Abstract
The nematode Caenorhabditis elegans is a well-known model organism for research on aging and life span, but very little is known about its ecology and natural history. The strain N2 is the standard wild-type C. elegans and arose from the progeny of a single hermaphrodite. Since N2 has passed through laboratory culture, the influence of inadvertent selection and genetic drift on C. elegans strains kept in culture is unclear. Because it seems that other wild-type strains have also been subject to lengthy laboratory culture, the life span and biodemography of wild-caught C. elegans is of interest. We recovered C. elegans from snails (Helix aspersa) in ca. 50% of the California locations where we made collections. In experiments with one of the wild-caught isolates, it differed in important demographic properties, mortality, fertility, .tness, and activity patterns, from the standard N2 strain, when both strains were evaluated in a common laboratory environment. The differences were not only statistically significant; they were also large enough to be biologically important. The differences are consistent with the hypothesis that N2 has adapted to laboratory conditions.
Keywords: Aging, Fitness, Life span, Matrix population models, Sensitivity, Elasticity, Wild-caught
1. Introduction
The nematode Caenorhabditis elegans is arguably the most completely characterized metazoan organism; as a result, it is widely used as a model system for studies of the biology and genetics of aging (Gems and Riddle, 1996, 2000; Johnson, 1984, 2003; Johnson and Hutchinson, 1993; Johnson et al., 2001; Kenyon et al., 1993; Klass, 1977; Walker et al., 2000); however, very little is known about its ecology, natural history, or life span in the wild (Hodgkin and Doniach, 1997; Gershon and Gershon, 2002; Reznick and Gershon, 1999). The evolution of life span and senescence (the increase of mortality rate with age) is a difficult evolutionary problem (Hamilton, 1966; Medawar, 1957; Rose, 1991; Williams, 1957). One explanation, termed antagonistic pleiotropy (Rose, 1991; Williams, 1957), considers senescence to be an indirect effect of selection for genes with favorable effects on fitness at early ages but negative effects at later ages. To understand the selective forces acting on life history characteristics that contribute to life span, longevity patterns must be evaluated by examining fitness. Fitness, in turn, can be understood only in the context of the vital rates that contribute to it, including survival, fertility, and the timing of events in the life cycle (Carey, 2003; Caswell, 2001; Charlesworth, 1994, 2000). Comparisons based only on survival, or only on fertility, or only on summary statistics (e.g., median lifetime instead of the survival schedule, or total brood size instead of the age schedule of reproduction) cannot be relied upon to indicate the direction, let alone the magnitude of fitness differences. And, of course, all these vital rates are affected by the ecological environment of the population, and, with a few exceptions (Barriere and Felix, 2005; Caswell-Chen et al., 2005; Haber et al., 2005), very little is known of the ecology of C. elegans (Hodgkin and Doniach, 1997).
The canonical wild-type strain (N2), which is used as a comparison for studies on the effects of longevity mutants, has been cultured for many generations in the laboratory. It was originally isolated in 1956 from mushroom compost in England by Warwick Nicholas (Hansen et al., 1960). It is descended from the progeny of a single hermaphrodite picked from a nutrient agar plate (Nicholas, personal communication). The strain was moved from the United Kingdom to the Berkeley, California laboratory of Ellsworth Dougherty, and from there sent to Sydney Brenner who grew it on Escherichia coli and isolated a hermaphrodite that gave rise to the N2 strain (Brenner, 1974). Thus, the N2 strain has been subject to selection through laboratory culture and has grown for an unknown (but apparently large) number of generations in conditions very different from those of its natural habitat (Gershon and Gershon, 2002). The extent to which the demographic properties of N2 have changed because of selection in the laboratory environment, and the implications of such changes for studies of aging, are not completely clear, but it has been suggested that laboratory stocks may have adapted to laboratory conditions through maximized fertility (Johnson and Hutchinson, 1993), or through shortened life span and reduced brood sizes (Gems and Riddle, 2000). Other important possibilities have been reviewed (Gershon and Gershon, 2002; Reznick and Gershon, 1999). Given the life history variation that exists among N2 strains (Gems and Riddle, 2000) and the variable history of time in culture for the non-N2 wild-type strains (Hodgkin and Doniach, 1997), demographic comparison of recently wild-caught C. elegans isolates with laboratory strains would seem to be useful in determining wild-type life history traits, and here we present an example of such an analysis.
Despite the large and growing literature on the genetics of aging in C. elegans, demographic analyses have been surprisingly limited in scope. Among the many excellent studies of survival, and the smaller number of studies of reproduction, there are almost no estimates of the population growth rate λ (or r = logλ); but see (Chen et al., 2006; Hodgkin and Barnes, 1991; Shook and Johnson, 1999; Vassilieva and Lynch, 1999; Vassilieva et al., 2000; Venette and Ferris, 1998). Of these, some are simply measurements of the increase in population size in batch culture (Hodgkin and Barnes, 1991; Venette and Ferris, 1998). While these data provide a sort of estimate of λ, they provide no information on the specific contributions of the vital rates (age-specific survival and reproduction) to fitness. Evolutionary interpretations of aging require estimates of the selection gradients on survival and fertility (i.e., the rate at which fitness changes as a result of changes in those traits), and on how the selection gradients change with age (Hamilton, 1966; Charlesworth, 2000), but these gradients have never been reported for C. elegans.
