Abstract
The Bmi-1 oncogene is overexpressed in a number of malignancies including breast cancer. In addition to Bmi-1, mammalian cells also express four other polycomb group (PcG) proteins that are closely related to Bmi-1. Virtually nothing is known about the role of these PcG proteins in oncogenesis. We have recently reported that Mel-18, a Bmi-1–related PcG protein, negatively regulates Bmi-1 expression, and that its expression negatively correlates with Bmi-1 in proliferating and senescing human fibroblasts. Here, we report that the expression of Bmi-1 and Mel-18 inversely correlates in a number of breast cancer cell lines and in a significant number of breast tumor samples. Overexpression of Mel-18 results in repression of Bmi-1 and reduction of the transformed phenotype in malignant breast cancer cells. Furthermore, the repression of Bmi-1 by Mel-18 is accompanied by the reduction of Akt/protein kinase B (PKB) activity in breast cancer cells. Similarly, Bmi-1 knockdown using RNA interference approach results in down-regulation of Akt/PKB activity and reduction in transformed phenotype of MCF7 cells. Importantly, we show that overexpression of constitutively active Akt overrides tumor-suppressive effect of Mel-18 overexpression and the knockdown of Bmi-1 expression. Thus, our studies suggest that Mel-18 and Bmi-1 may regulate the Akt pathway in breast cancer cells, and that Mel-18 functions as a tumor suppressor by repressing the expression of Bmi-1 and consequently down-regulating Akt activity.
Introduction
Polycomb group (PcG) proteins are chromatin-modifying proteins that play an important role in the development and cancer (1). Overexpression of certain PcG proteins, such as Bmi-1 and EZH2, has been linked to invasive breast and prostate cancer (2–4). Bmi-1 is also overexpressed in several other malignancies such as non–small-cell lung cancer (5), colorectal cancer (6), nasopharyngeal carcinoma (7), and oral cancer (8). Bmi-1 is known to be a key regulator of self-renewal of stem cells (1). In addition, recently, it was shown that Hedgehog signaling via Bmi-1 regulates self-renewal of normal and malignant human mammary stem cells (9).
After a finite number of cell divisions, most normal human cells undergo cellular senescence, whereby cells irreversibly cease to divide (10). Senescence constitutes a powerful barrier to oncogenesis (10). Bmi-1 has been shown to regulate cellular senescence and proliferation in rodent and human fibroblasts (11, 12). In addition, Bmi-1 can also bypass senescence and immortalize human mammary epithelial cells (HMEC; ref. 13). We have recently reported that Bmi-1 is negatively regulated by Mel-18 via repression of c-Myc, and that Mel-18 is overexpressed in senescent fibroblasts (14).
Here, we show that similar to human fibroblasts, expression of Mel-18 negatively correlates with Bmi-1 in a number of breast cancer cell lines and in a significant number of breast tumors. We also report that overexpression of Mel-18 in a commonly used breast cancer cell line MCF7 results in down-regulation of Bmi-1 and reduction of transformed phenotype. Furthermore, down-regulation of Bmi-1 by Mel-18 overexpression and knockdown of Bmi-1 expression by RNA interference (RNAi) approach is accompanied by down-regulation of Akt/protein kinase B (PKB) activity. We also show that overexpression of constitutively active Akt restores malignancy in MCF7 cells, in which Bmi-1 expression is reduced due to Mel-18 overexpression or Bmi-1 knockdown.
Materials and Methods
Cellular reagents, retroviral and short hairpin RNA vectors, virus production, and infection
MCF10A, MCF7, and other breast cancer cells were cultured as described (13). Retroviral vectors overexpressing Bmi-1 and Mel-18 and Bmi-1 short hairpin RNA (shRNA) are described earlier (14). A retroviral vector, pSRα-mAkt expressing constitutively active (myristylated) Akt (mAkt), was obtained from Dr. N. Hay (University of Illinois, Chicago, IL). Stable cell lines expressing Mel-18 or other gene of interest were generated by infection of the retroviral vectors expressing the particular gene as described (13, 14). The retroviruses were produced by transient transfection of the retroviral vector together with pIK packaging plasmid into tsa 54 packaging cell line as described (14). Soft-agar growth assay to determine the anchorage independence of cells was done as described (4).
