and Igf2r gene expression is consistent enhanced maternal performance.
Conclusion
To summarize, differential expression patterns of candidate genes in a number of pathways, including tight junction, MAPK and Wnt signalling pathways were associated with enhanced maternal performance of QSi5 mice relative to CBA mice. MAPK and Wnt pathways are both known regulators of ductal morphogenesis and alveolar development, while tight junction permeability is related to milk secretion,. Additionally, favourable expression patterns of imprinted genes in QSi5 mice may also contribute to a superior maternal performance phenotype.
Methods
Animals
Mice from QSi5 and CBA/CaH inbred strains were housed at the animal housing facility, Faculty of Veterinary Science, University of Sydney, and were maintained as per accepted guidelines. The room was maintained at a constant temperature of 21°C and the mice were exposed to a daily photoperiod of 12 hours (0600–1800). Mice had free access to water and a pellet diet (Rat and mouse cubes; Specialty Feeds, Glen Forrest WA 6071, Australia). 20 nulliparous QSi5 females and 19 nulliparous CBA females were mated to QSi5 and CBA males, respectively to assess lactation performance by neonatal pup growth method over the first two parities. Five nulliparous females of the QSi5 and CBA strains were similarly mated for transcript profiling of the mammary gland at day 9 of lactation, and five nulliparous females of each of the same strains were mated for histological analysis of mammary gland at day 12 of pregnancy.
Assessment of maternal performance
Females of breeding age from each strain of mice were pair-mated at eight weeks of age and observed for successful mating for three consecutive days. The appearance of the vaginal plug was recorded as day 0 of pregnancy and the day of parturition as day one of lactation. At the first day of lactation, the number of pups born alive (NBA) were recorded and immediately litter sizes were adjusted. Both maternal and cumulative litter weights were recorded for the first eight days of lactation between 1000–1100 hours each day [55]. Statistical differences in cumulative litter weight gain between the two strains across lactation was estimated using a similar litter sizes of 6 pups/dam for both strains. Maternal performance in both first and second parities was assessed by estimating the individual pup weight gain between day 1 and day 8 of lactation. Statistical significance between the two strains was estimated by a Student's unpaired t-test.
Histology and Morphometric Analysis
Left inguinal mammary glands harvested from day 12 pregnant mice were weighed and immediately used for preparing whole mounts. Mammary whole-mounts were prepared by spreading the mammary glands on a glass slide and fixing it in 10% formalin solution. Glands were treated in acetone before carmine alum (0.2% carmine, 0.5% aluminium sulfate) staining overnight. The whole-mount was dehydrated by using a graded ethanol series followed by xylene treatment for 60 min and storage in methyl salicylate [20]. Relative mammary gland weights for each animal were calculated by dividing the actual wet mammary gland weights by the maternal body weight. Quantitation of ductal side branching was performed by measuring the density of ductal branching using the NIH-ImageJ image analysis tool [56]. Similarly, TEB were counted as an estimate of alveolar density. Five representative fields of view from each gland were counted. Statistical differences between the two strains for relative mammary gland weights, ductal branching density and epithelial density were then estimated by a Student's unpaired t-test.
Transcript Profiling
The inguinal mammary glands (right) harvested from a total of ten mice, five from each strain, at lactation day 9 and were subsequently used for mRNA isolation. Upon removal, the gland was immediately snap-frozen in liquid nitrogen before storing at -80°C until further use. Total RNA was extracted from 20 mg of mammary tissue using Qiagen RNeasy mini kit (Qiagen, VIC, Australia) according to the manufacturer's instructions. RNA quality, concentration and integrity of samples were ascertained using Agilent 2100 Bioanalyser (Agilent Technologies, Foster City, CA). The cRNA targets were generated as recommended by Affymetrix (Santa Clara, USA) and hybridized to MOE 430A 2.0 GeneChips (Affymetrix). The GeneChips were subsequently scanned in GeneChip Scanner 3000 and finally the probe intensities were obtained using GCOS operating software (Affymetrix). Quantile normalization and calculation of probe-set intensity was performed using the robust-multi array average (RMA) function available from the "affy" package in R [57]. Both the normalized data and the raw "cel" files from this experiment are available at the NCBI GEO website under the series record GSE9668. Data filtering removed low intensity probesets close to background noise by selecting probesets having an un-logged intensity of over 20 in at least one of the conditions. Differential expression was assessed by ranked penalised t-statistics on the logged data using lm.series and e-bayes function in the "limma" package in R [58]. P-values were adjusted for multiple testing by the Benjamini and Hochberg (BH) method using the "multest" package in R. Genes with an adjusted P-value less than 0.05 can be interpreted as having a false discovery rate of 5%. Alternately, when genes were classified as differentially expressed using a less stringent unadjusted p < 0.01 threshold, FDR was calculated by a permutation based method. Differentially expressed genes were classified according to the Gene Ontology functional category based on molecular function using DAVID (The Database for Annotation, Visualization and Integrated Discovery) [59].