In this paper, we use some of these quantitative methods in a case study with a small cohort of wild-caught C. elegans to illustrate an approach to defining life span and fitness in wild-caught worms as compared to a standard laboratory strain, N2 (Bristol). The experiment we report uses a small cohort of wild-caught worms, and hence should be regarded as an initial foray rather than definitive, but in spite of this we document significant differences in mortality, fertility, fitness, and activity patterns between N2 and wild-caught C. elegans. The differences are consistent with the hypothesis that N2 has adapted to laboratory conditions. Our approach has potential applications to many other problems in the evolutionary biodemography of C. elegans.
2. Materials and methods
2.1. Experiments and culture methods
The experiments described here were performed with cohorts of individual nematodes. Wild-caught C. elegans, isolated from snails in Davis, California (Caswell-Chen et al., 2005) were obtained from surface-sterilized eggs (0.5% sodium hypochlorite for 3 min) placed on nematode growth medium (NGM) seeded with E. coli strain OP50 (Brenner, 1974) at 20 °C. Four days later, eggs were transferred to new NGM with OP50; in three days they developed into mature hermaphrodites. Newly hatched first-stage juveniles produced by these hermaphrodites, only three generations removed from nature, were used to initiate cohorts for demographic and behavioral experiments.
We obtained “wild-type” C. elegans (N2 var Bristol; DR subclone of CB original, Tc1 pattern I) from the Caenorhabditis Genetic Center at University of Minnesota, St. Paul in October 2000. The plate we received contained both males and hermaphrodites. Upon arrival, the stock was re-cultured to be maintained at −80 °C immediately. Experimental cohorts were two generations removed from this frozen culture maintained at −80 °C. To initiate cohorts, frozen stock was placed on NGM with E. coli strain OP50 at 20 °C. Four days later, the eggs laid on the plate were transferred onto new NGM with OP50, and in three days they developed into mature hermaphrodites laying eggs. First-stage juveniles, newly hatched from these eggs, were used to initiate experimental cohorts.
Cohorts of 20 wild-caught and 1000 N2 individuals were followed until the death of the last worm, with the 1000 worms followed 200 at a time. Experiments were conducted in the same laboratory using the same equipment under the same conditions, with the same lead personnel to provide experimental consistency and reduce possible environmental variation (Johnson and Hutchinson, 1993). We treat the data from the 1000-worm cohort as a reference data set (Chen et al., 2006; Müller et al., 2004). Combining the 200-worm subcohorts that comprise the cohort includes a component of variation due to the laboratory environment in the reference distribution. In spite of this component, our statistical procedures detected significant differences between N2 and wild-caught C. elegans.
Worms were transferred individually onto 60 × 15 mm NGM plates seeded with 1-day-old OP50 and then maintained in the dark at 20 °C in a constant temperature incubator. While laying eggs, worms were transferred each day to new NGM. To avoid mechanical damage, a small block of agar was cut from beneath the worm and transferred, with the worm, to new medium. After the worm had crawled off the agar block, the block was removed from the plate. Progeny were counted as juveniles emerging from eggs (one day after eggs were laid).
Survival was determined by observing worms for movement, if no movement was observed for 5–10 s, the plate was gently tapped to elicit movement. Absent motion, the worm was gently touched near the head with a small piece of agar and then a nematode pick (Chen et al., 2001). Worms that then failed to move were considered dead. Worm survival was evaluated daily. Adults that died because of the internal hatch of eggs (facultative vivipary) (Chen and Caswell-Chen, 2003, 2004) were recorded as dead and included in the analysis. Nematode movement was video-recorded for 2-min intervals every 2 days.
2.2. Demographic analysis
Our demographic methods are well-known and are described only briefly here; for details see (Carey, 2003; Caswell, 2001; Charlesworth, 1994). Age-specific survivorship l(x) was measured by the proportion of worms surviving to age x. Age-specific maternity m(x) was measured by the mean number of juvenile progeny produced per day at age x. The mortality rate µ(x) was calculated as
| (1) |
and the distribution of age at death as
| (2) |
The life expectancy at birth (the average days remaining to an individual at birth) is given by
| (3) |
In practice, it was calculated from the fundamental matrix (Caswell, 2006, Eq. 3.5). The cohort generation time (the mean age of the parents of offspring born to a cohort over its whole lifetime) is given by
| (4) |
we calculated it from the fundamental matrix and the maternity function (Caswell, 2006).
For analysis of population growth, age-classified, birth-flow population projection matrices with a projection interval of 1 day were calculated for each strain following (Caswell, 2001, Section 2.4). Population growth follows
| (5) |
with n(t) a vector giving the number of individuals in each age class at time t and A the projection matrix. For an age-classified model, A contains non-zero entries only on the first row (fertilities Fi) and the subdiagonal (survival probabilities Pi).