Immunologic reagents and methods
Bmi-1 was detected using either F6 mouse monoclonal antibody (mAb) from Upstate Cell Signaling Solutions or 1H6B10G7 mAb from Zymed. Mel-18 was detected by a rabbit polyclonal H-115 (Santa Cruz Biotechnology). For the analysis of the Akt pathway, phosphorylated Akt 1/2/3 (pAkt 1/2/3; Ser473; sc-7985-R), pAkt 1/2/3 (Thr308; sc-16646-R), Akt-1 (B-1; sc-5298), Akt-2 (F-7; sc-5270), glycogen synthase kinase-3β(GSK3β; sc-53931), and cyclin D1 (A-12; sc-8396) antibodies were obtained from Santa Cruz Biotechnology. Rabbit polyclonal against total Akt (#9272) and pGSK3β(#9336) were obtained from Cell Signaling Technology.
To determine Akt activity in synchronized cells, MCF7 cells were serum starved for 48 h and stimulated for 30 min by addition of 10% FCS. MCF10A cells were growth factor deprived using D3 medium (15) for 48 h and stimulated for 30 min by addition of D medium, which contains 12.5 ng/mL epidermal growth factor (15). For the inhibition of the phosphoinositide 3-kinase (PI3K) pathway, cells were pretreated with LY294002 (20 μmol/L) or Wortmannin (100 nmol/L; Calbiochem) for 1 h before the addition of complete medium. Western blot analyses of total cell extracts were done using antibodies that detect total Akt, pAkt, and various other proteins as described (13, 14).
Clinical samples and immunohistochemical and statistical analyses
A total of 61 invasive breast cancer tissue samples were collected from the archives of the Department of Pathology, Cancer Center, Sun Yat-sen University (Guangzhou, China). For the use of these clinical materials for research purposes, prior patients’ consent and approval from the Institute Research Ethics Committee were obtained. Bmi-1 and Mel-18 were detected in paraffin sections of breast cancer tissue as described (7). All slides were interpreted by two independent observers in a blinded fashion. For each sample, one score was given according to the percentage of positive cells as <5% of the cells (1 point), 6% to 35% of the cells (2 points), 36% to 70% of the cells (3 points), >71% of the cells (4 points). Another score was given according to the intensity of staining as negative staining (1 point), weak staining (2 points), moderate staining (3 points), and strong staining (4 points). A final score was then calculated by multiplying the above two scores. If the final score was ≥4, the tumor was considered positive; otherwise, the tumor was considered negative. All statistical analyses were done by using the SPSS 10.0 software package. The Spearman’s rank correlation was used to estimate the correlation between Bmi-1 and Mel-18 expression.
Results
Bmi-1 and Mel-18 expression inversely correlates in breast cancer cell lines and breast tumors
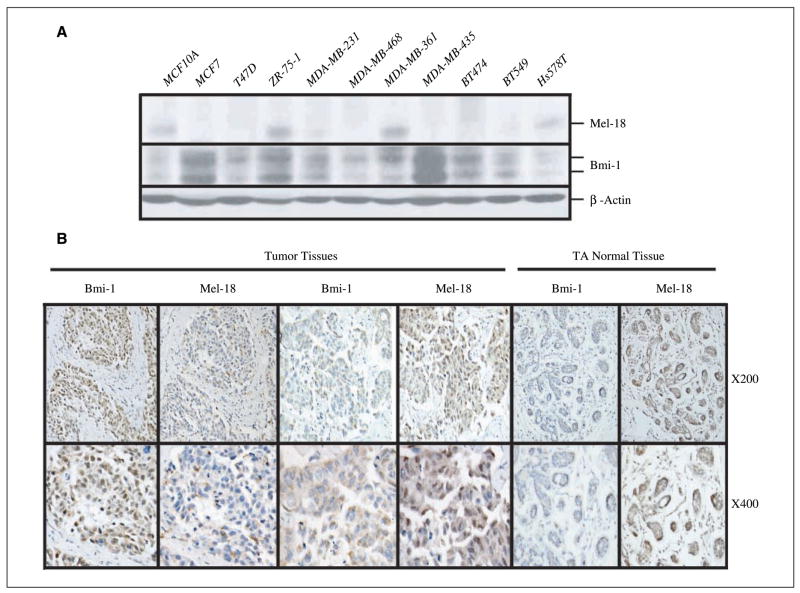
Our previous data in cultured human fibroblasts suggest an inverse correlation between Bmi-1 and Mel-18 expression; senescent cells show high expression of Mel-18, whereas proliferating cells show high expression of Bmi-1. These results suggested that breast cancer cell lines might express high Bmi-1 and low Mel-18. To probe this hypothesis, we analyzed expression of Bmi-1 and Mel-18 in several breast cancer cell lines (Fig. 1A). Our results suggested that compared with MCF10A, a normal immortal HMEC cell line, the majority of breast cancer cell lines (7 of 10) express high Bmi-1 and low Mel-18 (Fig. 1A).
