Abstract
Objective: MicroRNA (miRNA) expression is deregulated in many types of human cancers. We sought to investigate the expression patterns of miRNA in all major types of thyroid tumors, including tumors carrying distinct oncogenic mutations, and to explore the utility of miRNA profiling for the preoperative diagnosis of thyroid nodules.
Design: miRNA expression levels were detected in 60 surgically removed thyroid neoplastic and nonneoplastic samples and in 62 fine-needle aspiration (FNA) samples by RT-PCR using TaqMan MicroRNA Panel or individual miRNA sequence-specific primers. miRNA expression levels were calculated relative to normal thyroid tissue. All tumors were genotyped for most common mutations.
Results: Various histopathological types of thyroid tumors, including those deriving from the same cell type, showed significantly different profiles of miRNA expression. Oncocytic tumors, conventional follicular tumors, papillary carcinomas, and medullary carcinomas formed distinct clusters on the unsupervised hierarchical clustering analysis. Significant correlation between miRNA expression patterns and somatic mutations was observed in papillary carcinomas. A set of seven miRNAs (miR-187, miR-221, miR-222, miR-146b, miR-155, miR-224, and miR-197) that were most differentially overexpressed in thyroid tumors vs. hyperplastic nodules in the surgical samples was validated in the FNA samples, showing high accuracy of thyroid cancer detection.
Conclusions: In this study, we demonstrate that various histopathological types of thyroid tumors have distinct miRNA profiles, which further differ within the same tumor type, reflecting specific oncogenic mutations. A limited set of miRNAs can be used diagnostically with high accuracy to detect thyroid cancer in the surgical and preoperative FNA samples.
This study shows that a limited set of microRNAs can be used to detect thyroid cancer in surgical and pre-operative fine needle aspiration samples.
MicroRNAs (miRNAs) represent a recently identified class of endogenous noncoding RNAs that act as negative regulators of the protein-coding gene expression (1,2). miRNA dysregulation is common in cancer cells, and a rapidly growing number of studies provide evidence for miRNA involvement in carcinogenesis (3,4). miRNAs regulate the expression of well-known oncogenes and tumor suppressor genes (3,5). Specific subsets of overexpressed or down-regulated miRNAs have been identified in various cancer types, suggesting that aberrations in miRNA expression may be important in tumor development and progression (6,7,8).
Many miRNAs are expressed in a tissue-specific manner and exhibit expression profiles that are different between normal and neoplastic tissues and between tumors with distinct biological properties (6,9,10). Some data suggest that miRNA profiles allow reliable identification of the cell origin of tumors (11,12). However, it remains to be fully understood whether variable tumor types originating from the same cell type have different miRNA profiles and whether the profiles distinguish malignant tumors from benign hyperplastic/reactive processes.
In this regard, thyroid cancer represents an attractive model to study because it encompasses several histopathological tumor types originating from the same cell and tumors with distinct levels of differentiation. Most thyroid carcinomas originate from thyroid follicular cells and are subdivided into well-differentiated papillary carcinoma (PC) and follicular carcinoma (FC) (the latter further subclassified into conventional and oncocytic type) (13). Both PCs and FCs may progress to poorly differentiated carcinoma (PDC) or may completely lose differentiation to give rise to anaplastic carcinoma (AC). Follicular adenomas (FAs) are benign thyroid tumors and can be of either conventional type or oncocytic type. Less than 5% of cells within the thyroid gland are C-cells that give rise to medullary carcinoma (MC). Although several recent studies have assessed the miRNA expression profiles in specific types of thyroid cancer (14,15,16,17), miRNA expression signatures of all major types of thyroid neoplasms have not been analyzed and compared in a single study to our knowledge.
Significant information has been accumulated on carcinogenic mutations in thyroid cancer (18). Development of PCs, the most common thyroid malignant tumor, is known to involve the activation of the MAPK signaling pathway either as a result of BRAF or RAS point mutations or RET/PTC rearrangement. These mutually exclusive mutations are found in more than 70% of PCs and each of them is associated with the distinct phenotypical and biological properties of these tumors (19). FCs are known to harbor either RAS mutations or PAX8/PPARγ rearrangements, which are identified in 50–80% of conventional-type FCs and with lower prevalence in oncocytic tumors (20). It is not known, however, whether miRNA expression profiles are different among tumors carrying specific oncogenic mutations.
