Abstract
Background and objectives: Multidrug-resistant gram-negative bacteria are rapidly spreading throughout the world. The epidemiology of multidrug-resistant gram-negative bacteria in patients who require chronic hemodialysis has not been previously studied.
Design, setting, participants, & measurements: A prospective cohort study of an outpatient hemodialysis unit was conducted. Serial surveillance cultures for multidrug-resistant gram-negative bacteria, vancomycin-resistant enterococci, and methicillin-resistant Staphylococcus aureus were collected from patients who were undergoing chronic hemodialysis.
Results: Nineteen (28%) of the 67 enrolled patients were colonized with one or more antimicrobial-resistant bacteria at study enrollment. Eleven (16%), nine (13%), and three (5%) patients were colonized with multidrug-resistant gram-negative bacteria, vancomycin-resistant enterococci, and methicillin-resistant Staphylococcus aureus, respectively. Independent risk factors associated with harboring multidrug-resistant gram-negative bacteria at enrollment were residence in a long-term care facility and antibiotic exposure for ≥7 d in the previous 3 mo. Twenty-two (40%) of 55 patients who had follow-up cultures acquired at least one antimicrobial-resistant bacterium. A total of 20, 15, and 13% of patients acquired multidrug-resistant gram-negative bacteria, vancomycin-resistant enterococci, and methicillin-resistant Staphylococcus aureus, respectively. Antibiotic exposure was the only independent risk factor for multidrug-resistant gram-negative bacteria acquisition. Endogenous multidrug-resistant gram-negative bacteria acquisition was detected among 69% of acquired multidrug-resistant gram-negative bacterial strains.
Conclusions: The prevalence and acquisition of multidrug-resistant gram-negative bacteria surpassed that of vancomycin-resistant enterococci and methicillin-resistant Staphylococcus aureus. Endogenous acquisition, as opposed to patient-to-patient spread, was the predominant mechanism of acquisition. Residence in a long-term care facility and antibiotic exposure may be important factors promoting the spread of multidrug-resistant gram-negative bacteria among this patient population.
Rates of antibiotic-resistant bacteria are among the highest in patients who require chronic hemodialysis (1). Vancomycin-resistant enterococci (VRE) and methicillin-resistant Staphylococcus aureus (MRSA) are among the antimicrobial-resistant bacteria that have been intensely investigated in this patient population (1–3). In the past few years, however, a concerning increase in the prevalence of infections caused by multidrug-resistant gram-negative bacteria (MDRGN) has been documented in other patient populations (4–6). Infections that are caused by these MDRGN are associated with up to five times higher mortality rates compared with infections that are caused by susceptible gram-negative bacteria (7). Among chronic hemodialysis patients, approximately 25% of blood stream infections are caused by gram-negative bacteria (8), and this percentage is increasing steadily (9). Despite these rising rates, antimicrobial resistance among gram-negative bacteria has not been previously investigated in the population of those who undergo chronic hemodialysis.
A prospective surveillance study was therefore performed to describe the clinical epidemiology of MDRGN among patients who require chronic hemodialysis. The goals of this study were (1) to determine the percentage of chronic hemodialysis patients who are colonized with MDRGN and compare these rates with VRE and MRSA colonization, (2) to identify the risk factors that are associated with MDRGN colonization, and (3) to determine the mechanism of MDRGN acquisition. For achieving these goals, cultures were obtained at three intervals from patients who were receiving chronic hemodialysis at an outpatient unit to determine whether they were colonized with MDRGN, VRE, or MRSA at enrollment and whether they acquired these antimicrobial-resistant bacteria over time. To understand the transmission dynamics of these antimicrobial-resistant bacteria and identify clonal spread, we performed DNA fingerprinting using pulsed-field gel electrophoresis (PFGE).
Materials and Methods
The study was performed at an outpatient hemodialysis unit affiliated with Caritas St. Elizabeth Medical Center from March 1, 2005, to September 1, 2005. The unit has 16 beds centered around a nursing station and one isolation room. Each hemodialysis nurse provided care to two patients during each 3- to 4-h hemodialysis shift, with one circulating nurse supervisor. Infection control precautions for patients who were known to be colonized with VRE or MRSA followed the Centers for Disease Control and Prevention guidelines (10). There were no specific precautions for patients who were colonized or infected with MDRGN. During the study period, 85 patients received chronic hemodialysis at this unit. After obtaining approval from the study institution's internal review board, all patients who were receiving chronic hemodialysis at the study site were approached for study participation.
