Abstract
Tissue inhibitor of metalloproteinase 3 (TIMP-3) promoter methylation has been linked to loss of TIMP-3 expression in various cancers. In this study, we analyzed TIMP-3 gene methylation using MethyLight assay and TIMP-3 mRNA expression using reverse transcription-polymerase chain reaction analysis in 22 esophageal cancers, 27 gastric carcinomas, and 7 cancer cell lines. We also analyzed TIMP-3 protein expression by immunohistochemistry and its association with clinicopathological characteristics in two cohorts of gastric cancer comprising a total of 347 patients. The TIMP-3 gene was more commonly methylated in adenocarcinomas of the esophagus (9/13) and stomach (9/15) than in the corresponding nonneoplastic mucosa of the esophagus (1/8; P = .024) and stomach (2/14; P = .021). In gastric cancer patients, TIMP-3 was decreased in a diffuse-type gastric cancer and in cancers with poor differentiation and was associated with poor survival (P = .04). In summary, we observed frequent TIMP-3 promoter methylation in adenocarcinomas of the esophagus and stomach and the loss of TIMP-3 expression seems to be of clinical and prognostic relevance in these cancers.
Introduction
Despite the recent improvements in the diagnosis for adenocarcinomas of the esophagus and stomach, most patients are diagnosed at advanced stages in which the therapeutic options are limited, with a 5-year survival rate of less than 25% [1–5]. Currently, esophageal and gastric adenocarcinomas are believed to develop in a stepwise model of intestinal metaplasia leading to intraepithelial neoplasia (IEN) and, subsequently, adenocarcinoma. In the esophagus, adenocarcinomas develop from Barrett metaplasia that results from a long-standing history of reflux esophagitis. Approximately 0.5% of patients per year advance to IEN and Barrett cancer, while the underlying molecular changes are not well understood so far [6]. Similarly, in the stomach, the Correa model indicates a stepwise process leading to metaplastic, then early and advanced neoplastic lesions. However, whereas the etiology of Barrett metaplasia is linked to reflux esophagitis, in gastric cancer, Helicobacter pylori is the single most important risk factor.
Both diseases result from molecular alterations including activation of oncogenes and inactivation of tumor suppressor genes, which are crucial for the sequential development from premalignant lesions to adenocarcinomas [3,7,8]. Aberrant methylation of CpG islands frequently leads to inactivation and silencing of respective tumor suppressor genes, thus aberrant methylation and subsequent transcriptional silencing of various genes including APC, HPP1, COX-2, DAPK, p16, and p14 have been identified in esophageal and gastric adenocarcinomas [9–12]. Tissue inhibitor of metalloproteinase 3 (TIMP-3) is an inhibitor of metalloproteases (a group of peptidases involved in degradation of the extracellular matrix) and was shown to induce apoptosis and inhibit growth of tumor cells and is, thus, considered to be a tumor suppressor [13–15]. Recently, the role of TIMP-3 methylation was analyzed in esophageal and gastric cancers; however, the clinical implications of loss of TIMP-3 expression has not been addressed in these cancers [16–18].
Materials and Methods
Cell Lines and 5-aza-2′-deoxycytidine Treatment
Gastric adenocarcinoma cell lines (SNU1, AGS, MKN45, MKN28, and N87), esophageal adenocarcinoma (EAC; OE33), and squamous cell cancer (OE21) cell lines were obtained from American Type Culture Collection (Manassas, VA) and Japan Cell Bank (Tokyo, Japan) and were maintained in either F12K or RPMI-1640 media. Incubation with 5-aza-2′-deoxycytidine was carried out as previously described [19].
Reverse Transcription-Polymerase Chain Reaction Analysis
Conventional histologic diagnosis of serial sections was used to confirm the high density of cancer cells in the specimens before RNA extraction; total RNA was then extracted using Nucleospin RNA L Kit (Macherey-Nagel GmbH & Co. KG, Düren, Germany) according to the manufacturer's instructions [20]. Polymerase chain reaction was performed under the following conditions: initial denaturation at 92°C for 120 seconds, 35 cycles with denaturation at 92°C for 40 seconds, annealing at 56°C for 40 seconds, elongation at 72°C for 60 seconds, and finally at 72°C for 7 minutes. Polymerase chain reaction primers were designed to amplify a 457-bp cDNA fragment of the human TIMP-3 gene: sense: 5′-CTACACCATCAAGCAGATGAAG ATG-3′, antisense: 5′-GCTCAGGGGTCTGTGGCATTGAT-3′. TIMP-3 mRNA levels were quantified by densitometric scanning and normalization using β-actin cDNA fragments.