One of the main diagnostic problems in the thyroid field involves the preoperative assessment of thyroid nodules. Palpable nodules are common in the adult population, with an estimated prevalence in the United States in the range of 4–7% or 10–18 million affected individuals (21,22). Thyroid fine-needle aspiration (FNA) is an important method for preoperative evaluation of thyroid nodules, although in 10–20% of samples, the precise diagnosis cannot be reached, and they are reported as indeterminate or atypical (21). Most of these patients undergo surgery, although only 8–17% of surgically removed nodules are found to be malignant (21). Some improvement in the diagnostic accuracy can be achieved by additional testing of the FNA material for somatic mutations known to occur in thyroid tumors (23,24), although its sensitivity is limited because a significant proportion of PCs and FCs do not have any known mutations. Therefore, additional methods to improve the preoperative diagnosis are highly desirable and would result in a major impact on the clinical care.
In this study, we 1) determined and compared miRNA expression profiles of all major types of thyroid tumors, 2) explored the correlation between miRNA expression patterns and specific oncogenic mutations, and 3) determined the diagnostic utility of the detection of specific miRNAs in the preoperative assessment of thyroid nodules.
Materials and Methods
Thyroid tissue samples
Snap-frozen tissue from surgically removed thyroid samples was collected at the Department of Pathology, University of Cincinnati following the University of Cincinnati Institutional Review Board approval or obtained through the Cooperative Human Tissue Network. In total, 60 thyroid neoplastic and nonneoplastic samples were analyzed including 23 PCs [18 classical PCs and five follicular variant of PC (PC, FV)], nine FCs of conventional or oncocytic (Hürthle cell) type, eight FAs of conventional or oncocytic (Hürthle cell) type, four ACs, four PDCs, two MCs, five normal thyroid tissues, and five hyperplastic nodules. The age of patients ranged from 21–79 yr, and the female to male ratio was 3.3:1. All tumors were classified according to the widely accepted diagnostic histological criteria (13).
Thyroid FNA samples
Sixty-two thyroid FNA samples were collected as part of the prospective molecular study on thyroid FNA samples at the University of Cincinnati, which was approved by the University of Cincinnati Institutional Review Board. During the FNA procedure, half of the first or second pass of the aspirated material was directly collected into DNA/RNA preservative solution (Roche Molecular Biochemicals, Manheim, Germany) and frozen at −80 C.
RNA isolation
Total RNA was extracted from surgical specimens using Trizol reagent (Invitrogen, Carlsbad, CA) as previously described (25). RNA quality was assessed by 1% agarose gel electrophoresis in the presence of ethidium bromide. RNA samples that did not show intact 18S and 28S ribosomal bands were excluded from the study. Total nucleic acids were isolated from FNA samples using magnetic glass particles (Roche) according to the manufacturer’s protocol.
miRNA expression analysis
Quantitation of mature miRNA expression levels in thyroid tumors and normal thyroid tissue was performed by RT-PCR using TaqMan MicroRNA Assays Human Panel (Applied Biosystems Inc., Foster City, CA), which was designed to detect 158 human miRNAs. One tumor sample (PTC30) was assayed twice to test the reproducibility of the detection. A good correlation (0.912) in miRNA expression levels was found between the two runs. Expression of individual miRNAs was analyzed using miRNA sequence-specific primers (Applied Biosystems). Briefly, 10 ng total RNA was reverse transcribed using High-Capacity cDNA Archive kit (Applied Biosystems) followed by amplification on the ABI 7500 Real-Time PCR System (Applied Biosystems). All RT-PCR were performed in triplicate. Two endogenous controls were used for the normalization of RNA input: let7-a miRNA (according to the manufacturer’s protocol) and small nucleolar RNA RNU44 (Applied Biosystems). To evaluate the appropriateness of these endogenous controls for use in thyroid tissue, their expression levels were determined in 16 random thyroid tumor and normal samples. All samples demonstrated low variability in the expression levels of let7a and RNU44, validating their use as normalization controls. Two nonhuman miRNAs, ath-mir159a and cek-lin-4, were used as negative controls.
miRNA expression levels were calculated by relative quantitation using the ABI 7500 Real-Time PCR SDS 1.2 software (Applied Biosystems) and the fold expression changes were determined by 2−ΔΔCT method (26). The data are presented as the fold change of miRNA expression in tumors relative to normal thyroid tissues after normalization to an endogenous control (let7-a or RNU44).
Detection of mutations
Tumor DNA was tested for BRAF V600E, NRAS codon 61, HRAS codon 61, KRAS codons 12/13 mutations using real-time LightCycler PCR as previously reported (19). RET/PTC1, RET/PTC3, and PAX8/PPARγ rearrangements were detected from RNA by RT-PCR as previously reported (20,25).
Statistical analysis
Agglomerative hierarchical clustering between thyroid specimens was performed in R software. miRNA expression data underwent filtering by computation of the number of missingness for each miRNA. The purpose of the filtering was to remove miRNAs with no detectable expression across all thyroid specimens that may introduce noise to the clustering. A subset of 59 miRNAs remained after filtering and was used for hierarchical clustering.