Definitions
Multidrug resistance among gram-negative bacteria was defined as resistance to three or more of the following antimicrobials: Ceftazidime, ciprofloxacin, meropenem, gentamicin, ampicillin/sulbactam, or piperacillin/tazobactam. These antimicrobial were chosen because they are commonly prescribed by physicians.
Patient acquisition of MDRGN was defined as a patient in whom MDRGN was not recovered from any specimens that were collected at baseline but was recovered during one or more follow-up cultures. Endogenous MDRGN acquisition, implying emergence of multidrug resistance from a previous non-MDRGN strain, was defined as recovery of a non-MDRGN at enrollment among patients who acquired MDRGN. The non-MDRGN was required to be of the same species and have the identical PFGE pattern as the acquired MDRGN (11).
Specimen Collection
Specimens were collected from each patient at study enrollment and after approximately 2 and 4 mo after enrollment, during two follow-up periods. Specimens from the rectum, nares, and skin (a 5 × 5-cm area in the jugulodigastric and inguinal region) were obtained using Starplex cotton swabs (Etobicoke, ON, Canada) for the detection of VRE, MRSA, and MDRGN. One swab was used for both skin sites. For the rectal specimen, two swabs were collected. The first swab was processed for the identification and susceptibility testing of MDRGN and VRE as outlined next. The second swab was frozen and stored at −70°C for future recovery of non-MDRGN isolates to determine whether endogenous acquisition of MDRGN had occurred in patients with a positive follow-up culture for MDRGN.
Environmental cultures were obtained from a 5 × 5-cm area of the patient's dialysis machines (dialysis control screen) and over the arm rest of his or her chair during the patient's hemodialysis shift at enrollment and during the two follow-up periods using a Straplex cotton swab. Only one swab was used for both environmental sites. All specimens were sent to the microbiology laboratory and processed within 24 h of collection.
Data Collection
Medical charts were reviewed for patient demographic data, comorbidities, underlying renal disease, and residency in a long-term care facility (LTCF). The Charlson score was used to provide a composite score of comorbid conditions (12). A nonambulatory status, fecal incontinence, and presence of a draining wound or colostomy were prospectively recorded during the study. Data on antibiotic and hospital exposure ≥7 d in the previous 3 mo before enrollment were also collected. During the period between enrollment and each follow-up period, potential factors that would increase the likelihood of MDRGN acquisition were also collected, including antibiotic and hospital exposure, as defined previously, presence of a new wound or colostomy, and fecal incontinence.
Microbiological Methods
Identification and Susceptibility Testing.
For identification of MDRGN, skin and rectal swabs were inoculated in 2% thyoglycolate broth and incubated at 35°C. Specimens with evidence of growth within 24 to 48 h were streaked onto two MacConkey agar plates supplemented with 2 μg/ml levofloxacin and 2 μg/ml ceftazidime each and incubated again at 35°C. Supplementation with low concentrations of these antimicrobials prevented the growth of antimicrobial-susceptible gram-negative bacteria. After 24 to 48 h, gram-negative colonies were subcultured onto MacConkey agar to isolate pure colonies, and species identification was performed using API-Rapid Strep Strips (bioMerieux Vitex, Durham, NC), according to the manufacturer's instructions. Antibiotic susceptibility testing was performed by the disk diffusion susceptibility testing and interpreted according to Clinical and Laboratory Standards Institute guidelines for each species (13).
For identification of VRE, skin and rectal swabs were also plated onto colistin and nalidixic acid media supplemented with 8 μg of vancomycin and incubated for 24 to 48 h at 35°C. Enterococci isolates were identified according to standard microbiology procedures (14), and vancomycin resistance was determined by agar dilution method, using an minimum inhibitory concentration ≥8 μg/ml as the cutoff for vancomycin resistance.
For identification of MRSA, nasal swabs from each patient were plated on mannitol salt agar (Becton-Dickinson, Sparks, MD) and incubated for 24 h at 35°C. Colonies with morphology, gram stain, and catalase results suggestive of Staphylococcus aureus were confirmed by Staphaurex Plus rapid agglutination test (Remel, Lenexa, KS). Susceptibility to oxacillin was performed according to Clinical and Laboratory Standards Institute guidelines (15) assessing growth on Mueller-Hinton agar supplemented with 4% NaCl and 6 μg/ml oxacillin after incubation for 24 h at 35°C. Environmental samples were processed for MRSA, VRE, and MDRGN as outlined here.