Figure 1.
Mel-18 and Bmi-1 expression inversely correlates in breast cancer cell lines and breast tumors. A, Bmi-1 and Mel-18 expression in various breast cancer cell lines as detected by Western blot analysis. B, representative of two tumor samples: sample 1 expresses high Bmi-1 and low Mel-18, whereas sample 2 expresses high Mel-18 and low Bmi-1 expression. Tumor adjacent (TA) normal tissue of a biopsy sample with high Mel-18 and low Bmi-1. Tissues were stained with Bmi-1– or Mel-18– specific antibodies and counterstained with hematoxylin as described in Materials and Methods.
Because Bmi-1 is overexpressed in a large number of breast tumors (2, 3), and because its expression inversely correlates with Mel-18 expression in breast cancer cell lines, we hypothesized that Mel-18 down-regulation may lead to Bmi-1 up-regulation in breast tumors. To examine this possibility, we studied the expression of Mel-18 and Bmi-1 in 61 breast tumors by immunohistochemistry (Fig. 1B; Supplementary Fig. S1). By immunohistochemical analysis, 51 of 61 (83.6%) paraffin-embedded archival breast tumor biopsies showed a positive staining (score of ≥4) for Bmi-1, whereas 15 of 61 (24.5%) of the biopsies showed a positive staining (score of ≥4) of Mel 18. Of 15 Mel-18–positive and 51 Bmi-1–positive biopsies, only six were positive for both Bmi-1 and Mel-18 (Supplementary Table S1). The correlation between Bmi-1 and Mel 18 expression was further analyzed by Spearman correlation analysis, which showed a strong negative correlation (r = −0.673, P < 0.0001).
Overexpression of Mel-18 and knockdown of Bmi-1 expression reduce malignancy of breast cancer cells
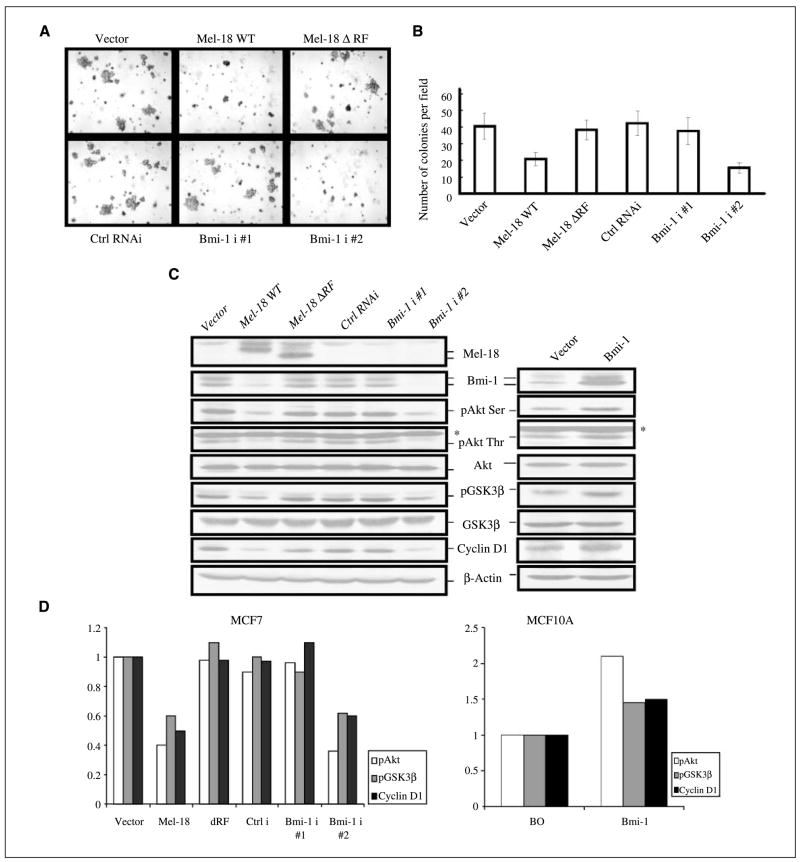
To examine the possibility that Mel-18 overexpression may reduce or revert the transformed phenotype of malignant cells, we determined the transformation potential of control and Mel-18–overexpressing MCF7 cells using anchorage independence growth assay. The results indicated that Mel-18 overexpression in MCF7 cells led to a decrease in colony formation in soft agar (Fig. 2A and B). The colonies in Mel-18–overexpressing MCF7 cells were less in frequency and also smaller in size (Fig. 2A, top). A RING finger mutant of Mel-18, which does not down-regulate Bmi-1 (14), did not inhibit soft agar colony formation when overexpressed in MCF7 cells (Fig. 2A, top).
Figure 2.