For class comparisons to identify differentially expressed miRNAs, a t test was first performed for each miRNA for two class comparisons,the and Benjamini-Hochberg procedure (27) was applied to control the false discovery rate at 5% (i.e. among detected miRNAs, 5% of them are false positives on average). For class predictions in the TaqMan MicroRNA Human Panel data, a nearest-shrunken-centroid method by Prediction Analysis of Microarray (28) software was applied. The software integrates selection of predictive miRNAs while constructing the prediction model and performs 10-fold cross-validation. Linear discriminant analysis was used for the class prediction of the set of individual miRNAs without feature selection.
For individually studied miRNAs, one-way ANOVA was used to detect the statistical significance of mutation effects for each miRNA. Principal component analysis (PCA) was applied to provide an unsupervised visualization and investigation of the relationship between miRNA expression and mutation type. For each pair of mutation-specific groups, PCA was performed to project the samples to the first principal component, and a simple t test was applied to test for significance of separation between the two groups.
Results
Overall miRNA expression in thyroid normal and tumor tissues
Initially, a set of five normal thyroid tissues, two hyperplastic nodules, and 24 thyroid tumors including nine PCs, five FCs, four FAs, two ACs, two PDCs, and two MCs was used to evaluate the expression of 158 human mature miRNAs. Overall, 148 (94%) of miRNAs were found to be expressed in normal and hyperplastic thyroid tissue. In the majority of thyroid tumors, 47 of 148 miRNAs (32%) were consistently up-regulated when compared with normal tissue, and 57 of 148 (38%) were down-regulated with more than a 2-fold change.
When the expression levels of miRNAs in all well-differentiated thyroid carcinomas (PCs, FCs, and MCs) were compared with those in less differentiated carcinomas (PDCs and ACs), 31 miRNAs showed higher expression levels in the well-differentiated carcinomas, whereas 27 miRNAs were expressed at higher levels in less differentiated carcinomas, showing no statistically significant difference between the numbers of up-regulated and down-regulated genes in the two groups (P = 0.69).
Expression of miRNAs in various types of thyroid tumors
To determine whether different histopathological types of thyroid tumors have distinct miRNA profiles, the unsupervised hierarchical clustering analysis of miRNA expression was performed. It revealed four major clusters: oncocytic follicular tumors (adenomas and carcinomas), conventional follicular tumors (adenomas and carcinomas), PCs and MCs (Fig. 1). The first three clusters were located closer to each other, whereas the MC cluster was at the greatest distance, consistent with their different cell type origins, i.e. thyroid follicular cells and C-cells. The oncocytic tumor cluster was most segregated of the three follicular cell-derived tumor clusters. Less differentiated tumors (PDCs and ACs) did not form distinct clusters and were situated either close or within the papillary or follicular clusters or separately, supporting their origin from the well-differentiated PCs and FCs and their propensity for profound dedifferentiation.
Figure 1.
Cluster dendrogram of miRNA expression of thyroid tumors showing four major clusters: oncocytic FC (OFC) and FA (OFA), conventional FC (CFC) and FA (CFA), PC, and MC.
Next, we searched for individual miRNAs that had the highest levels of overexpression in specific tumor types. The top 10 up-regulated miRNAs in each type of malignant and benign thyroid tumor are shown in Tables 1 and 2. There was virtually no overlap in the highly expressed miRNAs between MCs and the rest of the tumors, all of which derive from thyroid follicular cells. Seven miRNAs, miR-187, miR-221, miR-222, miR-181b, miR-146b, miR-155, and miR-224, were most consistently overexpressed in all follicular cell-derived carcinomas, although their expression levels varied significantly between individual tumor types. In ACs, these miRNAs were overexpressed at lower levels, whereas miR-302c, miR-205, and miR-137 showed most dramatic up-regulation. Additionally, miRNA-339, miR-183, and miR-197 were highly expressed in oncocytic follicular carcinomas. Among benign adenomas, tumors of conventional type and oncocytic type had quite distinct sets of mostly up-regulated miRNAs, with miR-200a being at the top of the list in conventional follicular adenomas and miR-31 in oncocytic adenomas.
Table 1.