Among patients who acquired an MDRGN, the frozen specimens that were obtained at enrollment were thawed and streaked onto unsupplemented MacConkey agar to isolate non-MDRGN. When the same species of gram-negative bacterium was recovered as the acquired MDRGN, PFGE was performed on these isolates.
Molecular Typing
For determination of the genetic relatedness of strains, PFGE was performed on all MDRGN, VRE, and MRSA isolates recovered from patients and the environment, as described previously (11,16). Criteria for interpretation followed standard guidelines (17).
Statistical Analyses
Categorical variables and continuous variables, dichotomized at the mean, were analyzed using the χ2 test. Statistical significance was defined as a two-sided P ≤ 0.1 in univariate analysis and a two-sided P ≤ 0.05 in multivariable analysis. Variables that were statistically significant on univariate analysis were included in a multivariable model. For determination of the independent risk factors associated with harboring MDRGN at enrollment (prevalence), a stepwise logistic regression model was created. Potential interaction terms to evaluate effect modification were also analyzed. To determine independent risk factors associated with MDRGN acquisition, patients who acquired an MDRGN during the follow-up period were matched to two patients who did not acquire MDRGN at any time during the follow-up period. For controlling for the time at risk for acquiring a MDRGN, cases were matched to controls for period at risk (number of days from enrollment to first positive follow-up cultures for cases, and number of days from enrollment to a negative follow-up culture for controls). Statistically significant variables on univariate analysis were included in a conditional logistic regression model. Statistical analysis was performed in STATA 7.0 (Stata Corp., College Station, TX).
Results
Prevalence of Antimicrobial-Resistant Bacteria
A total of 67 (79%) of 85 patients who received outpatient hemodialysis agreed to study participation and provided written informed consent. A total of 172 cultures were obtained at enrollment: 23 (34%) and six (9%) patients refused rectal and nares specimens, respectively.
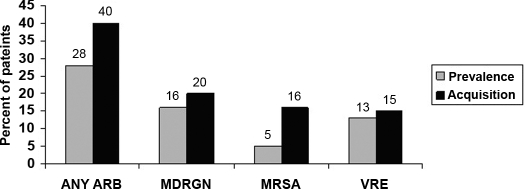
Among the 67 patients for whom at least one site was cultured, 19 (28%) were colonized with one or more antimicrobial-resistant bacteria at study enrollment. A total of 11 (16%), nine (13%), and three (5%) patients were colonized with MDRGN, VRE, and MRSA, respectively (Figure 1). Among patients who were colonized with MDRGN, four (36%) were co-colonized with VRE (three patients) and MRSA (one patient). Ten (91%) of the 11 MDRGN isolates were recovered from rectal cultures. The remaining MDRGN was recovered from a skin specimen.
Figure 1.
Percentage of patients who were colonized with antimicrobial-resistant bacteria at study enrollment and percentage of patients who were not colonized at baseline and acquired an antimicrobial-resistant bacterium during a 6-mo follow-up period. ARB, antimicrobial-resistant bacteria; MDRGN, multidrug-resistant gram-negative bacteria; MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
The species of MDRGN were as follows (number of isolates): Citrobacter species (n = 3), Klebsiella species (n = 3), Enterobacter species (n = 3), Escherichia coli (n = 1), and Pseudomonas aeruginosa (n = 1). Resistance to individual antibiotics among MDRGN isolates was as follows (percent of isolates): ampicillin/sulbactam (100%), ceftazidime (83%), piperacillin/tazobactam (75%), ciprofloxacin (58%), meropenem (25%), and gentamicin (17%). Six different co-resistant patterns were identified. The most common pattern was co-resistance to three antimicrobials (ceftazidime, ciprofloxacin, and ampicillin/sulbactam), present among three MDRGN isolates. Two co-resistance patterns to five different antimicrobials were identified among three (25%) MDRGN isolates: Two isolates were co-resistant to ceftazidime, gentamicin, meropenem, piperacillin/tazobactam, and ampicillin/sulbactam, and one isolate was co-resistant to ceftazidime, ciprofloxacin, meropenem, piperacillin/tazobactam, and ampicillin/sulbactam.