A population growing according to (5) eventually converges to a stable age distribution (the proportions of the different age classes) and grows exponentially at a rate λ. This growth rate integrates the entire age schedules of survival and fertility into a single measure which, with slight complications caused by diploid genetics, measures fitness in age-structured populations (Charlesworth, 1994, 2000). It is calculated as the dominant eigenvalue of A; the continuous-time rate of increase is r = logλ. The stable age distribution w is given by the right eigenvector of A corresponding to λ. The reproductive values of each age class are given by the entries of the corresponding left eigenvector v.
These quantities permit calculation of the sensitivity of λ to changes in entries of A:
| (6) |
Sometimes it is convenient to consider the effect of proportional rather than absolute changes; these are given by the elasticity of λ to aij:
| (7) |
Because natural selection responds to phenotypic changes that translate into changes in fitness, the sensitivities of λ to any trait are proportional to the selection gradient on that trait. The rate of change in a trait is proportional to the product of the selection gradient and the additive genetic variance in the trait (Lande, 1982).
Demographic data on two or more strains yields estimates of the differences in fitness among those strains. Such differences integrate all the inter-strain differences in survival and fertility. The contributions of each age-specific survival and fertility difference can be calculated using LTRE (life table response experiment) analysis (Caswell, 2001, Chap. 10). For age-classified models of the sort used here, the difference in fitness between wild-caught and N2 worms is
| (8) |
Each term in the first summation is the contribution of the difference in survival at one age to the fitness differential. Each term in the second summation is the contribution of the difference in fertility at one age to the fitness differential. The contributions are the product of the difference in a vital rate and the sensitivity of λ to that rate; even large inter-strain differences will make negligible contributions to the fitness differential if λ is very insensitive to that rate.
2.3. Statistical analysis
Confidence intervals were calculated using bootstrap resampling methods and hypothesis tests conducted using randomization methods. Bootstrap confidence intervals (Efron and Tibshirani, 1993) were computed on all estimated quantities, following (Caswell, 2001, Section 12.1). Each individual, with its age at death and its history of reproduction and/or movement, was treated as a unit. Bootstrap data sets were created by randomly sampling 1000 (for N2) or 20 (for wild-caught) individuals, with replacement, from the real data set. Each bootstrap data set was subjected to the same algorithm used for the real data; 95% confidence intervals were computed using the percentile method (because all quantities were nearly median-unbiased, no bias correction was applied). Results for all scalar quantities are shown in Table 1.
Table 1.
Estimates and 95% bootstrap confidence intervals for life expectancy at birth (e0), cohort generation time (G), and fitness (λ) for wild-caught and N2 strains of C. elegans
| Quantity | Estimate | Confidence interval |
|---|---|---|
| Wild-caught | ||
| Life expectancya (e0) | 16.10 | [11.72, 20.45] |
| Generation timeb (G) | 3.13 | [2.83, 3.47] |
| Fitnessc (λ) | 3.49 | [3.10, 3.80] |
| N2 | ||
| Life expectancy (e0) | 14.33 | [14.00, 14.65] |
| Generation time (G) | 3.85 | [3.84, 3.87] |
| Fitness (λ) | 3.85 | [3.83, 3.87] |
Life expectancy .
Generation time .
Fitness is the dominant eigenvalue of the population projection matrix A.
Significance tests were carried out using nonparametric randomization tests (Manly, 1997; Caswell, 2001, Section 12.3). Test statistics, measuring the differences between strains, were defined for each estimated quantity, as follows.
Life expectancy, λ, generation time: the absolute value of the difference between N2 and wild-caught C. elegans.
Survivorship: let l be the vector of age-specific survivorship. The test statistic was the ∞-norm of the difference between the two functions, |1 − l2|∞ = maxx|l1(x) − l2(x)|. Because survivorship is the complement of a cumulative probability distribution, this test statistic is equivalent to that used for the two-sample Kolmogorov–Smirnov test of the difference between two cumulative probability distributions.
Age at death: let d be the vector giving the probability of death at each age. The test statistic was the 1-norm of the difference between the two distributions, |d1 − d2|1 = ∑x|d1(x) − d2(x)|; this is a standard measure of the difference between two probability distributions.
Fertility and movement: let m be the vector giving age-specific fertility. The test statistic was the 2-norm of the difference between the two vectors, , which is appropriate since these are simply non-negative vectors. The same test statistic was used for the age-specific movement data.
To obtain the distribution of the test statistic under the null hypothesis, individuals (with their complete record of reproduction, movement, and age at death) were randomly permuted between treatments, maintaining sample size. The permuted data were subjected to the same analyses as the original data, and the appropriate test statistic calculated for each of 2000 permuted data sets. The statistical significance of the observed test statistic is the proportion of the permutation statistics greater than or equal to the observed value.
These tests naturally account for the difference in sample size between N2 and wild-caught C. elegans. Because the N2 and wild-caught cohorts were measured at different times (although in the same lab and under virtually identical conditions) the possibility of environmental differences cannot be ruled out. Given that the 1000-worm N2 cohort was obtained from five different experiments however, the randomization distribution of the test statistics under the null hypothesis includes a component due to environmental differences, which would bias our tests against significance, thus compensating to some degree for the possible environmental component of the inter-strain differences.