DNA Extraction and MethyLight Analysis of TIMP-3 Gene
Genomic DNA was extracted from tissues using the Nucleospin Tissue Kit (Macherey-Nagel GmbH & Co. KG) according to the manufacturer's instructions and was analyzed by the MethyLight technique after bisulfite conversion, as previously reported by Eads et al. [21,22]. Briefly, two locus-specific polymerase chain reaction primers flank an oligonucleotide probe with a 5′ fluorescent reporter dye (6FAM) and a 3′ quencher dye (BHQ-1). For this analysis, primers and probes are specifically designed to bind to bisulfiteconverted DNA, which generally span 7 to 10 CpG dinucleotides. The gene of interest is then amplified and normalized to a reference set [β-actin (ACTB)] to normalize for input DNA. TaqMan polymerase chain reaction reactions were performed in parallel with primers specific for the bisulfite-converted methylated sequence for a particular locus and with the ACTB reference primers. The ratio between the values was calculated in these two TaqMan analyses; using this approach, the degree of methylation at that locus was determined. The extent of methylation at a specific locus was determined by the following formula: [(gene/actb)sample: (gene/actb)SssI-treated genomic DNA] x 100. A cutoff value of 4% gave the best discrimination between normal and cancerous samples, as previously reported [21,22]. Therefore, samples with ≥4% fully methylated molecules were termed methylated, whereas samples with <4% were considered unmethylated. The primer and probe sequences for TIMP-3 (GenBank Accession No. U14394) are: forward primer (5′–3′): 5′-GCGTCGGAGGTTAAGGTTGTT-3′ reverse primer (5′–3′): 5′-CTCTCCAAAA TTACCGTACGCG-3′ probe sequence (5′–3′): 6FAM-AACTCGCTCGCCCG CCGAABHQ1 (Figure 1).
Figure 1.
Overview of the TIMP-3 amplicon locations. Map showing the amplicons used in this study in relation to the transcription start of the TIMP-3 gene. Whereas the whole CpG island spans more than 1200 bp, the amplicon for the MethyLight assay covers 13 CpG sites more than 155 bp proximal to the coding region. An extended amplicon for bisulfite sequencing covers 48 CpG sites (in total, 442 bp).
Cohorts for Molecular Analysis
The tissue samples were obtained by resection from patients (19 males, 3 females, median age: 59.9 years, range: 26–87 years) with esophageal cancer (squamous cell cancer: 9 cases, adenocarcinoma: 13 cases) and gastric cancer (20 males, 7 females; median age: 64.4 years, range: 26–86 years). In all patients with esophageal cancer and gastric cancer, tissue samples from cancer were obtained for molecular analysis; in 17 of these esophageal cancer cases and all gastric cancer cases, matched nonneoplastic tissues were also obtained for molecular analysis from a tumor-free location which was at least 2 cm distant from the tumor and confirmed to be without any tumor cell infiltration by histologic assessment. None of the patients with esophageal cancer or gastric cancer underwent a preoperative radio- or chemotherapy. In addition, tissue samples were also obtained from 14 patients with Barrett metaplasia (8 males, 6 females, median age: 69.3 years, range: 46–90 years) undergoing endoscopy for surveillance of the lesion and biopsies were taken for histologic and molecular analysis. In 10 and 12 of these cases, respectively, the matched cardia and antrum mucosa was also available for molecular analysis. Immediately after resection, tissue samples were put in liquid nitrogen and stored at -80°C or were fixed in formalin for histologic diagnosis (see below). The study was approved by the ethics committee of the University of Magdeburg and Charité, Berlin, Germany (EA1/06/2004).
Cohorts for Immunohistochemical Analysis
For the immunohistochemical analysis two cohorts were used: cohort 1 (Magdeburg) consisted of 176 gastric cancer patients treated by total or partial gastrectomy at the Otto-von-Guericke University between 1995 and 2005. Survival data were available from 68 patients of whom 38 died during follow-up. Median follow-up time for those patients was 20.1 months. Cohort 2 (Berlin) consisted of 171 gastric cancer patients undergoing gastrectomy at the Charité University Hospital between 1995 and 2002. In this cohort, survival data were available from 136 patients of whom 91 died during follow-up. The median follow-up time for the patients was 31.1 months. No patient was treated with radio- or chemotherapy preoperatively.
Tumors were classified according to Lauren [23] and guidelines [24] of the International Union against Cancer.
Tissue Microarray
For the evaluation of TIMP-3 expression in cohort 1, tissue microarrays (TMAs) were generated using a precision instrument (Beecher Instruments, Silver Spring, MD). Six tissue cylinders of 0.6 mm in diameter were punched from each tumor-bearing donor block and 12 tissue cylinders (six antrum and six corpus) from corresponding nonneoplastic mucosa, constructing 20 blocks of TMAs.
Immunohistochemistry
Immunohistochemical staining of TIMP-3 was performed as previously described [19]; TIMP-3 antibody was from Chemicon Europe (Hofheim, Germany). Evaluation of the slides was performed by two experienced pathologists (C. R. and W. W.) who were blinded toward patient characteristics, patient outcome, and results of the methylation analyses.
Staining Evaluation
For TMA evaluation (cohort 1), immunostaining was graded semiquantitatively as 0 (negative), 1 (weak), 2 (moderate), or 3 (strong) for each microarray spot and each tissue location (tumor, nonneoplastic mucosa) separately. Subsequently, the immunoreactivity scores were added for each case and each location and divided by the number of microarray spots studied. Thereby a mean immunoreactivity score was obtained for each individual patient and tissue site, i.e., tumor, normal antrum mucosa, and normal corpus mucosa.
For correlation analysis and survival analysis, we decided to use the easiest and most reproducible way of grouping the cancer cases as either negative when no staining whatsoever was present in any of the tissue spots evaluated or positive when a discernable staining was present in at least one tissue spot.