Ten most up-regulated miRNAs in various thyroid tumors: PCs, conventional and oncocytic FCs, and PDCs
| PCs
|
Conventional FCs
|
Oncocytic FCs
|
PDCs
|
||||
|---|---|---|---|---|---|---|---|
| miRNA | Fold | miRNA | Fold | miRNA | Fold | miRNA | Fold |
| miR-187 | 73.7 | miR-187 | 33.4 | miR-187 | 53.7 | miR-181b | 24.7 |
| miR-221 | 19.1 | miR-181b | 13.6 | miR-221 | 46.9 | miR-187 | 23.5 |
| miR-222 | 17.2 | miR-200a | 6.7 | miR-339 | 42.5 | miR-221 | 20.6 |
| miR-181b | 14.4 | miR-224 | 6.2 | miR-183 | 26.6 | miR-129 | 17.3 |
| miR-146b | 10.5 | miR-182 | 5.7 | miR-222 | 26.0 | miR-222 | 13.5 |
| miR-155 | 9.5 | miR-155 | 5.5 | miR-181b | 19.7 | miR-146b | 12.4 |
| miR-122a | 8.9 | miR-222 | 4.5 | miR-182 | 15.6 | miR-339 | 6.1 |
| miR-31 | 7.5 | miR-221 | 3.6 | miR-213 | 14.3 | miR-183 | 5.7 |
| miR-205 | 6.8 | miR-96 | 3.5 | miR-96 | 10.3 | miR-213 | 4.3 |
| miR-224 | 6.2 | miR-146b | 2.8 | miR-197 | 7.2 | miR-181a | 3.9 |
The fold change of miRNA expression was calculated relative to normal thyroid tissue. The bolded miRNAs signify those that were also significantly overexpressed in thyroid tumors as compared with hyperplastic nodules.
Table 2.
Ten most up-regulated miRNAs in various thyroid tumors: ACs, conventional and oncoytic FAs, and MCs
| ACs
|
Conventional FAs
|
Oncocytic FAs
|
MCs
|
||||
|---|---|---|---|---|---|---|---|
| miRNA | Fold | miRNA | Fold | miRNA | Fold | miRNA | Fold |
| miR-302c | 114.2 | miR-200a | 35.0 | miR-31 | 35.2 | miR-323 | 142.2 |
| miR-205 | 75.6 | miR-181b | 23.1 | miR-339 | 18.2 | miR-370 | 136.4 |
| miR-137 | 64.9 | miR-200b | 18.1 | miR-183 | 17.5 | miR-129 | 123.3 |
| miR-187 | 19.1 | miR-339 | 13.4 | miR-182 | 15.6 | miR-137 | 116.4 |
| miR-214 | 16.3 | miR-224 | 11.0 | miR-181b | 14.1 | miR-10a | 63.6 |
| miR-181b | 15.3 | miR-205 | 10.1 | miR-221 | 12.8 | miR-124a | 61.9 |
| miR-155 | 13.2 | miR-210 | 9.6 | miR-96 | 10.8 | miR-224 | 55.2 |
| miR-224 | 12.0 | miR-190 | 9.1 | miR-182 | 8.4 | miR-127 | 52.3 |
| miR-222 | 9.5 | miR-328 | 8.1 | miR-224 | 6.6 | miR-9 | 42.1 |
| miR-221 | 9.4 | miR-342 | 7.9 | miR-203 | 6.2 | miR-154 | 32.3 |
The fold change of miRNA expression was calculated relative to normal thyroid tissue. The bolded miRNAs signify those that were also significantly overexpressed in thyroid tumors as compared with hyperplastic nodules.
Expression of miRNAs in tumors carrying different oncogenic mutations
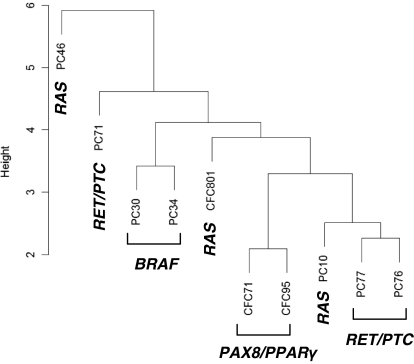
To explore whether miRNA expression profiles vary in tumors with different initiating mutations, all PCs and FCs were tested for the presence of BRAF and RAS point mutations and RET/PTC and PAX8/PPARγ rearrangements. Among PCs, two tumors were positive for RET/PTC1, one for RET/PTC3, two for BRAF, and two for NRAS, and two were found negative for any of these mutations. Both PCs positive for RAS mutation were the follicular variants. Among FCs, two were positive for PAX8/PPARγ and one for NRAS, and two were negative for these alterations. The unsupervised hierarchical clustering analysis of miRNA expression in the tumors positive for mutations was performed. It demonstrated that BRAF-, RET/PTC-, and PAX8/PPARγ-positive tumors formed individual clusters, whereas tumors with RAS mutations did not form a separate cluster and were positioned between other clusters (Fig. 2). These pointed toward the possible variation in miRNA signatures depending on the mutational status.
Figure 2.
Cluster dendrogram of miRNA expression in thyroid tumors with different mutations.