Pathway and mammalian phenotype ontology analysis
Pathway classification of the differentially expressed genes was performed using the Pathway Express tool available in Onto-Express [60]. Enriched pathways among the differentially expressed genes below a p-value of 0.1 were identified and those associated with mammary gland development are listed in Table 1. Hierarchical clustering of 25 differentially expressed Wnt signalling pathway genes and five replicate samples from each strain was performed using an Euclidean distance metric with average linkage (Spotfire DecisionSite 8.0) (Figure 5). Differentially expressed genes within these pathways were subsequently investigated for any mammalian phenotypes associated with maternal performance, such as post-natal growth retardation, post-natal lethality, mammary gland development and maternal/paternal imprinting by data mining the Mouse Genome informatics (MGI) database [61].
Quantitative RT-PCR
First strand cDNA synthesis was performed using MMLV reverse transcriptase (Invitrogen, VIC, Australia) according to manufacturer's instructions. PCR primers for all eight target genes used for validation and one housekeeping gene Cyclophilin A (CypA) were designed based on 100% homology to the RefSeq as well as mismatch to other genes. PCR primer (Sigma Genosys, Castle Hill, NSW, Australia) sequences and the product sizes are listed in Table 3. Quantitative PCR was performed using a master mix containing Platinum Taq DNA Polymerase (Invitrogen, VIC, Australia), 2.5 pmol of each primer and Sybr Green I (Molecular Probes, Eugene, OR, USA) using a Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science, NSW, Australia). Each reaction was performed in duplicate using a minimum of four samples from each strain and using cDNA from at least two separate cDNA synthesis reactions. Relative quantitation of the PCR products was performed by normalizing to an internal control, CypA. Statistical significance was determined by using Student's unpaired t-test and the observed fold-change differences between the two strains are reported.
Table 3.
Primer sequences used in real-time PCR. Table showing the forward (F) and reverse (R) primer sequences for 8 target genes and one housekeeping gene used in real-time PCR along with the amplicon sizes.
| Name | Primer sequence | Product size (bp) |
| Kif5b F | GCGGAGTGCAACATCAAAGTG | 127 |
| Kif5b R | CATAAGGCTTGGACGCGATCA | |
| Ptger2 F | ATCACCTTCGCCATATGCTC | 152 |
| Ptger2 R | GGTGGCCTAAGTATGGCAAA | |
| Grb10 F | GCCAAGCATGATGTCAAAGTCT | 183 |
| Grb10 R | GTCCTCCAGGCACCTCTCTA | |
| Dkkl1 F | ACTGAGGGTCTTGCTGCTGCT | 218 |
| Dkkl1 R | GGAAGTTCCTAGGAAGGTCTC | |
| Tnc F | ACGGCTACCACAGAAGCTG | 247 |
| Tnc R | CGCGGCTTATTCCATAGAGTTC | |
| Wnt4 F | CGAGGAGTGCCAATACCAGT | 138 |
| Wnt4 R | GCCACACCTGCTGAAGAGAT | |
| C-myc F | CACCAGCAGCGACTCTGAA | 99 |
| C-myc R | CCCGACTCCGACCTCTTG | |
| Ccnd1 F | CATCAAGTGTGACCCGGACTG | 116 |
| Ccnd1 R | CCTCCTCCTCAGTGGCCTTG | |
| CypA F | GAGCTGTTTGCAGACAAAGTTC | 125 |
| CypA R | CCCTGGCACATGAATCCTGG |
Authors' contributions
PR, IM and PW designed and implemented the study. PW and RT supervised the study and prepared the manuscript with PR. MGG and PT contributed expertise and guided PR's analysis of microarray data. CJO and CM provided resources to enable the study. All authors had direct input to the final version of the manuscript.
Supplementary Material
Gene lists of enriched biological pathway categories. Strain-specific differentially expressed genes between the QSi5 and CBA strains of mice belonging to the five enriched biological pathway categories are listed along with their P-values and fold changes.