Statistical Analysis
Statistics were performed with Statistical Package for the Social Sciences version 15.0 (SPSS Inc., Chicago, IL). The different clinicopathologic characteristics were used as nominal variables in the Fisher's exact test, χ2 test, and χ2 test for trends. Otherwise, one-way analysis of variance or Student's t test was used to determine statistical difference. The survival curves were plotted using the Kaplan-Meier method, and comparison of survival times was performed with the log-rank test. All tests were two-sided, and P < .05 was considered statistically significant.
Results
TIMP-3 Is Frequently Methylated in Human Esophageal and Gastric Cancers
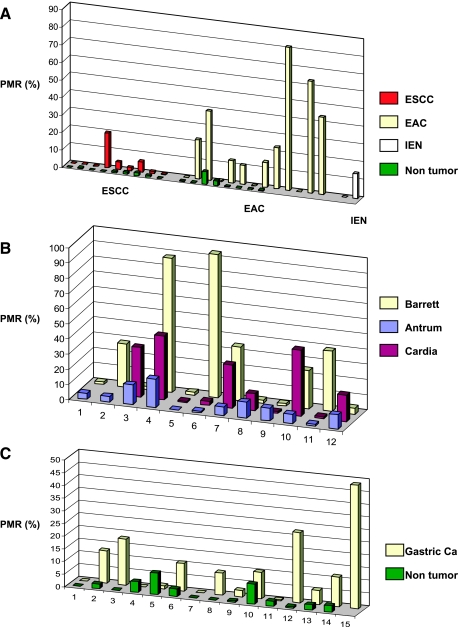
Using the MethyLight assay, promoter methylation of TIMP-3 in 13 EACs was assessed and compared to the corresponding normal esophageal mucosa in 8 patients. The cutoff of methylation was selected to be a percentage of methylation ratio (PMR) of ≥4% as previously described [21,22]. Interestingly, 9 EACs had a PMR ≥ 4% (9/13), whereas TIMP-3 methylation was only detected in 1 of 8 nonneoplastic esophageal samples (P = .024; Figure 2A). TIMP-3 methylation was less common in esophageal squamous cell cancers (ESCC; 2 of 9 cases), whereas all corresponding matched nonneoplastic mucosa samples exhibited a PMR < 4% (Figure 2A).
Figure 2.
TIMP-3 gene methylation in esophageal and gastric cancer. (A) TIMP-3 gene methylation in ESCC and EAC. Matched non-cancerous esophageal mucosa in green. Two cases with IEN in Barrett metaplasia included (IEN). (B) Levels of TIMP-3 methylation in antral and cardia compared to Barrett metaplasia. (C) TIMP-3 gene methylation in gastric cancers and matched noncancerous gastric mucosa. PMR (%) indicates percentage methylated reference.
We also observed TIMP-3 methylation in 1 of 2 cases with low-grade IEN in Barrett esophagus (Figure 2A).
Then, we studied matched samples from Barrett mucosa, cardia, and antrum mucosa. TIMP-3 methylation was frequently observed in Barrett mucosa (6/12), matched cardia (6/10), and antrum mucosa (7/12). Furthermore, when adjusting the methylation levels with regard to the quantity of methylated CpGs per sample and entity, a high degree of methylation was found in Barrett mucosa (mean ± SD, 27.2 ± 33.3%) compared with the matched cardia (18.1% ± 17.3%) and antrum (6.79% ± 5.47%; Figure 2B).
Next, we studied samples from 15 gastric cancer patients. Again, the MethyLight assay showed that TIMP-3 was more frequently methylated in the tumor (9/15) compared with the matched non-neoplastic mucosa (2/14, P = .021; Figure 2C).
TIMP-3 Methylation and Expression in Esophageal and Gastric Adenocarcinoma
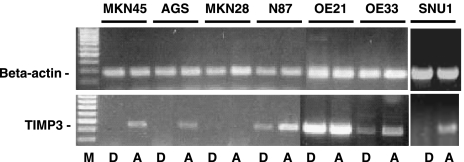
Next, we analyzed the frequency of TIMP-3 methylation and expression levels in esophageal and gastric adenocarcinoma cell lines. TIMP-3 expression was studied in 6 adenocarcinoma cell lines and 1 ESCC cell line (OE21) after incubation with the methylation inhibitor 5-aza-2′-deoxycytidine (Figure 3). Whereas in 4 gastric adenocarcinoma cell lines TIMP-3 mRNA was below the detection level, SNU1, AGS, and MKN45 exhibited readily detectable TIMP-3 mRNA levels after treatment with 5-aza-2′-deoxycytidine. MKN28 exhibited a loss of TIMP-3 mRNA expression despite 5-aza-2′-deoxycytidine treatment and was therefore excluded from the methylation analysis. In N87 gastric cancer cells and the EAC cell line OE33, TIMP-3 mRNA was constitutively expressed, and after treatment, mRNA levels further increased. In contrast, although the ESCC cell line (OE21) exhibited readily detectable expression of TIMP-3, 5-aza-2′-deoxycytidine treatment did not change the mRNA levels in these cells (Figure 3).
Figure 3.
TIMP-3 expression in cell lines. Changes of TIMP-3 mRNA levels in esophageal and gastric cancer cell lines after incubation with 5-aza-2′-deoxycytidine treatment. D indicates incubation with DMSO; A, incubation with 5-aza-2′-deoxycytidine.