Reduction of transformed phenotype of MCF7 cells by Mel-18 overexpression and knockdown of Bmi-1 expression. A, overexpression of Mel-18 and knockdown of Bmi-1 expression in MCF7 decreases colony formation in soft agar. Control or Mel-18–overexpressing MCF7 cells (top), and control (Ctrl RNAi) or Bmi-1 shRNAs (Bmi-1 i #1 and Bmi-1 i #2) cells (bottom) were plated in soft agar to determine the anchorage-independent growth as described in Materials and Methods. B, colonies from three different experiments were counted and plotted. C, left, Mel-18 and Bmi-1 regulate Akt activity. Bmi-1 knockdown by RNAi approach or its down-regulation by Mel-18 overexpression leads to reduction in pAkt as determined by Western blot (WB) analysis using both anti–phosphorylated Ser473 and anti–phosphorylated Thr308 Akt antibodies. Reduction in Akt activity results in corresponding decrease in pGSK3βand cyclin D1 protein levels. Mel-18, Bmi-1, total Akt, pAkt, pGSK3β, total GSK3β, cyclin D1, and β-Actin (loading control) were detected by Western blot analysis as described in Materials and Methods. *, nonspecific band reacting to pAkt (Thr308) antibody. Right, Bmi-1 overexpression up-regulates Akt activity in MCF10A cells. Bmi-1 was overexpressed in MCF10A cells using pBabe-Bmi-1 retrovirus, and vector control and Bmi-1–overexpressing cells were analyzed for the activation of the Akt/GSK3β/cyclin D1 pathway by Western blot analysis as described in Materials and Methods. D, quantification of Akt and GSK3β activity. The pAkt and pGSK3β signal in each lane was quantified by densitometric analysis using ImageJ 1.37 software (NIH, Bethesda, MD) and normalized to total Akt and total GSK3β signal of each lane, respectively, and plotted. Similarly, levels of cyclin D1 were quantified using densitometric analysis of signal present in each lane, normalized to β-actin signal of each lane, and plotted.
We also determined the anchorage-independent growth potential of MCF7 cells, which stably express Bmi-1 shRNAs. We used two Bmi-1 shRNAs (Bmi-1 i#1 and Bmi-1 i#2). Western blot analysis of Bmi-1 indicted that Bmi-1 i#2 efficiently knocked down Bmi-1 expression (Fig. 2C). Accordingly, we found that stable expression of Bmi-1 i#2 in MCF7 cells led to significant decrease in number of colonies in soft agar, indicating a decrease in transformed phenotype of these cells (Fig. 2A, bottom and Fig. 2B).