Differentially expressed imprinted genes. Strain-specific differentially expressed imprinted genes between the QSi5 and CBA strains of mice are listed along with their P-values and fold changes.
Acknowledgments
Acknowledgements
We thank Karen Barnes for help in preparing mammary whole mounts. PR was a recipient of a F.H Loxton Postgraduate Research Scholarship.
Contributor Information
Palaniappan Ramanathan, Email: palaniap@vetsci.usyd.edu.au.
Ian C Martin, Email: ianm@vetsci.usyd.edu.au.
Margaret Gardiner-Garden, Email: margar@gimr.garvan.unsw.edu.au.
Peter C Thomson, Email: petert@camden.usyd.edu.au.
Rosanne M Taylor, Email: rosannet@vetsci.usyd.edu.au.
Christopher J Ormandy, Email: c.ormandy@garvan.org.au.
Christopher Moran, Email: chrism@vetsci.usyd.edu.au.
Peter Williamson, Email: peter_williamson@usyd.edu.au.
References
- Riley LG, Zubair M, Thomson PC, Holt M, Xavier SP, Wynn PC, Sheehy PA. Lactational performance of Quackenbush Swiss line 5 mice. J Anim Sci. 2006;84:2118–2125. doi: 10.2527/jas.2005-609. [DOI] [PubMed] [Google Scholar]
- Hennighausen L, Robinson GW. Information networks in the mammary gland. Nat Rev Mol Cell Biol. 2005;6:715–725. doi: 10.1038/nrm1714. [DOI] [PubMed] [Google Scholar]
- Naylor MJ, Oakes SR, Gardiner-Garden M, Harris J, Blazek K, Ho TWC, Li FC, Wynick D, Walker AM, Ormandy CJ. Transcriptional Changes Underlying the Secretory Activation Phase of Mammary Gland Development. Mol Endocrinol. 2005;19:1868–1883. doi: 10.1210/me.2004-0254. [DOI] [PubMed] [Google Scholar]
- Rudolph MC, McManaman J, Phang T, Russell T, Kominsky DJ, Serkova NJ, Stein T, Anderson SM, Neville MC. Metabolic regulation in the lactating mammary gland: A lipid synthesizing machine. Physiol Genomics. 2006;14:14. doi: 10.1152/physiolgenomics.00020.2006. [DOI] [PubMed] [Google Scholar]
- Holt M, Nicholas FW, James JW, Moran C, Martin IC. Development of a highly fecund inbred strain of mice. Mamm Genome. 2004;15:951–959. doi: 10.1007/s00335-004-3030-8. [DOI] [PubMed] [Google Scholar]
- Knight CH, Maltz E, Docherty AH. Milk yield and composition in mice: effects of litter size and lactation number. Comp Biochem Physiol A. 1986;84:127–133. doi: 10.1016/0300-9629(86)90054-X. [DOI] [PubMed] [Google Scholar]
- Kouros-Mehr H, Werb Z. Candidate regulators of mammary branching morphogenesis identified by genome-wide transcript analysis. Dev Dyn. 2006;235:3404–3412. doi: 10.1002/dvdy.20978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepera SB, McCrea PD, Rosen JM. A beta-catenin survival signal is required for normal lobular development in the mammary gland. J Cell Sci. 2003;116:1137–1149. doi: 10.1242/jcs.00334. [DOI] [PubMed] [Google Scholar]
- Chu EY, Hens J, Andl T, Kairo A, Yamaguchi TP, Brisken C, Glick A, Wysolmerski JJ, Millar SE. Canonical WNT signaling promotes mammary placode development and is essential for initiation of mammary gland morphogenesis. Development. 2004;131:4819–4829. doi: 10.1242/dev.01347. [DOI] [PubMed] [Google Scholar]
- Rudolph MC, McManaman JL, Hunter L, Phang T, Neville MC. Functional development of the mammary gland: use of expression profiling and trajectory clustering to reveal changes in gene expression during pregnancy, lactation, and involution. J Mammary Gland Biol Neoplasia. 2003;8:287–307. doi: 10.1023/B:JOMG.0000010030.73983.57. [DOI] [PubMed] [Google Scholar]
- Clarkson RW, Boland MP, Kritikou EA, Lee JM, Freeman TC, Tiffen PG, Watson CJ. The genes induced by signal transducer and activators of transcription (STAT)3 and STAT5 in mammary epithelial cells define the roles of these STATs in mammary development. Mol Endocrinol. 2006;20:675–85. Epub 2005 Nov 17.. doi: 10.1210/me.2005-0392. [DOI] [PubMed] [Google Scholar]