Using the MethyLight assay cell lines SNU1, OE33, AGS, MKN45, and N87 showed TIMP-3 methylation before and after 5-aza-2′-deoxycytidine treatment (not shown). Similarly, bisulfite sequencing of the amplicons revealed no base pair changes and confirmed the MethyLight assay results (Figure 4). However, sequencing of a longer fragment of the TIMP-3 (spanning 94-bp upstream and 85-bp downstream from the included original amplicon) for SNU1 revealed a CpG 71-bp upstream of the analyzed region, which was demethylated after 5-aza-2′-deoxycytidine treatment (Figure 4).
Figure 4.
Bisulfite sequencing of TIMP-3 promoter region. Results from sequencing of different clones from cancer cells incubated with DMSO or 5-aza-2′-deoxycytidine. Sequences of the amplicons used in methylation analysis of five cell lines after incubation with 5-aza-2′-deoxycytidine treatment and DMSO-treated controls. Reference sequence refers to the reference sequence for the TIMP-3 gene according to GenBank. Single CpG site that turned to unmethylated after incubation with 5-aza-2′-deoxycytidine marked by vertical rectangle (position 993).
In 9 of 27 cases with gastric cancer, TIMP-3 mRNA was downregulated or lost in cancers compared with matched nonmalignant mucosa. TIMP-3 mRNA levels were also decreased in cancerous tissues in 3 of 8 cases with EAC when compared with matched noncancerous tissues. Similarly, in 4 of 9 patients with ESCC, reduction of TIMP-3 mRNA levels was present in neoplastic tissues compared with matched normal esophageal mucosa (not shown). We observed no statistical significant association of TIMP-3 promoter methylation with loss or down-regulation of mRNA expression in esophageal and gastric adenocarcinomas.
Association of Loss of TIMP-3 Protein Expression with Clinical Characteristics of Gastric Cancer Progression
To explore the role of putative TIMP-3 losses in the development and progression of adenocarcinoma of the stomach, we obtained tumor samples from 347 patients with gastric cancer and assessed the association of TIMP-3 expression in tumor cells with clinicopathologic characteristics of these patients. Complete loss of TIMP- expression was noted in 17% (cohort 1) and 27.5% (cohort 2) of adenocarcinomas, respectively (Table 1). The remaining tumors in both cohorts showed preserved expression of the protein, but expression varied to a considerable extent (Figure 5).
Table 1.
TIMP-3 Expression in Both Cohorts and Correlation of TIMP-3 Expression with Clinicopathological Features.
| Cohort 1 | Cohort 2 | |||||||
| All Cases | TIMP-3-Negative | TIMP-3-Positive | P | All Cases | TIMP-3-Negative | TIMP-3-Positive | P | |
| All cases | 176 (100%) | 30 (17%) | 146 (83%) | 171 (100%) | 47 (27.5%) | 124 (72.5%) | ||
| Age | ||||||||
| ≤65 years | 84 (47.7%) | 14 (16.7%) | 70 (83.3%) | 1.000* | 94 (55%) | 25 (26.6%) | 69 (73.4%) | 0.864* |
| >65 years | 92 (52.3%) | 16 (17.4%) | 76 (82.6%) | 77 (45%) | 22 (28.6%) | 55 (71.4%) | ||
| Tumor stage | ||||||||
| T1 | 19 (10.8%) | 2 (10.5%) | 17 (89.5%) | 0.142† | 8 (4.6%) | 0 (0%) | 8 (100%) | 0.312† |
| T2 | 75 (42.6%) | 11 (14.7%) | 64 (85.3%) | 80 (46.8%) | 22 (27.5%) | 58 (72.5%) | ||
| T3 | 72 (40.9%) | 14 (19.4%) | 58 (80.6%) | 68 (39.8%) | 21 (30.9%) | 47 (69.1%) | ||
| T4 | 10 (5.7%) | 3 (30%) | 7 (70%) | 15 (8.8%) | 4 (26.7%) | 11 (73.3%) | ||
| Nodal status | ||||||||
| N0 | 49 (28.3%) | 5 (10.2%) | 44 (89.8%) | 0.090† | 36 (21.1%) | 8 (22.2%) | 28 (77.8%) | 0.208† |
| N1 | 64 (37%) | 12 (18.8%) | 52 (81.2%) | 64 (37.4%) | 17 (26.6%) | 47 (73.4%) | ||
| N2 | 37 (21.4%) | 5 (13.5%) | 32 (86.5%) | 51 (29.8%) | 14 (27.5%) | 37 (72.5%) | ||
| N3 | 23 (13.3%) | 7 (30.4%) | 16 (69.6%) | 20 (11.7%) | 8 (40%) | 12 (60%) | ||
| State of metastasis | ||||||||
| M0 | 150 (85.2%) | 22 (14.7%) | 128 (85.3%) | 0.052* | 156 (91.2%) | 44 (28.2%) | 112 (71.8%) | 0.763* |
| M1 | 26 (14.8%) | 8 (30.8%) | 18 (69.2%) | 15 (8.8%) | 3 (20%) | 12 (80%) | ||
| Grade | ||||||||
| G1 | 3 (1.7%) | 0 (0%) | 3 (100%) | 0.003† | 2 (1.2%) | 0 (0%) | 2 (100%) | 0.012† |
| G2 | 49 (27.8%) | 2 (4.1%) | 47 (95.9%) | 43 (25.1%) | 6 (14%) | 37 (86%) | ||
| G3 | 124 (70.5%) | 28 (22.6%) | 96 (77.4%) | 126 (73.7%) | 41 (32.5%) | 85 (67.5%) | ||
| Lauren | ||||||||
| Intestinal | 107 (63.3%) | 12 (11.2%) | 95 (88.8%) | 0.006‡ | 90 (52.6%) | 13 (14.4%) | 77 (85.6%) | <0.001‡ |
| Mixed | 0 (0%) | / | / | 69 (40.4%) | 27 (39.1%) | 42 (60.9%) | ||
| Diffuse | 62 (36.7%) | 18 (29%) | 44 (71%) | 12 (7%) | 7 (58.3%) | 5 (41.7%) | ||
P values were calculated for each cross-table correlation separately in the respective subgroups.