A total of eight of the 11 patients who were colonized with MDRGN at baseline had one or more follow-up cultures. The same species of MDRGN, which was identified at baseline, was also recovered from follow-up cultures among three (37.5%) patients.
Risk Factors for MDRGN Colonization
Demographics and clinical characteristics of patients with and without MDRGN colonization are presented in Table 1. More than 25 different diagnoses of underlying renal disease were documented, the most common of which was diabetic nephropathy, present in 22% of patients. Only two patients had fecal incontinence, and none had a colostomy. On logistic regression, antibiotic exposure ≥7 d in the previous 3 mo (odds ratio [OR] 5.7; 95% confidence interval [CI] 1.2 to 27.1; P = 0.03) and residence in an LTCF (OR 7.8; 95% CI 1.6 to 37.1; P = 0.01) were independently associated with MDRGN colonization.
Table 1.
Demographic and clinical characteristics of long-term hemodialysis patients with and without MDRGN at enrollmenta
| Variable | Patients Colonized with MDRGN (%; n = 11) | Patients not Colonized with MDRGN (%; n = 56) | Unadjusted OR (95% CI) | P |
|---|---|---|---|---|
| Age ≥65 yr | 7 (63) | 31 (55) | 1.40 (0.30 to 7.30) | 0.600 |
| Male | 6 (55) | 36 (66) | 0.60 (0.10 to 2.90) | 0.500 |
| White | 10 (91) | 35 (63) | 6.00 (0.70 to 27.30) | 0.070 |
| ≥3 yr on hemodialysis | 5 (45) | 29 (52) | 0.80 (0.20 to 3.50) | 0.700 |
| Previous renal transplant | 1 (9) | 3 (5) | 1.80 (0.03 to 25.00) | 0.100 |
| Resident of an LTCF | 6 (55) | 6 (11) | 10.00 (1.80 to 54.00) | 0.001 |
| Charlson score ≥5 | 9 (82) | 32 (57) | 3.40 (0.60 to 34.30) | 0.100 |
| Diabetes | 6 (55) | 20 (36) | 2.20 (0.50 to 10.10) | 0.050 |
| Nonambulatory | 5 (45) | 7 (13) | 5.80 (1.10 to 29.90) | 0.009 |
| Draining wound | 2 (18) | 5 (9) | 2.30 (0.20 to 16.50) | 0.400 |
| Hospitalization ≥7 db | 7 (64) | 13 (23) | 5.80 (1.20 to 30.50) | 0.007 |
| Antibiotic exposure ≥7 db | 8 (73) | 15 (27) | 7.20 (1.50 to 47.00) | 0.003 |
CI, confidence interval; LTCF, long-term care facility; MDRGN, multidrug-resistant Gram-negative bacteria; OR, odds ratio.
Mean number of days for study population.
Acquisition of Antimicrobial-Resistant Bacteria
A total of 55 (82%) patients had one or more sets of follow-up cultures: 50 (75%) patients during the first follow-up surveillance cultures and 55 (82%) patients during the second set of follow-up cultures. Twenty-two (40%) of the 55 patients for whom at least one set of follow-up cultures was obtained acquired one or more antimicrobial-resistant pathogens. A total of 11 (20%) patients acquired MDRGN, eight (15%) acquired VRE, and seven (13%) acquired MRSA de novo (Figure 1). Two patients acquired both VRE and MRSA, and three patients acquired both MRSA and MDRGN. Five, four, and two MDRGN were recovered from rectal, skin, and nares cultures, respectively.
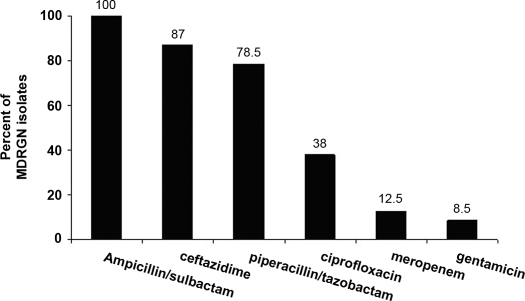
The species of acquired MDRGN were as follows (number of isolates): Enterobacter species (n = 5), E. coli (n = 2), Citrobacter species (n = 2), Providencia species (n = 1), and Hafnia species (n = 1). Resistance to individual antibiotics was as follows (percentage of isolates): ampicillin/sulbactam (100%), ceftazidime (91%), piperacillin/tazobactam (82%), ciprofloxacin (18%), gentamicin (9%), and meropenem (0%). Figure 2 shows the percentage of MDRGN isolates to individual antimicrobials among all 23 MDRGN isolates (those recovered at enrollment and on follow-up cultures). Among the acquired MDRGN, three different three-drug-resistant patterns were identified, the most common of which was resistance to ceftazidime, piperacillin/tazobactam, and ampicillin/tazobactam (nine isolates).