Mel-18 and Bmi-1 regulate Akt activity in breast cancer cells
To determine the mechanism of inhibition of colony formation in soft agar and growth inhibition by Mel-18 overexpression or knockdown of Bmi-1 expression, we examined various growth regulators in these cells. Our results showed that Mel-18 overexpression did not affect p53 or its target p21 and pRb (Supplementary Fig. S2). Because Akt activity is constitutively high in many cancer cells, including breast cancer cells, we hypothesized that Mel-18 overexpression or Bmi-1 knockdown may reduce transforming phenotype via down-regulation of Akt pathway. To examine this possibility, we determined total Akt and pAkt by Western blot analysis. Our results showed that Bmi-1 down-regulation by Mel-18 overexpression or RNAi approach leads to substantial reduction in pAkt (Ser473 and Thr308) in MCF7 cells, suggesting that Bmi-1 regulates Akt activity (Fig. 2C; Supplementary Fig. S2). Our results also showed that total Akt levels remained unaffected by inhibition of Bmi-1 expression.
To further confirm the down-regulation of Akt activity by Bmi-1 knockdown or Mel-18 overexpression, we determined the expression of downstream targets of Akt pathway. GSK3β is known to be phosphorylated at Ser9 and inactivated by activated Akt (16). Inactivation of GSK3β by Akt mediated phosphorylation at Ser9 also results in cyclin D1 up-regulation (16). Hence, we determined GSK3β and cyclin D1 expression in control, Mel-18–overexpressing cells, and Bmi-1 knockdown cells. Consistent with reduction of Akt activity, Western blot analysis of cells with reduced expression of Bmi-1 due to Mel-18 overexpression or Bmi-1 knockdown showed decreased levels of pGSK3β and down-regulation of cyclin D1 (Fig. 2C, left and Fig. 2D). In MCF7 cells, activation of Akt depends on the presence of estradiol (E2) in the serum, which can be removed by charcoal stripping. Using regular serum (contains E2) and charcoal-stripped serum (no E2), we confirmed that Mel-18 overexpression or Bmi-1 knockdown inhibits activation of Akt (Supplementary Fig. S3), which depends on the presence of E2 in serum.
We also confirmed regulation of Akt activity by Bmi-1 using overexpression studies (Fig. 2C, right and Fig. 2D). Consistent with Bmi-1 knockdown studies, Bmi-1 overexpression led to up-regulation of Akt activity as determined by Western blot analysis using pAkt and pGSK3β antibodies (Fig. 2C, right and Fig. 2D). To determine the mechanism of Akt regulation by Bmi-1, we used PI3K inhibitors LY294002 and Wortmannin. Pretreatment of cells with these inhibitors strongly attenuated Akt activity in both control and Bmi-1–overexpressing cells (Supplementary Fig. S4), indicating that Bmi-1 regulates Akt activity via the PI3K pathway.