Fisher's exact test.
χ2 test for trends.
χ2 test.
Figure 5.
TIMP-3 expression in gastric cancer. Immunohistochemical detection of TIMP-3 in nonneoplastic gastric foveloar epithelium (A) and well-differentiated intestinal-type cancer (B). Poorly differentiated intestinal-type cancer (C) and diffuse-type gastric cancers (D) frequently demonstrated loss of TIMP-3 expression. Hematoxylin counterstain. Original magnifications, x200.
No clear-cut correlations of expression of the protein with TNM-stage were seen. However, locally advanced metastasized tumors tended to show higher rates of TIMP-3 loss when compared with their locally restricted counterparts (Table 2). Interestingly, a significant loss of TIMP-3 expression was evident in diffuse-type gastric cancers in comparison to intestinal-type tumors in cohort 1 (P = .006), which was confirmed in cohort 2 (P < .001; Table 1). Consequently, in both cohorts, TIMP-3 expression correlated with tumor grade, with poorly differentiated tumors being significantly more likely to be TIMP-3-negative than well-differentiated ones (cohort 1: P = .003; cohort 2: P = .012; Table 1).
Table 2.
Clinicopathological Characteristics of Gastric Cancer and Survival in Two Cohorts.
| Cohort 1 | Cohort 2 | |||||||||
| Cases | Events | Mean Survival (months) | SE | Log-Rank Test (P) | Cases | Events | Mean Survival (months) | SE | Log-rank test (P) | |
| Age at diagnosis | ||||||||||
| ≤65 years | 33 | 17 | 46.7 | 9.0 | 0.296 | 74 | 46 | 35.6 | 4.1 | 0.053 |
| >65 years | 35 | 21 | 25.7 | 5.8 | 62 | 45 | 28.8 | 4.6 | ||
| Tumor stage | ||||||||||
| pT1 | 4 | 1 | 20.3 | 5.7 | 0.442 | 6 | 3 | 48.2 | 9.9 | <0.001 |
| pT2 | 33 | 15 | 40.3 | 7.6 | 65 | 35 | 42.4 | 4.7 | ||
| pT3 | 28 | 20 | 30.8 | 7.6 | 53 | 42 | 23.2 | 4.2 | ||
| pT4 | 3 | 2 | 13.4 | 4.1 | 12 | 11 | 7.8 | 2.5 | ||
| Nodal status | ||||||||||
| pN0 | 18 | 4 | 47.5 | 7.3 | 0.001 | 27 | 15 | 39.3 | 6.5 | 0.001 |
| pN1 | 28 | 16 | 39.1 | 8.4 | 54 | 33 | 38.5 | 4.9 | ||
| pN2 | 10 | 8 | 16.9 | 7.8 | 40 | 31 | 23.7 | 4.7 | ||
| pN3 | 10 | 8 | 10.3 | 3.3 | 15 | 12 | 11.5 | 4.4 | ||
| State of metastasis | ||||||||||
| M0 | 56 | 27 | 44.6 | 7.2 | 0.001 | 124 | 82 | 33.6 | 3.4 | 0.144 |
| pM1 | 12 | 11 | 11.7 | 3.5 | 12 | 9 | 14.3 | 4.1 | ||
| Grade | ||||||||||
| G1 | / | / | / | / | 0.135 | 1 | 0 | / | / | 0.198 |
| G2 | 22 | 9 | 39.8 | 9.0 | 39 | 23 | 41.5 | 6.3 | ||
| G3 | 46 | 29 | 34.5 | 6.8 | 96 | 68 | 25.6 | 2.8 | ||
| TIMP-3 expression | ||||||||||
| Negative | 14 | 11 | 22.2 | 6.6 | 0.309 | 35 | 27 | 19.4 | 3.7 | 0.040 |
| Positive | 54 | 27 | 43.4 | 7.4 | 101 | 64 | 36.5 | 3.8 | ||
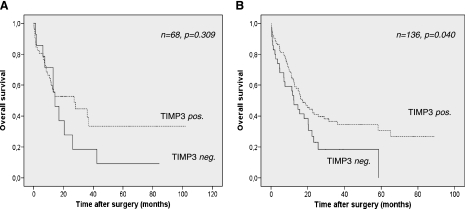
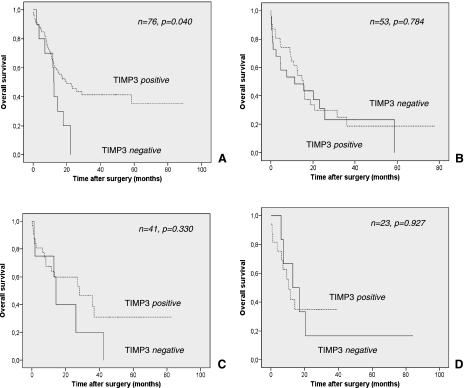
In cohort 1, in which only limited data on patient survival were available, TIMP-3 expression failed to show a statistical significant association with overall survival time (P = .309; Figure 5). However, those patients with TIMP-3-negative tumors tended toward shortened mean overall survival time (22.2 vs 43.4 months; Table 2). In the second larger cohort, again disadvantages in overall survival time were evident in the TIMP-3-negative patient group (19.4 vs 36.5 months) when compared with patients with preserved tumoral TIMP-3 expression (Table 2). Interestingly, in this more representative tumor cohort, these survival differences proved to be statistically significant (P = .040; Table 2 and Figure 6). After stratification for Lauren type of gastric cancer, only intestinal-type gastric cancers exhibited a significant difference in survival with regard to the expression or loss of TIMP-3 expression (Figure 7). To test for independent prognostic significance of TIMP-3 expression, we performed a multivariate survival analysis including age, stage, nodal status, state of distant metastasis, and grade. In this analysis, we failed to prove independent statistical significance of our marker (HR = 0.69, P = .128).