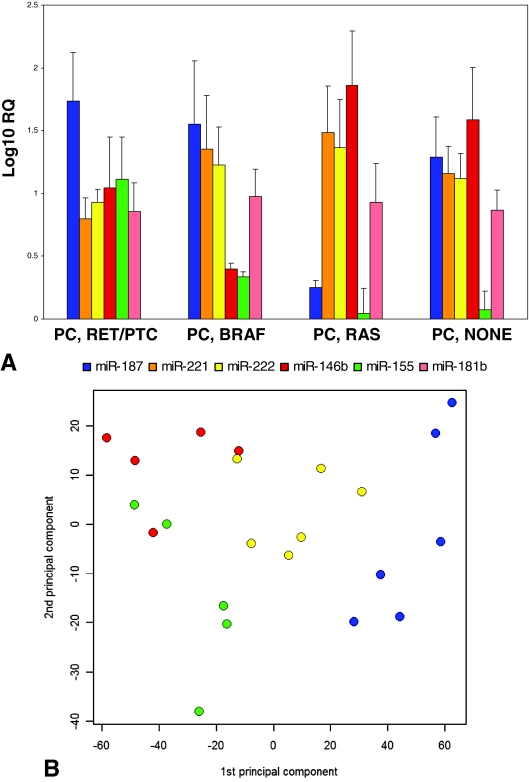
To explore this possibility further, an additional 14 PCs with BRAF, RAS, RET/PTC, or none of these mutations were analyzed for the expression of six miRNAs with the highest overall expression levels in PCs: miR-187, miR-221, miR-222, miR-181b, miR-146b, and miR-155 (Fig. 3A). The one-way ANOVA analysis of all 23 PCs revealed significant differences in the expression levels of five of these miRNAs between the groups (P = 0.09 to P < 0.0001), whereas the expression of miR-181b was not significantly different (P = 0.95). PCA was used for an unsupervised assessment of the relationship between mutation type and expression levels of the six miRNAs. It revealed a strong relationship between miRNA expression and mutational status (Fig. 3B). Similar results were obtained using pairwise comparison between the individual groups, which revealed significant differences in the expression profiles between all group pairs (P = 0.003 or lower) except for the RET/PTC and RAS groups (P = 0.46).
Figure 3.
A, Expression levels of selected miRNAs in PCs with various mutations (mean ± se); B, PCA of log-transformed data for all mutational group probe sets. Blue, BRAF mutants; green, RAS mutants; red, RET/PTC mutants; yellow, tumors with no mutation.
Diagnostic utility of miRNA detection in clinical thyroid samples
The major challenge in the preoperative FNA diagnosis of thyroid nodules is to differentiate between the follicular-patterned thyroid cancers and hyperplastic nodules, which are common in the general population (21,22). Therefore, a diagnostically valid assay must distinguish thyroid tumors not only from normal thyroid tissue but also from hyperplastic nodules.
To explore the diagnostic utility of miRNA profiling in this setting, we selected a subset of miRNAs based on 1) at least 2-fold overexpression in thyroid cancers compared with hyperplastic nodules (Tables 1 and 2, data in bold), and 2) their up-regulation in different types of thyroid cancer. A subset of seven miRNAs (miR-187, miR-221, miR-222, miR-146b, miR-155, miR-224, and miR-197) was selected. Expression levels of these miRNAs were evaluated in surgical samples from additional 14 malignant thyroid tumors (six PCs, four FCs, two ACs, and two PDCs) and three hyperplastic nodules, showing different patterns of expression in all types of thyroid cancers compared with hyperplastic nodules (Fig. 4A). This set of miRNAs was highly effective in separating thyroid cancer from hyperplastic nodules in all surgical samples studied, because supervised prediction analysis by linear discriminant analysis misdiagnosed only one (2.4%) of 41 malignant tumors and hyperplastic nodules.
Figure 4.
A, Expression levels of selected miRNAs in various types of thyroid cancer based on the analysis of surgically removed tumors (mean ± se); B, expression of selected miRNAs in thyroid preoperative FNA samples from patients who subsequently underwent surgery. The surgical pathology diagnoses are shown at the bottom. CFC, Conventional FC; HN, hyperplastic nodules; OFC, oncocytic FC.