3. Results
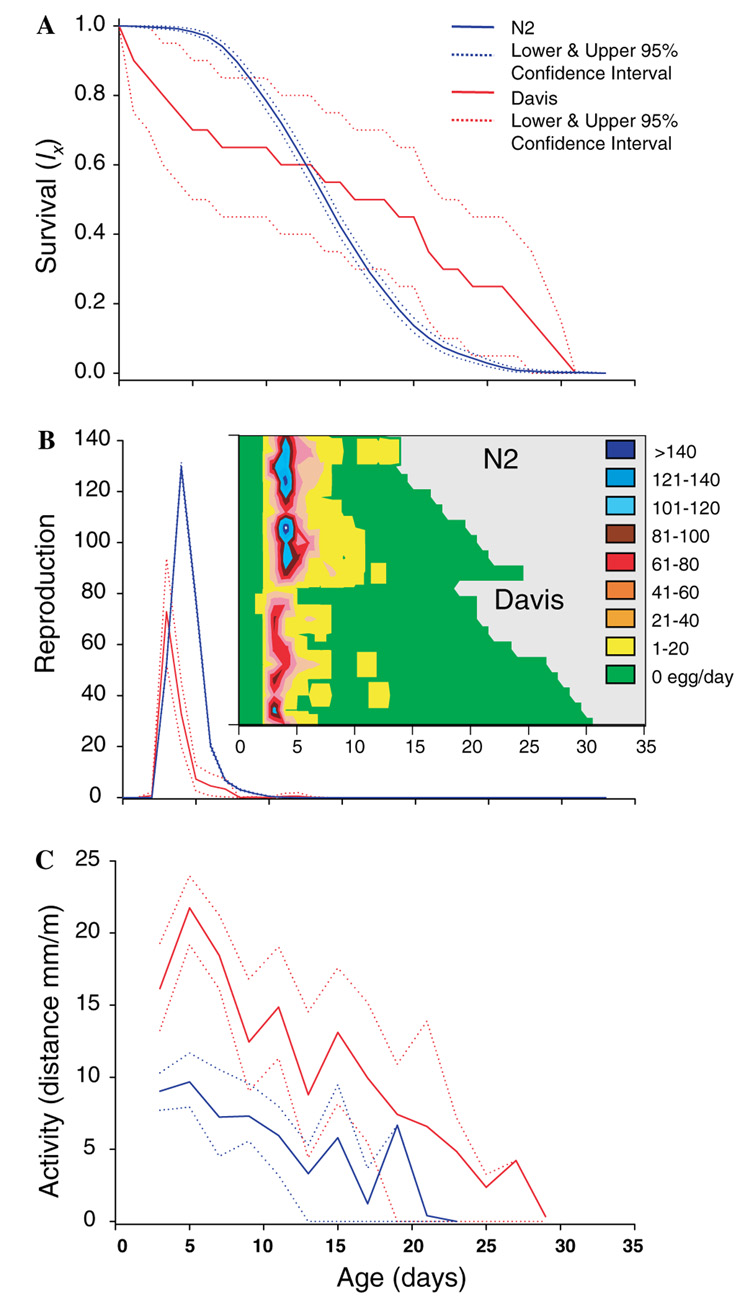
Wild-caught and N2 worms exhibit significantly different survivorship (Fig. 1A; P = 0.0145). Wild-caught worms had lower early survival and higher late survival (the difference in distribution of age at death was significant, P ≤ 5 × 10−4). However, because of the survival crossover apparent in Fig. 1A, life expectancy did not differ (wild-caught 16.1 days, N2 14.3 days, P = 0.1300). The fertility of wild-caught worms was lower than, and shifted earlier relative to, N2 (Fig. 1B; P ≤ 5 × 10−4). The generation time of wild-caught worms was about 20% shorter than that of N2 worms (wild-caught 3.13 days, N2 3.85 days, P ≤ 5 × 10−4).
Fig. 1.
(A) Survivorship measured in cohorts of 20 wild-caught and 1000 N2 worms. Estimates and 95% bootstrap confidence intervals are shown. (B) Fertility (eggs per individual per day) in cohorts of 20 wild-caught and 1000 N2 worms. Estimates with 95% bootstrap confidence intervals. Survival with color-coded representation of reproduction for a cohort of 20 N2 and 20 wild-caught worms is shown in the inset. (C) Movement (mm per minute) in cohorts of 10 wild-caught and 10 N2 worms, recorded using a digital video camera. Estimates and 95% bootstrap confidence intervals are shown.