Figure 2.
Resistance to individual antimicrobials among 22 multidrug-resistant gram-negative isolates recovered at enrollment and on follow-up cultures.
Risk Factors for Acquisition of MDRGN
Eleven case patients were matched to 22 control subjects by period at risk for acquiring an MDRGN (number of days from first culture obtained at enrollment and first positive follow-up culture for MDRGN among case patients, and negative follow-up culture for control subjects). On univariate analysis, patients who acquired MDRGN were more likely to have diabetes, a draining wound at enrollment, and exposure to antibiotics during the follow-up period (days between the first negative cultures obtained at baseline and the first positive culture for MDRGN; Table 2). No patients had a colostomy or change in ambulatory status during the follow-up period. Only one case patient had fecal incontinence, and three patients (two case patients and one control subject) were admitted to an LTCF during the follow-up period. On conditional logistic regressing, antibiotic exposure for ≥7 d during the follow-up period was the only independent risk factor associated with acquisition of an MDRGN (OR 5.3; 95% CI 1.1 to 27; P = 0.04).
Table 2.
Demographic and clinical characteristics of long-term hemodialysis patients who did or did not acquire MDRGN during the 6-mo study period
| Variable | Patients Who Acquired MDRGN (%; n = 11) | Patients Who Did not Acquire MDRGN (%; n = 22) | Unadjusted OR (95% CI) | P |
|---|---|---|---|---|
| Age ≥66 yr | 5 (45) | 15 (68) | 0.300 (0.060 to 1.800) | 0.200 |
| Male | 4 (36) | 7 (32) | 1.200 (0.300 to 4.600) | 0.800 |
| White | 4 (36) | 16 (72) | 0.200 (0.004 to 1.200) | 0.070 |
| ≥3 yr on hemodialysis | 6 (55) | 10 (45) | 1.500 (0.300 to 7.500) | 0.600 |
| Previous renal transplant | 0 (0) | 1 (5) | – | 0.500 |
| Resident of an LTCF at baseline | 3 (27) | 3 (14) | 3.400 (0.300 to 36.000) | 0.300 |
| Charlson score ≥4 | 6 (55) | 13 (59) | 1.500 (0.400 to 5.800) | 0.800 |
| Diabetes | 6 (55) | 1 (5) | 10.200 (1.200 to 85.400) | 0.002 |
| Nonambulatory | 2 (18) | 5 (23) | 0.800 (0.100 to 4.500) | 0.800 |
| Draining wound at baseline | 4 (36) | 1 (50 | 5.500 (0.600 to 58.000) | 0.030 |
| During follow-up period | ||||
| hospitalization ≥7 d | 5 (45) | 4 (21) | 3.300 (0.600 to 17.900) | 0.200 |
| antibiotic exposure ≥7 d | 6 (55) | 3 (14) | 5.300 (1.100 to 26.800) | 0.010 |
| developed a new wound | 3 (27) | 1 (5) | 6.000 (0.600 to 58.000) | 0.060 |
Endogenous Acquisition of MDRGN
Among the 11 patients who acquired MDRGN, two had only nasal and skin cultures obtained at baseline. Because only rectal specimens were saved and frozen, baseline cultures were not available for these two patients. Among the nine patients for whom baseline frozen specimens were available, non-MDRGN of the same species as the acquired MDRGN was recovered from baseline specimens among seven (78%) patients. Six (69%) of the non-MDRGN and MDRGN isolate pairs were indistinguishable by PFGE (data not shown).
Environmental Contamination
A total of 172 environmental cultures were collected (one swab for two sites per patient per follow-up period [67 at baseline and 50 and 55 during the first and second follow-up periods, respectively). Antimicrobial-resistant bacteria were isolated from 16 (9%) of 172 environmental cultures. Ten isolates were identified as MDRGN (Stenotrophomonas species [six isolates], Klebsiella species [two isolates], E. coli [one isolate], and Serratia species [one isolate]), and six isolates as VRE. MRSA was not recovered from environmental samples.