Then, the diagnostic accuracy of this panel was validated in 62 consecutive thyroid FNA samples. Thirteen of these patients underwent surgery based on cytological diagnosis (four positive and eight atypical cytology) or clinical suspicion (one case), yielding the diagnosis of malignant tumor in eight cases (seven PCs and one FC of oncocytic type) and hyperplastic nodule in five cases. Expression of miR-187, miR-221, miR-222, miR-146b, miR-155, miR-224, and miR-197 was analyzed in these FNA samples without knowing the surgical pathology diagnosis. All of the eight malignant cases revealed more than 2-fold overexpression of one to six miRNAs tested (Fig. 4B). Among them, seven PCs showed at least three up-regulated miRNAs, whereas one case of oncocytic FC demonstrated up-regulation of miR-221 only. Among the four hyperplastic nodules that were surgically removed based on atypical cytology, only one case showed more than 2-fold overexpression of one of these miRNAs (miR-197). The other 49 samples from patients that did not undergo surgery revealed no up-regulation of any of these miRNAs in 46 cases, up-regulation of one miRNA in two cases, and strong up-regulation of three miRNAs (miR-221, miR-222, and miR-146b) in one case. The latter case had atypical cytology but no surgery during the follow-up and therefore was removed from further analysis. All other cases had negative cytology and no further evidence of thyroid disease on follow-up and were assumed to be negative for malignancy. Based on these data, the panel of seven miRNAs allowed discrimination between malignant tumors and hyperplastic nodules with high accuracy. Specifically, when at least one miRNA was overexpressed more than 2-fold, the sensitivity of tumor detection was 100%, specificity 94%, and accuracy 95%. When three or more miRNAs were up-regulated, the sensitivity of tumor detection was 88%, specificity 100%, and accuracy 98%.
Discussion
In this study, we demonstrate that various histopathological types of thyroid tumors derived from the same cell have distinct miRNA profiles, which further differ within the same tumor type reflecting specific oncogenic mutations found in these tumors. A limited set of miRNAs can be used diagnostically with high accuracy to detect thyroid cancer in the surgical and preoperative FNA samples.
From the early stages of their discovery, it has been known that many miRNAs are expressed in a tissue-specific manner (1,2). Therefore, it was not surprising to find markedly different profiles of miRNA expression between C-cell-derived thyroid MCs and all other thyroid tumors that derives from follicular cells. However, the results of this study show that miRNA profiles have significant variability between different histopathological types of thyroid tumors that originate from the same cell.
The importance of this information for classification of thyroid tumors and refining their scheme of progression is at least 2-fold. First, it supports the independent notion that oncocytic tumors represent a distinct class of thyroid neoplasms, which has been a subject of a longstanding debate in the field. Although the most recent World Health Organization classification of thyroid tumors designates oncocytic thyroid adenomas and carcinomas as a variant of follicular tumors (13), some histological features and mutational profiles (29) support their independent origin. The results of miRNA analysis provide more evidence to support the latter notion, because it showed that oncocytic tumors had a distinct set of up-regulated miRNA and clustered separately from conventional follicular tumors. Second, the clusters of follicular tumor and oncocytic tumor included both adenomas and carcinomas, supporting the possibility of stepwise progression for each tumor type.
Some reports have suggested that overall, most miRNAs are expressed at lower levels in tumors and tumor-derived cell lines compared with normal tissues and in poorly differentiated tumors compared with their well-differentiated counterparts (11,30). Other studies have not confirmed such a tendency (12). We did not observe this trend in thyroid tumors, where a significant proportion of miRNAs was found to be up-regulated and overexpressed at high levels in tumors compared with normal thyroid tissue. Likewise, we did not observe significant differences in miRNA expression in tumors depending on their differentiation status.
Most interestingly, we observed that miRNA expression profiles had substantial variability within specific tumor types. The first study of miRNA expression in thyroid PCs has reported the overall highest up-regulation of miR-221, miR-222, and miR-146b (14). We confirmed these data but show that these miRNAs are not equally expressed in all tumors of this type and mostly up-regulated in tumors carrying BRAF mutation. Another study has shown significant up-regulation of miR-221, miR-222, and miR-181b in PCs compared with normal thyroid tissue (15). Our data suggest that of these three markers, miR-181b is overexpressed in virtually all types of follicular cell-derived thyroid tumors and also in thyroid hyperplastic nodules. We also identified several additional miRNAs that are highly up-regulated in PCs. One of them, miR-187, appears to be the most up-regulated miRNA in tumors harboring RET/PTC rearrangement and RAS mutations but is expressed at significantly lower levels in tumors with BRAF mutation. miR-224, in contrast, is a pan-PC marker because it is overexpressed, albeit at lower levels, in all PCs.