- Lydon JP, DeMayo FJ, Funk CR, Mani SK, Hughes AR, Montgomery CA, Jr., Shyamala G, Conneely OM, O'Malley BW. Mice lacking progesterone receptor exhibit pleiotropic reproductive abnormalities. Genes Dev. 1995;9:2266–2278. doi: 10.1101/gad.9.18.2266. [DOI] [PubMed] [Google Scholar]
- Shyamala G, Yang X, Silberstein G, Barcellos-Hoff MH, Dale E. Transgenic mice carrying an imbalance in the native ratio of A to B forms of progesterone receptor exhibit developmental abnormalities in mammary glands. PNAS. 1998;95:696–701. doi: 10.1073/pnas.95.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- Vomachka AJ, Pratt SL, Lockefeer JA, Horseman ND. Prolactin gene-disruption arrests mammary gland development and retards T-antigen-induced tumor growth. Oncogene. 2000;19:1077–1084. doi: 10.1038/sj.onc.1203348. [DOI] [PubMed] [Google Scholar]
- Wiseman BS, Werb Z. Stromal effects on mammary gland development and breast cancer. Science. 2002;296:1046–1049. doi: 10.1126/science.1067431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naylor MJ, Ormandy CJ. Mouse strain-specific patterns of mammary epithelial ductal side branching are elicited by stromal factors. Developmental Dynamics. 2002;225:100–105. doi: 10.1002/dvdy.10133. [DOI] [PubMed] [Google Scholar]
- Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- Weber-Hall SJ, Phippard DJ, Niemeyer CC, Dale TC. Developmental and hormonal regulation of Wnt gene expression in the mouse mammary gland. Differentiation. 1994;57:205–214. doi: 10.1046/j.1432-0436.1994.5730205.x. [DOI] [PubMed] [Google Scholar]
- Bradbury JM, Edwards PA, Niemeyer CC, Dale TC. Wnt-4 expression induces a pregnancy-like growth pattern in reconstituted mammary glands in virgin mice. Dev Biol. 1995;170:553–563. doi: 10.1006/dbio.1995.1236. [DOI] [PubMed] [Google Scholar]
- Brisken C, Heineman A, Chavarria T, Elenbaas B, Tan J, Dey SK, McMahon JA, McMahon AP, Weinberg RA. Essential function of Wnt-4 in mammary gland development downstream of progesterone signaling. Genes Dev. 2000;14:650–654. [PMC free article] [PubMed] [Google Scholar]
- Atwood CS, Hovey RC, Glover JP, Chepko G, Ginsburg E, Robison WG, Vonderhaar BK. Progesterone induces side-branching of the ductal epithelium in the mammary glands of peripubertal mice. J Endocrinol. 2000;167:39–52. doi: 10.1677/joe.0.1670039. [DOI] [PubMed] [Google Scholar]
- Robinson GW, Hennighausen L, Johnson PF. Side-branching in the mammary gland: the progesterone-Wnt connection. Genes Dev. 2000;14:889–894. [PubMed] [Google Scholar]
- Humphreys RC, Lydon J, O'Malley BW, Rosen JM. Mammary gland development is mediated by both stromal and epithelial progesterone receptors. Mol Endocrinol. 1997;11:801–811. doi: 10.1210/me.11.6.801. [DOI] [PubMed] [Google Scholar]
- Farago M, Dominguez I, Landesman-Bollag E, Xu X, Rosner A, Cardiff RD, Seldin DC. Kinase-inactive glycogen synthase kinase 3beta promotes Wnt signaling and mammary tumorigenesis. Cancer Res. 2005;65:5792–5801. doi: 10.1158/0008-5472.CAN-05-1021. [DOI] [PubMed] [Google Scholar]
- Song DH, Sussman DJ, Seldin DC. Endogenous protein kinase CK2 participates in Wnt signaling in mammary epithelial cells. J Biol Chem. 2000;275:23790–23797. doi: 10.1074/jbc.M909107199. [DOI] [PubMed] [Google Scholar]
- Song DH, Dominguez I, Mizuno J, Kaut M, Mohr SC, Seldin DC. CK2 phosphorylation of the armadillo repeat region of beta-catenin potentiates Wnt signaling. J Biol Chem. 2003;278:24018–24025. doi: 10.1074/jbc.M212260200. [DOI] [PubMed] [Google Scholar]
- Gao ZH, Seeling JM, Hill V, Yochum A, Virshup DM. Casein kinase I phosphorylates and destabilizes the beta-catenin degradation complex. Proc Natl Acad Sci U S A. 2002;99:1182–1187. doi: 10.1073/pnas.032468199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman S, Silberstein GB, Daniel CW. Ductal morphogenesis in the mouse mammary gland: evidence supporting a role for epidermal growth factor. Dev Biol. 1988;127:304–315. doi: 10.1016/0012-1606(88)90317-X. [DOI] [PubMed] [Google Scholar]