Molecular Typing of Antimicrobial-Resistant Bacteria
The PFGE patterns of the five acquired MDR-Enterobacter species and the two MDR-Citrobacter species isolates differed by seven or more bands and were considered genetically unrelated. The two MDR-E. coli isolates were also unrelated to each other and to the one strain of MDR-E .coli recovered from environmental cultures. All eight acquired VRE isolates were unrelated to each other. Environmental contamination with the same VRE strain that colonized a patient was detected among two (12%) of a total of 17 patients who were colonized with VRE either at enrollment or at follow-up. Among the acquired MRSA isolates, six of the seven strains had indistinguishable PFGE patterns and were considered genetically related (data not shown).
Discussion
To the best of our knowledge, this is the first study to document the emerging threat of MDRGN among chronic hemodialysis patients with rates of both MDRGN colonization at enrollment and acquisition surpassing those of VRE and MRSA. In this study, MDRGN were recovered from 16% of chronic hemodialysis patients at enrollment, compared with 13 and 5% of patients with VRE and MRSA, respectively. The percentage of patients who acquired an antimicrobial-resistant bacterium during the study period was also substantial. A total of 40% of patients acquired at least one new antimicrobial-resistant pathogen during the 4-mo study period. Acquisition of MDRGN was the highest with 20% of patients becoming colonized de novo, compared with 15 and 13% of patients acquiring VRE and MRSA, respectively. These findings have important implications, because current measures aimed at preventing the spread of antimicrobial-resistant bacteria have focused on VRE and MRSA (10).
Although this study did not address infections that are caused by MDRGN, colonization is a necessary prerequisite for subsequent infection. Translocation of MDRGN across the intestinal wall into the blood stream and fecal contamination of vascular devices lead to infections (18,19). Thus, patients who are colonized with MDRGN are at greater risk for subsequently developing an infection with these bacteria. In one study, 15% of hospitalized patients who were colonized with MDRGN developed a bacteremia caused by the same colonizing strain of MDRGN (20). The co-resistance to multiple antimicrobials among MDRGN severely limits the therapeutic options that are available to physicians for treating infections that are caused by MDRGN. In this study, a great majority of MDRGN were resistant to broad-spectrum antimicrobials, with 100% resistant to ampicillin/sulbactam, 83% resistant to ceftazidime, and 78.5% resistant to piperacillin/tazobactam. Of even greater concern is the finding that 25% of MDRGN recovered from patients at enrollment were resistant to five different antimicrobial groups.
The subgroup of chronic hemodialysis patients who were at highest risk for harboring MDRGN at enrollment were patients who resided in an LTCF and those with antibiotic exposure in the previous 3 mo. Recent data support that residents of LTCF are among the major reservoirs of MDRGN and substantially contribute to the influx of MDRGN into the hospital setting (5,21–23). Dialysis initiation among the elderly is rapidly rising, with the greatest increase among octogenarians (24). It is therefore likely that the number of patients who require chronic hemodialysis and reside in an LTCF will also increase, providing further opportunities for the emergence and spread of MDRGN among this patient population.
Acquisition of antimicrobial-resistant gram-negative bacteria can occur through two mechanisms (11). Exogenous acquisition occurs through patient-to-patient spread through contaminated health care workers or environmental surfaces, similar to the transmission dynamics of VRE and MRSA. A second mechanism of acquisition is through endogenous acquisition. This term refers to the emergence of resistance in a previously susceptible gram-negative bacterium through antimicrobial pressure. In this study, 69% of MDRGN were acquired via the endogenous route. The documentation of antibiotic exposure as a risk factor for acquisition and the absence of environmental contamination with MDRGN further support this route of acquisition. In contrast, the clonality of MRSA strains in this study suggests exogenous transmission between patients. Clonal spread of VRE within the dialysis unit was not detected. Acquisition of VRE among these patients may have occurred during a previous hospitalization, a setting that has been frequently implicated in VRE spread between dialysis patients (2,16,25).