Overall, we observed significant correlation between the mutational status of PCs and miRNA expression. The oncogenic mutations in PCs, RET/PTC, BRAF, and RAS are all capable of activation of the MAPK pathway and rarely overlap in the same tumor, suggesting that activation of a single effector of this pathway is sufficient for transformation (31). However, each of these mutations is associated with distinct coding gene expression profiles, phenotypical characteristics, and biological properties of PCs (19,32,33). Of those, the association between BRAF mutation and tumor recurrence and treatment failure has produced the most substantial effect on clinical management of patients with thyroid cancer (34). Previous attempts have been made to identify the dysregulated miRNAs in thyroid cell lines carrying mutant BRAF and RET/PTC1 in comparison with the normal thyroid cell line (35,36). Herein, we show that miRNA expression profiles of human thyroid tumors carrying BRAF and other mutations are substantially different. Although the nature of the correlation between BRAF mutation and miRNA expression remains unclear, this finding suggests that miRNA profiling may also be potentially used as a prognostic marker for thyroid cancer.
Weber et al. (17) have found significant overexpression of miR-197 and miR-346 in FCs. In our series, up-regulation of miR-197 was predominantly seen in the oncocytic type of FC, whereas miR-346 was not studied. We identified several other miRNAs up-regulated in these tumors, with miR-187 showing the highest levels of overexpression in both conventional and oncocytic FCs.
The results obtained by the analysis of surgically removed tumors allowed us to assemble a small panel of miRNAs for the preoperative diagnosis of thyroid nodules. Using a large series of well-characterized FNA samples, we demonstrated the feasibility of miRNA testing in FNA samples and provided evidence for high diagnostic potential of a limited panel of miRNAs in cancer diagnosis. Despite overall lower levels of miRNA overexpression in the FNA samples compared with surgical material, the panel provided high diagnostic accuracy. Although our findings need to be further confirmed in a larger series of samples, they lay the foundation for the use of miRNA profiling as an effective diagnostic tool for the preoperative assessment of thyroid nodules.
Footnotes
This work was supported by National Institutes of Health Grant R01 CA88041 (to Y.E.N.). G.C.T. is partially supported by the National Institutes of Health (1 KL2 RR024154-02).
Disclosure Statement: The authors have nothing to disclose.
First Published Online February 12, 2008
Abbreviations: AC, Anaplastic carcinoma; FA, follicular adenoma; FC, follicular carcinoma; FNA, fine-needle aspiration; MC, medullary carcinoma; miRNA, microRNA; PC, papillary carcinoma; PCA, principal component analysis; PDC, poorly differentiated carcinoma.
References
- Bartel DP 2004 MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 [DOI] [PubMed] [Google Scholar]
- Ambros V 2004 The functions of animal microRNAs. Nature 431:350–355 [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ 2006 Oncomirs: microRNAs with a role in cancer. Nat Rev 6:259–269 [DOI] [PubMed] [Google Scholar]
- Negrini M, Ferracin M, Sabbioni S, Croce CM 2007 MicroRNAs in human cancer: from research to therapy. J Cell Sci 120:1833–1840 [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM 2006 MicroRNA signatures in human cancers. Nat Rev 6:857–866 [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC 2006 Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9:189–198 [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, Menard S, Palazzo JP, Rosenberg A, Musiani P, Volinia S, Nenci I, Calin GA, Querzoli P, Negrini M, Croce CM 2005 MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65:7065–7070 [DOI] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS 2005 MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65:6029–6033 [DOI] [PubMed] [Google Scholar]
- Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, Iuliano R, Palumbo T, Pichiorri F, Roldo C, Garzon R, Sevignani C, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM 2005 A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med 353:1793–1801 [DOI] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T 2007 MicroRNA expression profiling in prostate cancer. Cancer Res 67:6130–6135 [DOI] [PubMed] [Google Scholar]
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR 2005 MicroRNA expression profiles classify human cancers. Nature 435:834–838 [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM 2006 A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA 103:2257–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLellis RA, Lloyd RV, Heitz PU, Eng C, eds 2004 World Health Organization classification of tumours: pathology and genetics of tumours of endocrine organs. Lyon, France: IARC Press [Google Scholar]