- Kenney NJ, Bowman A, Korach KS, Barrett JC, Salomon DS. Effect of exogenous epidermal-like growth factors on mammary gland development and differentiation in the estrogen receptor-alpha knockout (ERKO) mouse. Breast Cancer Res Treat. 2003;79:161–173. doi: 10.1023/A:1023938510508. [DOI] [PubMed] [Google Scholar]
- Fowler KJ, Walker F, Alexander W, Hibbs ML, Nice EC, Bohmer RM, Mann GB, Thumwood C, Maglitto R, Danks JA, et al. A mutation in the epidermal growth factor receptor in waved-2 mice has a profound effect on receptor biochemistry that results in impaired lactation. Proc Natl Acad Sci U S A. 1995;92:1465–1469. doi: 10.1073/pnas.92.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Paterson AJ, Chin E, Nabell LM, Kudlow JE. Targeted expression of a dominant negative epidermal growth factor receptor in the mammary gland of transgenic mice inhibits pubertal mammary duct development. Mol Endocrinol. 1997;11:1766–1781. doi: 10.1210/me.11.12.1766. [DOI] [PubMed] [Google Scholar]
- Civenni G, Holbro T, Hynes NE. Wnt1 and Wnt5a induce cyclin D1 expression through ErbB1 transactivation in HC11 mammary epithelial cells. EMBO Rep. 2003;4:166–171. doi: 10.1038/sj.embor.embor735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musgrove EA. Wnt signalling via the epidermal growth factor receptor: a role in breast cancer? Breast Cancer Res. 2004;6:65–68. doi: 10.1186/bcr737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DA, Neville MC. Tight junction regulation in the mammary gland. J Mammary Gland Biol Neoplasia. 1998;3:233–246. doi: 10.1023/A:1018707309361. [DOI] [PubMed] [Google Scholar]
- Umeda K, Ikenouchi J, Katahira-Tayama S, Furuse K, Sasaki H, Nakayama M, Matsui T, Tsukita S, Furuse M, Tsukita S. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126:741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- Rubenstein NM, Guan Y, Woo PL, Firestone GL. Glucocorticoid down-regulation of RhoA is required for the steroid-induced organization of the junctional complex and tight junction formation in rat mammary epithelial tumor cells. J Biol Chem. 2003;278:10353–60. Epub 2003 Jan 13.. doi: 10.1074/jbc.M213121200. [DOI] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends Genet. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Isles AR, Holland AJ. Imprinted genes and mother-offspring interactions. Early Hum Dev. 2005;81:73–77. doi: 10.1016/j.earlhumdev.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Alexander WS, Starr R, Fenner JE, Scott CL, Handman E, Sprigg NS, Corbin JE, Cornish AL, Darwiche R, Owczarek CM, Kay TW, Nicola NA, Hertzog PJ, Metcalf D, Hilton DJ. SOCS1 is a critical inhibitor of interferon gamma signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/S0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- Curley JP, Barton S, Surani A, Keverne EB. Coadaptation in mother and infant regulated by a paternally expressed imprinted gene. Proc Biol Sci. 2004;271:1303–1309. doi: 10.1098/rspb.2004.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, Pavlidis P, Journot L. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Shiura H, Miyoshi N, Konishi A, Wakisaka-Saito N, Suzuki R, Muguruma K, Kohda T, Wakana S, Yokoyama M, Ishino F, Kaneko-Ishino T. Meg1/Grb10 overexpression causes postnatal growth retardation and insulin resistance via negative modulation of the IGF1R and IR cascades. Biochem Biophys Res Commun. 2005;329:909–916. doi: 10.1016/j.bbrc.2005.02.047. [DOI] [PubMed] [Google Scholar]
- Ludwig T, Eggenschwiler J, Fisher P, D'Ercole AJ, Davenport ML, Efstratiadis A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev Biol. 1996;177:517–535. doi: 10.1006/dbio.1996.0182. [DOI] [PubMed] [Google Scholar]