Figure 6.
Overall survival in dependence of TIMP-3 expression. Kaplan-Meier curves depicting overall survival in patients stratified for TIMP-3 expression in cohorts 1 (A) and 2 (B). P values were calculated with a log-rank test.
Figure 7.
Overall survival in diffuse and intestinal-type gastric cancer with regard to TIMP-3 expression. Kaplan-Meier curves depicting overall survival in patients stratified for TIMP-3 expression in cohorts 2 (A, intestinal-type; B, diffuse-type) and 1 (C, intestinal-type; D, diffuse-type). P values were calculated with a log-rank test.
Discussion
The family of TIMPs consists of four members that specifically inhibit the proteolytic activity of matrix metalloproteinases [25]. TIMP-3 is secreted as a 24-kDa protein that, in contrast to other TIMP family members, also binds to the extracellular matrix. TIMP-3 is considered a tumor suppressor [26], because TIMP-3 can inhibit tumor growth, angiogenesis, and invasion and can promote apoptosis. Accordingly, its expression is frequently down-regulated or lost in tumor cells [13,14,27]. The TIMP-3 gene is located on chromosome 22q12.3, a region in which loss of heterozygosity is frequently observed in various cancers, such as pancreatic endocrine tumors and secondary glioblastoma [28–30]. Interestingly, a recent study by Bachmann et al. [15] reported that silencing of TIMP-3 gene is also associated with 5′ CpG island hypermethylation in kidney, brain, and colon cancers. Using the MethyLight assay, we found frequent CpG island methylation of the TIMP-3 gene promoter in adenocarcinomas of esophagus and stomach. This observation confirms previous studies by Eads et al. [21] and Kang et al. [11], who also reported TIMP-3 methylation in their series of esophageal or gastric adenocarcinomas. In our analysis, only few matched normal esophageal mucosa samples in these patients exhibited TIMP-3 gene methylation. Further analysis in Barrett esophagus, the precancerous lesion of EAC, revealed a high frequency of TIMP-3 gene methylation, indicating that early precancerous lesions, such as Barrett metaplasia harbor TIMP-3 gene methylation. Thus, this epigenetic alteration is a common event in adenocarcinomas of the stomach and esophagus and seems to be an early molecular alteration in the stepwise sequence leading to adenocarcinomas of the upper gastrointestinal tract. Consistent with our findings, recent reports by Kang et al. [11] and Darnton et al. [12] also demonstrated an increasing frequency of TIMP-3 gene methylation along the pathogenesis of adenocarcinomas of the stomach and esophagus.
The loss or down-regulation of TIMP-3 expression has been linked to TIMP-3 gene methylation in adenocarcinomas of the esophagus and stomach in previous reports [12,31]. We also analyzed the TIMP-3 mRNA levels after incubation with the methylation inhibitor 5-aza-2′-deoxycytidine in cancer cell lines. Apart from MKN28, TIMP-3 mRNA levels were up-regulated or restored in all cell lines, thus confirming previous observations [12,31]. However, by bisulifte sequencing, we did not observe a significant alteration of the methylation status of the TIMP-3 gene. Therefore, we sequenced a longer fragment of the TIMP-3 gene (spanning 94-bp upstream and 85-bp downstream from the included original amplicon). In SNU1 cells, we found a CpG 71-bp upstream of the analyzed region, which was demethylated after 5-aza-2′-deoxycytidine treatment.
Loss of TIMP-3 expression by reverse transcription-polymerase chain reaction analysis was not consistently observed in gastric and esophageal cancers despite the high frequency of TIMP-3 gene methylation in our series of esophageal and gastric adenocarcinomas. We believe that the stromal expression of TIMP-3 may mask the loss of TIMP-3 expression that we observed in our immunohistochemical analysis [12]. So from the analysis of the cancer tissues, we cannot clearly state that TIMP-3 methylation in gastric or esophageal cancers leads to gene silencing, although data from other studies point to this association [16–18].