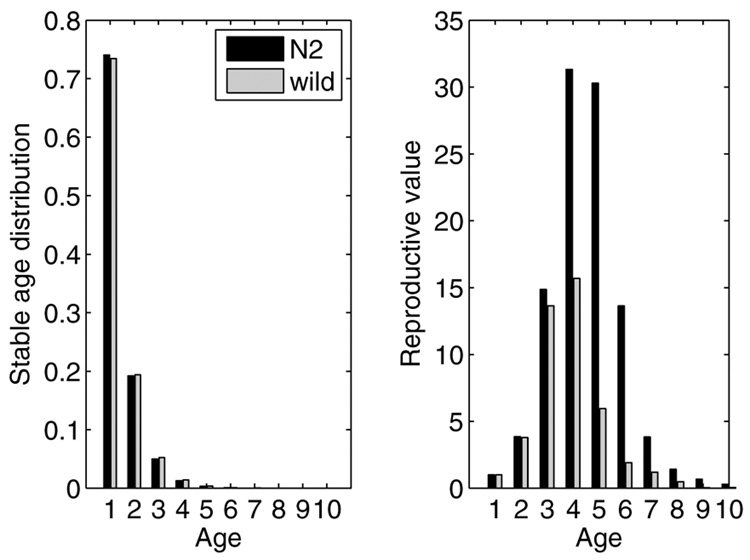
The population growth rate of the wild-caught isolate is significantly lower than that of N2 (wild-caught λ = 3.49, N2 λ = 3.85, P ≤ 5 × 10−4). The stable age distributions of the two strains are very similar, and because of the high growth rate in the laboratory environment they are both dominated (>99%) by individuals from 1 to 4 days old (Fig. 2). The reproductive value distributions are similar in shape, but the relative value of individuals aged 4–9 days is higher in N2 than in wild-caught (Fig. 2).
Fig. 2.
The stable age distribution w and the age-specific reproductive value schedule v for N2 and wild-caught C. elegans.
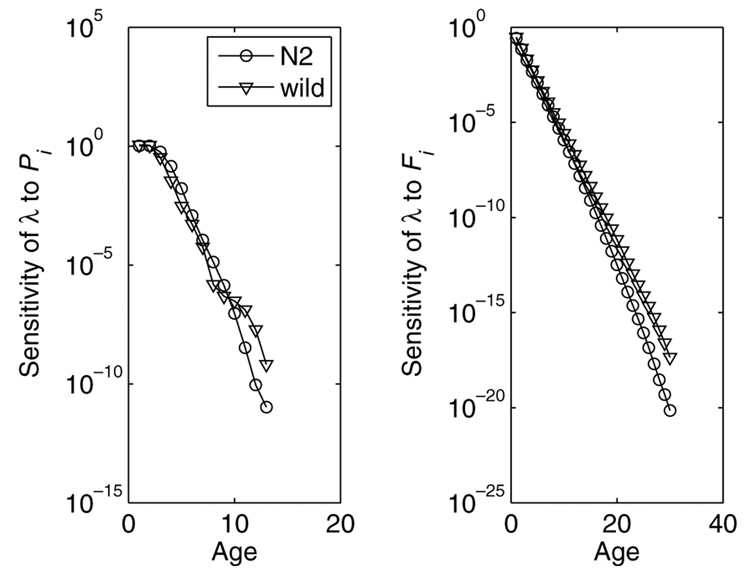
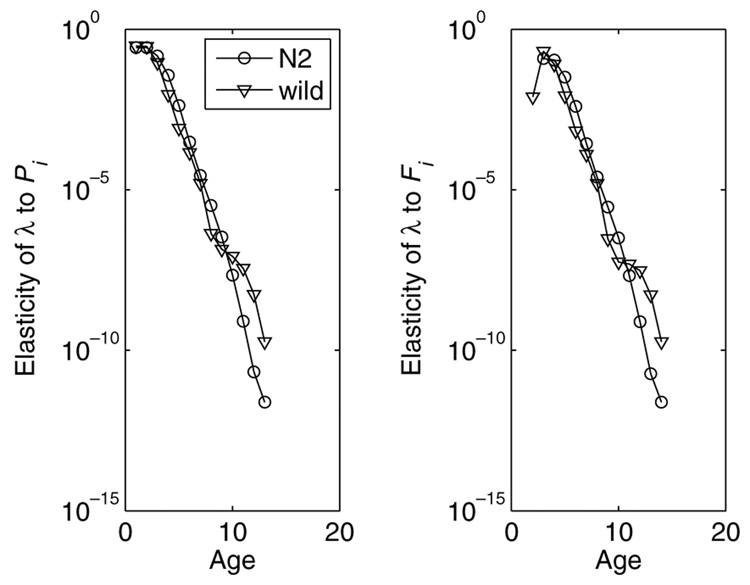
The sensitivities and elasticities of λ to changes in survival and fertility show similar patterns in both strains (Fig. 3 and Fig. 4). The sensitivities to Pi and Fi fall off. nearly exponentially with age, as does the elasticity to Pi. The elasticity to Fi first increases (up to age 3) and then declines.
Fig. 3.
The sensitivity of λ to changes in age-specific survival probability (Pi) and age-specific fertility (Fi) for N2 and wild-caught C. elegans.
Fig. 4.
The elasticity of λ to changes in age-specific survival probability (Pi) and age-specific fertility (Fi) for N2 and wild-caught C. elegans.