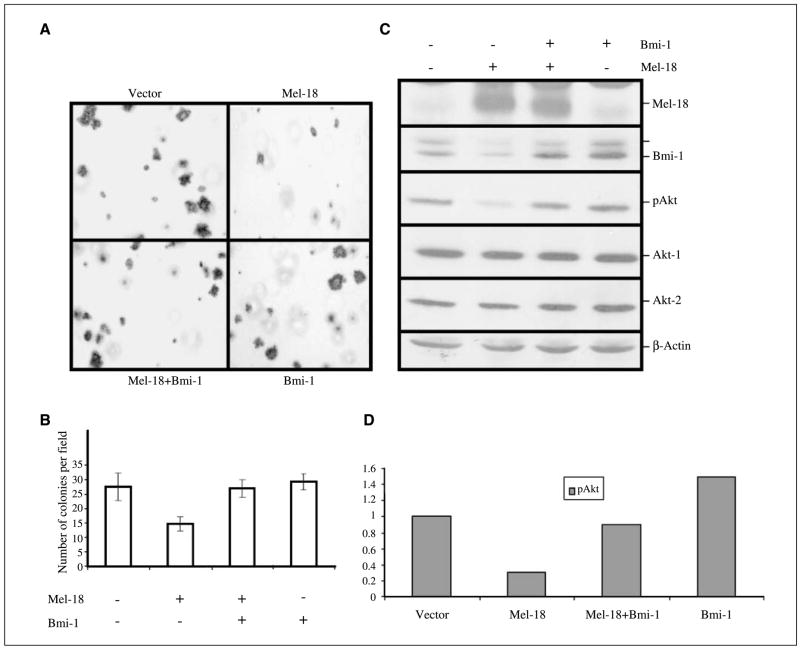
Exogenous Bmi-1 expression restores Akt activity and anchorage-independent growth in Mel-18–overexpressing MCF7 cells
Next, we examined whether exogenous expression of Bmi-1 using a retroviral promoter, which is not repressed by Mel-18, can restore Akt activity and full anchorage-independent growth in Mel-18–overexpressing MCF7 cells. The anchorage-independent growth of vector-infected control, Mel-18–overexpressing and Bmi-1–overexpressing MCF7 cells, and MCF7 cells expressing both Bmi-1 and Mel-18 was determined using soft-agar assays. The results (Fig. 3A and B) indicated that exogenous Bmi-1 could indeed restore anchorage-independent growth in Mel-18–overexpressing MCF7 cells. Western blot analysis of cells expressing both Mel-18 and Bmi-1 suggested that Bmi-1 could restore Akt activity in MCF7 cells (Fig. 3C and D).
Figure 3.
Exogenous Bmi-1 restores Akt activity and anchorage-independent growth potential of Mel-18–overexpressing cells. A, MCF7 cells were infected with a control retrovirus or Bmi-1–overexpressing retrovirus. Cells were selected in hygromycin and super-infected with Mel-18–expressing retrovirus. After selection, vector, Mel-18, Bmi-1, and Mel-18 and Bmi-1 coexpressing cells were analyzed for colony formation in soft agar. B, numbers of colonies growing in soft agar were quantified per field, and data were plotted. C, Western blot analysis of cells expressing Mel-18, Bmi-1, or Bmi-1 and Mel-18 and control cells was done to confirm overexpression as well as restoration of Akt activity as described in Materials and Methods. D, quantification of Akt activity in control vector and Mel-18, Mel-18 + Bmi-1, and Bmi-1–overexpressing cells (as indicated). Akt activity was quantified as described in Fig. 2D.
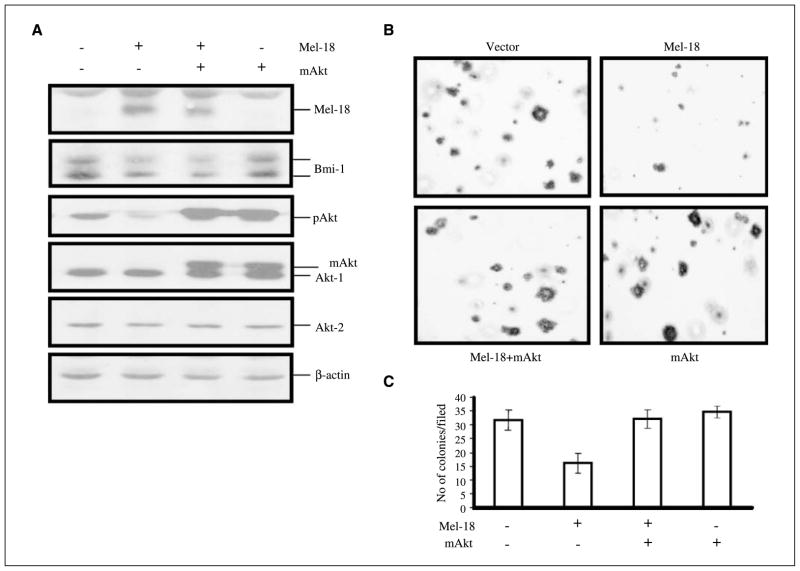
Exogenously expressed mAkt restores full transformed phenotype in Mel-18 overexpressing MCF7 cells
To test the hypothesis that Mel-18 overexpression or Bmi-1 knockdown reduces the transformed phenotype of MCF7 cells by down-regulating Akt activity, we co-overexpressed activated Akt (mAkt) in MCF7 cells with Mel-18 or Bmi-1 shRNA. MCF7 cells were selected for co-overexpression using different antibiotic resistance markers and analyzed for the overexpression of mAkt. Western blot analysis indicated that overexpression of mAkt resulted in high pAkt proteins indicative of activated Akt (Fig. 4A; Supplementary Fig. S5A). Consistent with Akt acting downstream of Bmi-1, mAkt overexpression did not result in Bmi-1 up-regulation. Next, using soft agar assay, anchorage-independent growth potential of control cells and cells expressing Mel-18, mAkt, or both was examined. Results indicated that mAkt fully restores anchorage-independent growth of MCF7 cells expressing Mel-18 (Fig. 4B and C) or Bmi-1 shRNA (Supplementary Fig. S5A–C), without perturbing Bmi-1 expression Collectively, these data indicate that Mel-18 and Bmi-1 shRNA inhibit colony formation in MCF7 cells via down-regulation of Akt activity.