By immunohistochemical analysis, we observed a high frequency of loss of TIMP-3 expression in human gastric cancers. Interestingly, loss of TIMP-3 expression was especially frequent in diffuse-type cancers and cancers with poor differentiation. All diffuse-type gastric cancers and high-grade intestinal-type tumors (G3) are characterized by a strikingly infiltrative invasion pattern with single tumor cells and small groups of cells invading between preexisting gastric wall structures. Well and moderately differentiated intestinal-type cancers, in contrast, tend to show a less-infiltrative cohesive pushing invasion pattern. Our observation that loss of TIMP-3 expression is associated with the former elegantly underscores the importance of this metalloproteinase inhibitor in the regulation of tissue invasion in vivo. It seems likely that a loss of this key enzyme leads to a shift toward enhanced proteolytic activity of the tumor cells which in turn facilitates single-cell tissue invasion.
Finally, we assessed the clinical impact of loss of TIMP-3 expression in esophageal and gastric cancers. Interestingly, gastric cancer patients with loss of TIMP-3 expression experienced a shorter survival rate compared to cancers expressing TIMP-3. These data indicate that loss or reduction of TIMP-3 expression is of prognostic significance in gastric adenocarcinoma, and thus points to a role of TIMP-3 in the progression of these cancers.
Footnotes
This work was supported by grants from the Else Kröner-Fresenius-Stiftung (Homburg, Germany) and Astra-Zeneca (Wedel, Germany) awarded to M. Ebert, and by the Rudolf Bartling Foundation awarded to C. Röcken and M. Ebert.
References
- 1.Pera M, Cameron AJ, Trastek VF, Carpenter HA, Zinsmeister AR. Increasing incidence of adenocarcinoma of the esophagus and esophagogastric junction. Gastroenterology. 1993;104:510–513. doi: 10.1016/0016-5085(93)90420-h. [DOI] [PubMed] [Google Scholar]
- 2.Pera M, Manterola C, Vidal O, Grande L. Epidemiology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:151–159. doi: 10.1002/jso.20357. [DOI] [PubMed] [Google Scholar]
- 3.Souza RF, Spechler SF. Concepts in the prevention of adenocarcinoma of the distal esophagus and proximal stomach. CA Cancer J Clin. 2005;55:334–351. doi: 10.3322/canjclin.55.6.334. [DOI] [PubMed] [Google Scholar]
- 4.Koppert LB, Wijnhoven BP, van Dekken H, Tilanus HW, Dinjens WN. The molecular biology of esophageal adenocarcinoma. J Surg Oncol. 2005;92:169–190. doi: 10.1002/jso.20359. [DOI] [PubMed] [Google Scholar]
- 5.Berardi R, Scartozzi M, Romagnoli E, Antognoli S, Cascinu S. Gastric cancer treatment: a systematic review. Oncol Rep. 2004;11:911–916. [PubMed] [Google Scholar]
- 6.Shaheen NJ, Green B, Medapalli RK, Mitchell KL, Wei JT, Schmitz SM, West LM, Brown A, Noble M, Sultan S, et al. The perception of cancer risk in patients with prevalent Barrett's esophagus enrolled in an endoscopic surveillance program. Gastroenterology. 2005;129:429–436. doi: 10.1016/j.gastro.2005.05.055. [DOI] [PubMed] [Google Scholar]
- 7.Maley CC. Multistage carcinogenesis in Barrett's esophagus. Cancer Lett. 2007;245:22–32. doi: 10.1016/j.canlet.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 8.Leung SY, Yuen ST, Chung LP, Chu KM, Chan AS, Ho JC. hMLH1 promoter methylation and lack of hMLH1 expression in sporadic gastric carcinomas with high-frequency microsatellite instability. Cancer Res. 1999;59:159–164. [PubMed] [Google Scholar]
- 9.Shibata DM, Sato F, Mori Y, Perry K, Yin J, Wang S, Xu Y, Olaru A, Selaru F, Spring K, et al. Hypermethylation of HPP1 is associated with hMLH1 hypermethylation in gastric adenocarcinomas. Cancer Res. 2002;62:5637–5640. [PubMed] [Google Scholar]
- 10.Sato F, Meltzer SJ. CpG island hypermethylation in progression of esophageal and gastric cancer. Cancer. 2006;106:483–493. doi: 10.1002/cncr.21657. [DOI] [PubMed] [Google Scholar]
- 11.Kang GH, Lee S, Kim JS, Jung HY. Profile of aberrant CpG island methylation along multistep gastric carcinogenesis. Lab Invest. 2003;83:519–526. doi: 10.1097/01.lab.0000064704.53132.65. [DOI] [PubMed] [Google Scholar]
- 12.Darnton SJ, Hardie LJ, Muc RS, Wild CP, Casson AG. Tissue inhibitor of metalloproteinase-3 (TIMP-3) gene is methylated in the development of esophageal adenocarcinoma: loss of expression correlates with poor prognosis. Int J Cancer. 2005;115:351–358. doi: 10.1002/ijc.20830. [DOI] [PubMed] [Google Scholar]
- 13.Anand-Apte B, Bao L, Smith R, Iwata K, Olsen BR, Zetter B, Apte SS. A review of tissue inhibitor of metalloproteinases-3 (TIMP-3) and experimental analysis of its effect on primary tumor growth. Biochem Cell Biol. 1996;74:853–862. doi: 10.1139/o96-090. [DOI] [PubMed] [Google Scholar]