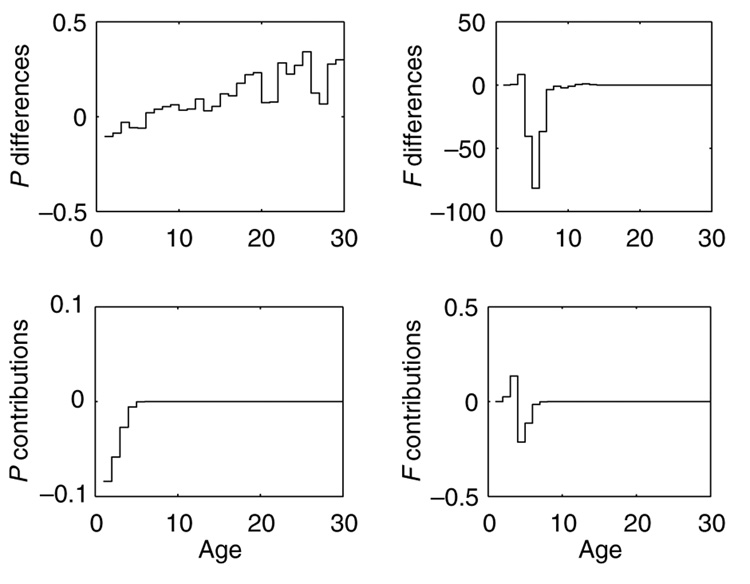
The fitness differential λw − λN2 = −0.36 is due almost entirely to differences in the vital rates between ages 1 and 7; the large survival differences between the strains at later ages make no detectable contribution (Fig. 5). The contributions of survival differences are smaller in magnitude than those of fertility differences, and are consistently negative. The contributions from fertility differences are positive early, and then negative later, reflecting the shift in reproduction in wild-caught relative to N2. Summing the contributions reveals nearly equal contributions to survival (−0.176) and fertility (−0.183).
Fig. 5.
LTRE (life table response experiment) analysis of the fitness differences between wild-caught and N2 C. elegans. Above: the differences in age-specific survival and age-specific fertility between the strains. Below: the contributions of the differences in survival and fertility to the difference in fitness, measured as λw − λN2.
Given that behavior is important because of its use in aging studies as a biomarker of senescence, we also compared distance moved per unit time of N2 and wild-caught worms as they aged. The wild-caught worms were more active, and remained active longer, than did the N2 worms (Fig. 1C; P = 0.0038). We also observed the percentage of time spent moving, and with age, wild-caught nematodes remained active longer.
4. Discussion
Caenorhabditis elegans can be reliably collected from snails (Helix aspersa) (Caswell-Chen et al., 2005), and we used this method to obtain wild-caught isolates of C. elegans in California. We recovered C. elegans from H. aspersa in locations in northern and southern California, including some areas from which C. elegans has not previously been recorded. Locations included Berkeley, Davis, and Palo Alto, in snails collected from urban street yards, gardens, and outdoor areas or parks (Caswell-Chen et al., 2005). Interestingly, H. aspersa is an invasive species, and was introduced to San Jose, California from France in ca. 1850 (Basinger, 1931; Stearns, 1900). We propose that because H. aspersa shows metapopulation structure (Arnaud, 2003), it is probably significant in moving C. elegans among locations, and the question of how C. elegans moves among locations arises relative to understanding gene flow (Barriere and Felix, 2005; Haber et al., 2005). Among individual snails from Davis and San Diego, C. elegans occurred in 26% of the snails examined, and among bulk collected snails at other locations, 30% yielded C. elegans. We repeatedly isolated C. elegans from snails obtained from Davis over one year. Of the California locations we have sampled for snails, 56% have yielded C. elegans, suggesting that collecting H. aspersa represents a method for predictably collecting “wild” C. elegans that will be useful to the C. elegans research community.
The association of C. elegans with snails may contribute to the widespread distribution of the worm and its seeming association with human habitation (Barriere and Felix, 2005), as snails move in human commerce as food, with plant material in agriculture, and predators may move snails for considerable distances. It is not clear whether the association is simply phoretic or if other aspects are involved.
Our study of a single small cohort of wild-caught C. elegans found significant differences from the standard N2 strain in important demographic properties, when both of them were evaluated in a common laboratory environment. A larger-scale comparison, applying the methods used here to larger cohorts of a greater number of wild-caught isolates, would permit more definitive conclusions about differences in survival, fertility, age-specific selection gradients, and fitness. Our results are the first, but definitely not the last word. Even so, the differences we found are not only statistically significant; they are also large enough to be biologically important. The difference in fitness in our experiments would result in a decline in the abundance of the wild-caught isolate, relative to N2, of 10% per day. That is enough to reduce the relative frequency of wild-caught from 0.5 to 0.03 within 10 generations (≈35 days). This is comparable to the fitness differences we have found in similar experiments comparing N2 with the longevity mutants clk-1 and daf-2 (Chen et al., 2006). However, the fitness differences in those experiments arise from different sources. The differences between N2 and clk-1 and N2 and daf-2 are almost completely due to early fertility (Chen et al., 2006), whereas the differences reported here between N2 and wild-caught show equal contributions from survival and fertility.