- Nakae J, Kido Y, Accili D. Distinct and overlapping functions of insulin and IGF-I receptors. Endocr Rev. 2001;22:818–835. doi: 10.1210/er.22.6.818. [DOI] [PubMed] [Google Scholar]
- Dufresne AM, Smith RJ. The adapter protein GRB10 is an endogenous negative regulator of insulin-like growth factor signaling. Endocrinology. 2005;146:4399–4409. doi: 10.1210/en.2005-0150. [DOI] [PubMed] [Google Scholar]
- McClurg P, Pletcher MT, Wiltshire T, Su AI. Comparative analysis of haplotype association mapping algorithms. BMC Bioinformatics. 2006;7:61. doi: 10.1186/1471-2105-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark HF, Gurney AL, Abaya E, Baker K, Baldwin D, Brush J, Chen J, Chow B, Chui C, Crowley C, Currell B, Deuel B, Dowd P, Eaton D, Foster J, Grimaldi C, Gu Q, Hass PE, Heldens S, Huang A, Kim HS, Klimowski L, Jin Y, Johnson S, Lee J, Lewis L, Liao D, Mark M, Robbie E, Sanchez C, Schoenfeld J, Seshagiri S, Simmons L, Singh J, Smith V, Stinson J, Vagts A, Vandlen R, Watanabe C, Wieand D, Woods K, Xie MH, Yansura D, Yi S, Yu G, Yuan J, Zhang M, Zhang Z, Goddard A, Wood WI, Godowski P, Gray A. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13:2265–70. Epub 2003 Sep 15.. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadsell DL, Torres DT, Lawrence NA, George J, Parlow AF, Lee AV, Fiorotto ML. Overexpression of des(1-3) insulin-like growth factor 1 in the mammary glands of transgenic mice delays the loss of milk production with prolonged lactation. Biol Reprod. 2005;73:1116–25. Epub 2005 Aug 3.. doi: 10.1095/biolreprod.105.043992. [DOI] [PubMed] [Google Scholar]
- Kleinberg DL, Feldman M, Ruan W. IGF-I: an essential factor in terminal end bud formation and ductal morphogenesis. J Mammary Gland Biol Neoplasia. 2000;5:7–17. doi: 10.1023/A:1009507030633. [DOI] [PubMed] [Google Scholar]
- Richards RG, Klotz DM, Walker MP, Diaugustine RP. Mammary gland branching morphogenesis is diminished in mice with a deficiency of insulin-like growth factor-I (IGF-I), but not in mice with a liver-specific deletion of IGF-I. Endocrinology. 2004;145:3106–3110. doi: 10.1210/en.2003-1112. [DOI] [PubMed] [Google Scholar]
- Bonnette SG, Hadsell DL. Targeted disruption of the IGF-I receptor gene decreases cellular proliferation in mammary terminal end buds. Endocrinology. 2001;142:4937–4945. doi: 10.1210/en.142.11.4937. [DOI] [PubMed] [Google Scholar]
- Brisken C, Ayyanan A, Doppler W. Prolactin signaling and Stat5: going their own separate ways? Breast Cancer Res. 2002;4:209–212. doi: 10.1186/bcr543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovey RC, Harris J, Hadsell DL, Lee AV, Ormandy CJ, Vonderhaar BK. Local insulin-like growth factor-II mediates prolactin-induced mammary gland development. Mol Endocrinol. 2003;17:460–471. doi: 10.1210/me.2002-0214. [DOI] [PubMed] [Google Scholar]
- Knight CH, Peaker M. Mammary development in mice: effects of hemihysterectomy in pregnancy and of litter size post partum. J Physiol. 1982;327:17–27. doi: 10.1113/jphysiol.1982.sp014216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Image Procesing and Analysis in Java [http://rsb.info.nih.gov/ij/]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- Khatri P, Sellamuthu S, Malhotra P, Amin K, Done A, Draghici S. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 2005;33:W762–5. doi: 10.1093/nar/gki472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–7. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene lists of enriched biological pathway categories. Strain-specific differentially expressed genes between the QSi5 and CBA strains of mice belonging to the five enriched biological pathway categories are listed along with their P-values and fold changes.
Differentially expressed imprinted genes. Strain-specific differentially expressed imprinted genes between the QSi5 and CBA strains of mice are listed along with their P-values and fold changes.