Figure 4.
Exogenous overexpression of activated Akt (mAkt) restores anchorage-independent growth potential of Mel-18–overexpressing MCF7 cells. A, mAkt was stably expressed in Mel-18–overexpressing cells using a retroviral expression vector as described in Materials and Methods. Cell expressing Mel-18, mAkt, and Mel-18 together with mAkt were analyzed for expression of activated (phosphorylated) Akt by Western blot analysis. B, soft agar assay was done to determine anchorage-independent growth potential of MCF7-derived cells done as described in Materials and Methods. Representative photograph of colonies of control MCF7 (vector) and MCF7 derivatives (as indicated) growing in soft agar. C, colonies of control MCF7 and MCF7 expressing Mel-18, mAkt, or Mel-18 and mAkt (as indicated) growing in soft agar were counted and plotted from three different experiments.
Discussion
Our cell culture data showing an inverse correlation between Bmi-1 and Mel-18 expression prompted us to examine if indeed this inverse correlation exists in vivo in breast tumors. Bmi-1 is overexpressed in invasive breast cancer; hence, we reasoned that in such breast tumors where Bmi-1 is highly expressed, Mel-18 expression might be low. Indeed, we found a strong negative correlation between Mel-18 and Bmi-1 expression in invasive breast cancer, which favored high Bmi-1 and low Mel-18 expression. A recent report did not find a negative correlation between Bmi-1 and Mel-18 expression in primary breast cancer samples (17). These authors also did not find negative correlation between Bmi-1 and p16/ARF expression, which has been shown in other cancers such as non–small-cell lung cancer (5) and colorectal cancer (6), and several in vivo and culture studies. At present, the reasons of discrepancy between the work published by Silva et al (17) and other studies (5, 6) and our data presented here is unclear. It may reflect tumor heterogeneity in the samples, different stages of tumor progression, and methods of detection and data analysis. All breast cancer samples used in our study were from late-stage invasive breast tumors, most of which had relatively undetectable to low Mel-18 expression compared with Bmi-1 expression as determined by immunohistochemistry. Based on these results, we suspect that this inverse correlation may persist with other cancer types. Analysis of Mel-18 and Bmi-1 coexpression in a large cohort of breast tumors and other cancers remains to be explored. Nonetheless, our studies suggest that Mel-18 is a physiologic regulator of Bmi-1 expression in breast epithelial cells.
It is interesting to note that Akt activity is up-regulated in a number of cancers including breast cancer (18, 19). Bmi-1 is thought to promote oncogenesis primarily by down-regulating the expression of the p16Ink4a/ARF locus (20). However, most breast cancer cells, including MCF7 cells that were used in this study, express very little, if any, p16, owing to p16 promoter methylation and/or deletion of the Ink4a/ARF locus. Our previous studies (13) and data presented here suggest that Bmi-1 can also promote oncogenesis via p16-independent mechanisms. In particular, Bmi-1 seems to regulate Akt activity in breast cancer cells and breast epithelial cells. Although the detailed mechanism of regulation of Akt activity by Bmi-1 remained to be elucidated, our PI3K inhibitor data and Akt phosphorylation studies suggest that Bmi-1 regulates Akt activity by up-regulating PI3K/3-phosphoinoisitide–dependent kinase-1 pathway. In conclusion, our studies suggest that polycomb proteins, in particular Bmi-1 and Mel-18, can regulate Akt activity in normal breast epithelial and breast cancer cells.