Both wild-caught and N2 C. elegans exhibit the dramatic (orders of magnitude) decline in selection gradients on survival and fertility with age. This decline has long been recognized as one of the keys to the evolution of senescence (Hamilton, 1966; Charlesworth, 1994, 2000); it implies that small positive changes in survival or fertility early in life can more than compensate for large deleterious changes later in life. Part of the reason for the magnitude of this decline is the extremely high rate of population growth exhibited by C. elegans under these laboratory conditions. Care is needed in making evolutionary inferences from such ecologically unrealistic conditions (e.g., E. coli as food, superabundant food, constant food, predator-free environment, etc.).
The biodemographic differences between wild-caught C. elegans and the N2 strain might result from the different geographical origins (and associated genetic divergence) between N2 and our California worms. Alternatively, they might result from adaptation by N2 to the laboratory environment (taken as a whole, including the food source). For example, it has been suggested that E. coli may have a negative influence on C. elegans (Gershon and Gershon, 2002; Johnson, 1984), and so wild-caught isolates may be more sensitive to such an influence and respond through altered behavior. The latter hypothesis is supported by the pattern of movement; wild-caught worms were more active, and remained more active as they aged, than N2 worms (Fig. 1C; P = 0.0038). The influence of alternative bacterial food sources on C. elegans fitness under natural conditions may provide additional insights on selection for life history characters.
Because environment and genetic background may influence gene expression patterns (Gems and Riddle, 2000; Shook and Johnson, 1999), further comparisons of aging and demography of C. elegans, in nature and in the laboratory, will provide an important frame of reference for detailed genetic analyses of aging in laboratory worm strains (Gershon and Gershon, 2002).
Acknowledgements
Thanks to Tom Johnson for helpful discussions on this subject and to two anonymous reviewers for helpful comments, to Steve Nadler, Greg Douhan, and John Chitambar for assistance in identification of worms, to the Caenorhabditis Genetic Center at University of Minnesota, St. Paul, for N2 C. elegans used in the research. Funding by NIH/NIA Grant #P01 AG022500-01 and NSF Grant DEB-0343820 is gratefully acknowledged.
References
- Arnaud J-F. Metapopulation genetic structure and migration pathways in the land snail Helix aspersa: influence of landscape heterogeneity. Landsc. Ecol. 2003;18:333–346. [Google Scholar]
- Barriere A, Felix M-A. High local genetic diversity and low outcossing rate in Caenorhabditis elegans natural populations. Curr. Biol. 2005;15:1176–1184. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- Basinger AJ. The European brown snail in California. Univ. Calif. Ag. Exp. Sta. Bull. 1931;515:3–22. [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey JR. Longevity: The Biology and Demography of Life Span. Princeton: Princeton University Press; 2003. [Google Scholar]
- Caswell H. Matrix Population Models: Construction, Analysis, and Interpretation. second ed. Sunderland, Massachusetts: Sinauer; 2001. [Google Scholar]
- Caswell H. Applications of Markov chains in demography. In: Langville AN, Stewart WJ, editors. MAM2006: Markov Anniversary Meeting. North Carolina: Boson Books, Raleigh; 2006. pp. 319–334. [Google Scholar]
- Caswell-Chen EP, Chen J, Lewis EE, Douhan GW, Nadler SA, Carey JR. Revising the standard wisdom of C. elegans natural history: ecology of longevity. Sci. Aging Knowl. Environ. 2005;40:pe30. doi: 10.1126/sageke.2005.40.pe30. [DOI: 10.1126 sageke.2005.40.pe30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Evolution in Age-structured Populations. Cambridge: Cambridge University Press; 1994. [Google Scholar]
- Charlesworth B. Fisher, Medawar, Hamilton and the evolution of aging. Genetics. 2000;156:927–931. doi: 10.1093/genetics/156.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Caswell-Chen EP. Why Caenorhabditis elegans adults sacrice their bodies to progeny. Nematology. 2003;5:641–645. [Google Scholar]
- Chen J, Caswell-Chen EP. Facultative vivipary is a life-history trait in C. elegans. J. Nematol. 2004;36:107–113. [PMC free article] [PubMed] [Google Scholar]
- Chen J, Carey JR, Ferris H. Comparative demography of isogenic populations of Caenorhabditis elegans. Exp. Gerontol. 2001;36:431–440. doi: 10.1016/s0531-5565(00)00225-4. [DOI] [PubMed] [Google Scholar]
- Chen J, Senturk D, Wang J-L, Müller, H-G, Carey JR, Caswell H, Caswell-Chen EP. Demographic demonstration of the fitness cost of extended longevity in C. elegans. J. Gerontol. 2006 doi: 10.1093/gerona/62.2.126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman & Hall; 1993. [Google Scholar]