- He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S, Kloos RT, Croce CM, de la Chapelle A 2005 The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA 102:19075–19080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallante P, Visone R, Ferracin M, Ferraro A, Berlingieri MT, Troncone G, Chiappetta G, Liu CG, Santoro M, Negrini M, Croce CM, Fusco A 2006 MicroRNA deregulation in human thyroid papillary carcinomas. Endocr Relat Cancer 13:497–508 [DOI] [PubMed] [Google Scholar]
- Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, Liu CG, Petrocca F, Troncone G, Calin GA, Scarpa A, Colato C, Tallini G, Santoro M, Croce CM, Fusco A 2007 Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene 26:7590–7595 [DOI] [PubMed] [Google Scholar]
- Weber F, Teresi RE, Broelsch CE, Frilling A, Eng C 2006 A limited set of human MicroRNA is deregulated in follicular thyroid carcinoma. J Clin Endocrinol Metab 91:3584–3591 [DOI] [PubMed] [Google Scholar]
- Kondo T, Ezzat S, Asa SL 2006 Pathogenetic mechanisms in thyroid follicular-cell neoplasia. Nat Rev 6:292–306 [DOI] [PubMed] [Google Scholar]
- Adeniran AJ, Zhu Z, Gandhi M, Steward DL, Fidler JP, Giordano TJ, Biddinger PW, Nikiforov YE 2006 Correlation between genetic alterations and microscopic features, clinical manifestations, and prognostic characteristics of thyroid papillary carcinomas. Am J Surg Pathol 30:216–222 [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Lynch RA, Biddinger PW, Alexander EK, Dorn GW, 2nd, Tallini G, Kroll TG, Nikiforov YE 2003 RAS point mutations and PAX8-PPARγ rearrangement in thyroid tumors: evidence for distinct molecular pathways in thyroid follicular carcinoma. J Clin Endocrinol Metab 88:2318–2326 [DOI] [PubMed] [Google Scholar]
- Mazzaferri EL 1992 Thyroid cancer in thyroid nodules: finding a needle in the haystack. Am J Med 93:359–362 [DOI] [PubMed] [Google Scholar]
- Mazzaferri EL 1993 Management of a solitary thyroid nodule. N Engl J Med 328:553–559 [DOI] [PubMed] [Google Scholar]
- Salvatore G, Giannini R, Faviana P, Caleo A, Migliaccio I, Fagin JA, Nikiforov YE, Troncone G, Palombini L, Basolo F, Santoro M 2004 Analysis of BRAF point mutation and RET/PTC rearrangement refines the fine-needle aspiration diagnosis of papillary thyroid carcinoma. J Clin Endocrinol Metab 89:5175–5180 [DOI] [PubMed] [Google Scholar]
- Xing M, Tufano RP, Tufaro AP, Basaria S, Ewertz M, Rosenbaum E, Byrne PJ, Wang J, Sidransky D, Ladenson PW 2004 Detection of BRAF mutation on fine needle aspiration biopsy specimens: a new diagnostic tool for papillary thyroid cancer. J Clin Endocrinol Metab 89:2867–2872 [DOI] [PubMed] [Google Scholar]
- Nikiforova MN, Caudill CM, Biddinger P, Nikiforov YE 2002 Prevalence of RET/PTC rearrangements in Hashimoto’s thyroiditis and papillary thyroid carcinomas. Int J Surg Pathol 10:15–22 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 25:402–408 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I 2001 Controlling the false discovery rate in behavior genetics research. Behav Brain Res 125:279–284 [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Hastie T, Narasimhan B, Chu G 2002 Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc Natl Acad Sci USA 99:6567–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforova MN, Biddinger PW, Caudill CM, Kroll TG, Nikiforov YE 2002 PAX8-PPARγ rearrangement in thyroid tumors: RT-PCR and immunohistochemical analyses. Am J Surg Pathol 26:1016–1023 [DOI] [PubMed] [Google Scholar]
- Gaur A, Jewell DA, Liang Y, Ridzon D, Moore JH, Chen C, Ambros VR, Israel MA 2007 Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res 67:2456–2468 [DOI] [PubMed] [Google Scholar]
- Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA 2003 High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 63:1454–1457 [PubMed] [Google Scholar]
- Xing M, Westra WH, Tufano RP, Cohen Y, Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, Tolaney S, Holt EH, Hui P, Umbricht CB, Basaria S, Ewertz M, Tufaro AP, Califano JA, Ringel MD, Zeiger MA, Sidransky D, Ladenson PW 2005 BRAF mutation predicts a poorer clinical prognosis for papillary thyroid cancer. J Clin Endocrinol Metab 90:6373–6379 [DOI] [PubMed] [Google Scholar]
- Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K, Gauger P, Doherty G, Thompson NW, Hanash S, Koenig RJ, Nikiforov YE 2005 Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene 24:6646–6656 [DOI] [PubMed] [Google Scholar]
- Xing M 2005 BRAF mutation in thyroid cancer. Endocr Relat Cancer 12:245–262 [DOI] [PubMed] [Google Scholar]
- Cahill S, Smyth P, Denning K, Flavin R, Li J, Potratz A, Guenther SM, Henfrey R, O’Leary JJ, Sheils O 2007 Effect of BRAFV600E mutation on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer 6:21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill S, Smyth P, Finn SP, Denning K, Flavin R, O’Regan EM, Li J, Potratz A, Guenther SM, Henfrey R, O’Leary JJ, Sheils O 2006 Effect of ret/PTC 1 rearrangement on transcription and post-transcriptional regulation in a papillary thyroid carcinoma model. Mol Cancer 5:70 [DOI] [PMC free article] [PubMed] [Google Scholar]