- 14.Ahonen M, Baker AH, Kahari VM. Adenovirus-mediated gene delivery of tissue inhibitor of metalloproteinases-3 inhibits invasion and induces apoptosis in melanoma cells. Cancer Res. 1998;58:2310–2315. [PubMed] [Google Scholar]
- 15.Bachman KE, Herman JG, Corn PG, Merlo A, Costello JF, Cavenee WK, Baylin SB, Graff JR. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 16.Brock MV, Gou M, Akiyama Y, Muller A, Wu TT, Montgomery E, Deasel M, Germonpré P, Rubinson L, Heitmiller RF, et al. Prognostic importance of promoter hypermethylation of multiple genes in esophageal adenocarcinoma. Clin Cancer Res. 2003;9:2912–2919. [PubMed] [Google Scholar]
- 17.Clément G, Braunschweig R, Pasquier N, Bosman FT, Benhattar J. Methylation of APC, TIMP3, and TERT: a new predictive marker to distinguish Barrett's oesophagus patients at risk for malignant transformation. J Pathol. 2006;208:100–107. doi: 10.1002/path.1884. [DOI] [PubMed] [Google Scholar]
- 18.Schulmann K, Sterian A, Berki A, Yin J, Sato F, Xu Y, Olaru A, Wang S, Mori Y, Deacu E, et al. Inactivation of p16, RUNX3, and HPP1 occurs early in Barrett's-associated neoplastic progression and predicts progression risk. Oncogene. 2005;24:4138–4148. doi: 10.1038/sj.onc.1208598. [DOI] [PubMed] [Google Scholar]
- 19.Ebert MP, Yu J, Hoffmann J, Rocco A, Röcken C, Kahmann S, Müller O, Korc M, Sung JJ, Malfertheiner P. Loss of beta-catenin expression in metastatic gastric cancer. J Clin Oncol. 2003;2:1708–1714. doi: 10.1200/JCO.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 20.Ebert M, Yokoyama M, Kobrin MS, Friess H, Lopez ME, Büchler MW, Johnson GR, Korc M. Induction and expression of amphiregulin in human pancreatic cancer. Cancer Res. 1994;54:3959–3962. [PubMed] [Google Scholar]
- 21.Eads CA, Lord RV, Wickramasinghe K, Long TI, Kurumboor SK, Bernstein L, Peters JH, DeMeester SR, DeMeester TR, Skinner KA, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 22.Eads CA, Lord RV, Kurumboor SK, Wickramasinghe K, Skinner ML, Long TI, Peters JH, DeMeester TR, Danenberg KD, Danenberg PV, et al. Fields of aberrant CpG island hypermethylation in Barrett's esophagus and associated adenocarcinoma. Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 23.Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma. An attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965;64:31–49. doi: 10.1111/apm.1965.64.1.31. [DOI] [PubMed] [Google Scholar]
- 24.Wittekind C, Sodin LH. TNM Classification of Malignant Tumours. 6th ed. New York NY: Wiley-Liss; 2002. [Google Scholar]
- 25.Blavier L, Henriet P, Imren S, Declerck YA. Tissue inhibitors of matrix metalloproteinases in cancer. Ann NY Acad Sci. 1999;878:108–119. doi: 10.1111/j.1749-6632.1999.tb07677.x. [DOI] [PubMed] [Google Scholar]
- 26.Ahonen M, Poukkula M, Baker AH, Kashiwagi M, Nagase H, Eriksson JE, Kähäri VM. Tissue inhibitor of metalloproteinases-3 induces apoptosis in melanoma cells by stabilization of death receptors. Oncogene. 2003;22:2121–2134. doi: 10.1038/sj.onc.1206292. [DOI] [PubMed] [Google Scholar]
- 27.Anand-Apte B, Pepper MS, Voest E, Montesano R, Olsen B, Murphy G, Apte SS, Zetter B. Inhibition of angiogenesis by tissue inhibitor of metalloproteinase-3. Invest Ophthalmol Vis Sci. 1997;38:817–823. [PubMed] [Google Scholar]
- 28.Wild A, Langer P, Ramaswamy A, Chaloupka B, Bartsch DK. A novel insulinoma tumor suppressor gene locus on chromosome 22q with potential prognostic implications. J Clin Endocrinol Metab. 2001;86:5782–5787. doi: 10.1210/jcem.86.12.8089. [DOI] [PubMed] [Google Scholar]
- 29.Wild A, Langer P, Celik I, Chaloupka B, Bartsch DK. Chromosome 22q in pancreatic endocrine tumors: identification of a homozygous deletion and potential prognostic associations of allelic deletions. Eur J Endocrinol. 2002;147:507–513. doi: 10.1530/eje.0.1470507. [DOI] [PubMed] [Google Scholar]
- 30.Nakamura M, Ishida E, Shimada K, Kishi M, Nakase H, Sakaki T, Konishi N. Frequent LOH on 22q12.3 and TIMP-3 inactivation occur in the progression to secondary glioblastomas. Lab Invest. 2005;85:165–175. doi: 10.1038/labinvest.3700223. [DOI] [PubMed] [Google Scholar]
- 31.Kang SH, Choi HH, Kim SG, Jong HS, Kim NK, Kim SJ, Bang YJ. Transcriptional inactivation of the tissue inhibitor of metalloproteinase-3 gene by DNA hypermethylation of the 5′-CpG island in human gastric cancer cell lines. Int J Cancer. 2000;86:632–635. doi: 10.1002/(sici)1097-0215(20000601)86:5<632::aid-ijc5>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]