- Gems D, Riddle DL. Longevity in Caenorhabditis elegans reduced by mating but not gamete production. Nature. 1996;379:723–725. doi: 10.1038/379723a0. [DOI] [PubMed] [Google Scholar]
- Gems D, Riddle DL. Defining wild-type life span in Caenorhabditis elegans. J. Gerontol. A Biol. Sci. Med. Sci. 2000;55(5):B215–B219. doi: 10.1093/gerona/55.5.b215. [DOI] [PubMed] [Google Scholar]
- Gershon H, Gershon D. Caenorhabditis elegans—a paradigm for aging research: advantages and limitations. Mech. Ageing Dev. 2002;123:261–274. doi: 10.1016/s0047-6374(01)00401-8. [DOI] [PubMed] [Google Scholar]
- Haber M, Schuüngel M, Putz A, Muüller, S, Hasert B, Schulenburg H. Evolutionary history of Caenorhabditis elegans inferred from microsatellites: Evidence for spatial and temporal genetic differentiation and the occurrence of outbreeding. Mol. Biol. Evol. 2005;22:160–173. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. The moulding of senescence by natural selection. J. Theor. Biol. 1966;12:12–45. doi: 10.1016/0022-5193(66)90184-6. [DOI] [PubMed] [Google Scholar]
- Hansen EL, Yarwood EA, Nicholas WL, Sayre FW. Differential nutritional requirements for reproduction of two strains of Caenorhabditis elegans in axenic culture. Nematologica. 1960;5:27–31. [Google Scholar]
- Hodgkin J, Barnes TM. More is not better: brood size and population growth in a self-fertilizing nematode. Proc. R. Soc. Lond. B. 1991;246:19–24. doi: 10.1098/rspb.1991.0119. [DOI] [PubMed] [Google Scholar]
- Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE. Analysis of the biological basis of aging in the nematode, with special emphasis on Caenorhabditis elegans. In: Mitchell DH, Johnson TE, editors. Invertebrate Models in Aging Research. Boca Raton, Florida: CRC Press; 1984. pp. 59–93. [Google Scholar]
- Johnson TE. Advantages and disadvantages of Caenorhabditis elegans for aging research. Exp. Gerontol. 2003;38:1329–1332. doi: 10.1016/j.exger.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Johnson TE, Hutchinson EW. Absence of strong heterosis for life span and other life history traits in Caenorhabditis elegans. Genetics. 1993;134:465–474. doi: 10.1093/genetics/134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson TE, Wu D, Tedesco P, Dames S, Vaupel JW. Age-specific demographic profiles of longevity mutants in Caenorhabditis elegans show segmental effects. J. Gerontol. A Biol. Sci. Med. Sci. 2001;56:B331–B339. doi: 10.1093/gerona/56.8.b331. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Klass MR. Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech. Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Lande R. A quantitative genetic theory of life history evolution. Ecology. 1982;63:607–615. [Google Scholar]
- Manly BFJ. Randomization, bootstrap and Monte Carlo Methods in Biology. New York: Chapman and Hall; 1997. [Google Scholar]
- Medawar PB. The Uniqueness of the Individual. London: Methuen; 1957. [Google Scholar]
- Muüller H-G, Wang J-L, Carey JR, Caswell-Chen EP, Chen J, Papadopoulos N, Yao F. Demographic window to aging in the wild: constructing life tables and estimating survival functions from marked individuals of unknown age. Aging Cell. 2004;3:125–131. doi: 10.1111/j.1474-9728.2004.00096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznick AZ, Gershon D. Experimentation with Nematodes. In: Yu BP, editor. Methods in Aging Research. Boca Raton, Florida: CRC Press; 1999. [Google Scholar]
- Rose M. Evolutionary Biology of Aging. New York: Oxford University Press; 1991. [Google Scholar]
- Shook DR, Johnson TE. Quantitative traitloci affecting survival and fertility-related traits in Caenorhabditis elegans show genotype-environment interactions, pleiotropy and epistasis. Genetics. 1999;153:1233–1243. doi: 10.1093/genetics/153.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns REC. Exotic mollusca in California. Science. 1900;11:655–659. doi: 10.1126/science.11.278.655. [DOI] [PubMed] [Google Scholar]
- Vassilieva LL, Lynch M. The rate of spontaneous mutation for life-history traits in Caenorhabditis elegans. Genetics. 1999;151:119–129. doi: 10.1093/genetics/151.1.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilieva LL, Hook AM, Lynch M. The fitness effects of spontaneous mutations in Caenorhabditis elegans. Evolution. 2000;54:1234–1246. doi: 10.1111/j.0014-3820.2000.tb00557.x. [DOI] [PubMed] [Google Scholar]
- Venette RC, Ferris H. Influence of bacterial type and density on population growth of bacterial-feeding nematodes. Soil Biol. Biochem. 1998;30:949–960. [Google Scholar]
- Walker DW, McColl G, Jenkins NL, Harris J, Lithgow GJ. Evolution of lifespan in C. elegans. Nature. 2000;405:296–297. doi: 10.1038/35012693. [DOI] [PubMed] [Google Scholar]
- Williams GC. Pleiotropy, natural selection, and the evolution of senescence. Evolution. 1957;11:398–411. [Google Scholar]