Supplementary Material
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Acknowledgments
Grant support: National Cancer Institute grant RO1CA 094150 and U.S. Department of Defense grant DAMD17-02-1-0509 (G.P. Dimri).
We thank Dr. N. Hay for providing mAkt expression vector.
References
- 1.Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M. Stem cells and cancer; the polycomb connection. Cell. 2004;118:409–18. doi: 10.1016/j.cell.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 2.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. J Clin Invest. 2005;115:1503–21. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JH, Yoon SY, Jeong SH, et al. Overexpression of Bmi-1 oncoprotein correlates with axillary lymph node metastases in invasive ductal breast cancer. Breast. 2004;13:383–8. doi: 10.1016/j.breast.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Kleer CG, Cao Q, Varambally S, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vonlanthen S, Heighway J, Altermatt HJ, et al. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–6. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim JH, Yoon SY, Kim CN, et al. The Bmi-1 oncoprotein is overexpressed in human colorectal cancer and correlates with the reduced p16INK4a/p14ARF proteins. Cancer Lett. 2004;203:217–24. doi: 10.1016/j.canlet.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Song LB, Zeng MS, Liao WT, et al. Bmi-1 is a novel molecular marker of nasopharyngeal carcinoma progression and immortalizes primary human nasopharyngeal epithelial cells. Cancer Res. 2006;66:6225–32. doi: 10.1158/0008-5472.CAN-06-0094. [DOI] [PubMed] [Google Scholar]
- 8.Kang MK, Kim RH, Kim SJ, et al. Elevated Bmi-1 expression is associated with dysplastic cell transformation during oral carcinogenesis and is required for cancer cell replication and survival. Br J Cancer. 2007;96:126–33. doi: 10.1038/sj.bjc.6603529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu S, Dontu G, Mantle ID, et al. Hedgehog signaling and Bmi-1 regulate self-renewal of normal and malignant human mammary stem cells. Cancer Res. 2006;66:6063–71. doi: 10.1158/0008-5472.CAN-06-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dimri GP. What has senescence got to do with cancer? Cancer Cell. 2005;7:505–12. doi: 10.1016/j.ccr.2005.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Itahana K, Zou Y, Itahana Y, et al. Control of the replicative life span of human fibroblasts by p16 and the polycomb protein Bmi-1. Mol Cell Biol. 2003;23:389–401. doi: 10.1128/MCB.23.1.389-401.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–8. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 13.Dimri GP, Martinez JL, Jacobs JJ, et al. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002;62:4736–45. [PubMed] [Google Scholar]
- 14.Guo WJ, Datta S, Band V, Dimri GP. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536–46. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Band V, Zajchowski D, Kulesa V, Sager R. Human papilloma virus DNAs immortalize normal human mammary epithelial cells and reduce their growth factor requirements. Proc Natl Acad Sci U S A. 1990;87:463–7. doi: 10.1073/pnas.87.1.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:339–45. [PubMed] [Google Scholar]
- 17.Silva J, Garcia JM, Pena C, et al. Implication of polycomb members Bmi-1, Mel-18, and Hpc-2 in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc expression in primary breast carcinomas. Clin Cancer Res. 2006;12:6929–36. doi: 10.1158/1078-0432.CCR-06-0788. [DOI] [PubMed] [Google Scholar]
- 18.Hay N. The Akt-mTOR tango and its relevance to cancer. Cancer Cell. 2005;8:179–83. doi: 10.1016/j.ccr.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 19.Bose S, Chandran S, Mirocha JM, Bose N. The Akt pathway in human breast cancer: a tissue-array-based analysis. Mod Pathol. 2006;19:238–45. doi: 10.1038/modpathol.3800525. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–90. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).