There are several limitations of this study that warrant further discussion. First, 34% of patients refused rectal cultures and had cultures obtained only from the nares or skin. Thus, colonization with antimicrobial-resistant bacteria may have been underestimated. Similarly, if colonization at enrollment was not detected, then the rate of acquisition may have been overestimated. Obtaining serial specimens over multiple days may have increased the accuracy of detecting colonization. Second, for limiting the number of specimens, only a subset of environmental surfaces were sampled and health care worker contamination was not assessed. Thus, this study cannot fully elucidate their role in the transmission dynamics of these antimicrobial-resistant bacteria. Third, the definition of multidrug resistance used in this study was based on clinical practices and focused on commonly prescribed antimicrobials. At present, a standardized definition for multidrug resistance among gram-negative bacteria has not been developed (26). Different definitions may produce different results. Fourth, our results showing the correlation between residency in an LTCF and harboring MDRGN may not be generalizable to outpatient chronic hemodialysis units that do not provide care to patients in LTCF. In these units, the prevalence of MDRGN may be lower.
With the emergence of MDRGN throughout the world, health care institutions are now faced with developing preventive strategies aimed at limiting their spread. In the hospital setting, certain institutions place patients who harbor MDRGN on contact precautions and require active surveillance when an outbreak is detected (27). In this study, the role of antimicrobial exposure emphasizes the ongoing need to use antibiotics judiciously.
There is a need for substantial future research focusing on the epidemiology of MDRGN in the outpatient dialysis unit. Because the majority of studies have focused on infections that are caused by gram-positive bacteria, especially VRE and MRSA, a first step would require future studies also to focus on infections that are caused by gram-negative bacteria and report their antimicrobial susceptibility profile.
Disclosures
E.M.C.D. has received educational grant support form Merck Pharmaceuticals.
Acknowledgments
This study was supported by grant R21 DK077312-01 from the National Institute of Diabetes and Digestive and Kidney Diseases.
This study was presented in part at the Society for Healthcare Epidemiology of America, Chicago, IL, March 18–21, 2006.
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.D’Agata EMC: Antimicrobial-resistant gram-positive bacteria in the chronic hemodialysis population. Clin Infect Dis 35: 1212–1218, 2002 [DOI] [PubMed] [Google Scholar]
- 2.Hadley AC, Karchmer TB, Russell GB, McBride DG, Freedman BI: The prevalence of resistant bacterial colonization in chronic hemodialysis patients. Am J Nephrol 27: 352–359, 2007 [DOI] [PubMed] [Google Scholar]
- 3.Finelli L, Miller JT, Tokars JI, Alter MJ, Arduino MJ: National surveillance of dialysis-associated diseases in the United States, 2002. Semin Dial 18: 52–61, 2005 [DOI] [PubMed] [Google Scholar]
- 4.D’Agata EMC: Rapidly rising prevalence of nosocomial multidrug-resistant gram-negative bacilli: A 9-year surveillance study. Infect Control Hosp Epidemiol 25: 842–846, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Pop-Vicas AE, D’Agata EM: The rising influx of multidrug-resistant gram-negative bacilli into a tertiary care hospital. Clin Infect Dis 40: 1792–1798, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Bradford PA: Extended-spectrum beta-lactamases in the 21st century: Characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev 14: 933–951, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwaber MJ, Navon-Venezia S, Kaye KS, Ben-Ami R, Schwartz D, Carmeli Y: Clinical and economic impact of bacteremia with extended- spectrum-beta-lactamase-producing Enterobacter. Antimicrob Agents Chemother 50: 1257–1262, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marr KA, Sexton DJ, Conlon PJ, Corey GR, Schwab SJ, Kirkland KB: Catheter-related bacteremia and outcome of attempted catheters salvage in patients undergoing hemodialysis. Ann Intern Med 137: 275–280, 1997 [DOI] [PubMed] [Google Scholar]
- 9.National Institutes of Health: 2000 Annual Data Report: US Renal Data System, Bethesda, US Department of Health and Human Services, National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2000
- 10.Centers for Disease Control and Prevention: Recommendations for preventing transmission of infections among chronic hemodialysis patients. MMWR Recomm Rep 50: 1–43, 2001 [PubMed] [Google Scholar]
- 11.D’Agata E, Venkataraman L, DeGirolami P, Samore M: Molecular epidemiology of acquisition of ceftazidime-resistant gram-negative bacilli in a nonoutbreak setting. J Clin Microbiol 35: 2602–2605, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charlson M, Pompei P, Ales K, MacKenzie CR: A new method of classifying prognostic comorbidity in longitudinal studies: Development and validation. J Chron Dis 40: 373–383, 1987 [DOI] [PubMed] [Google Scholar]
- 13.Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved standard M2–A9, Wayne, PA, Clinical and Laboratory Standards Institute, 2006
- 14.Facklam RR, Sahm DF: Enterococcus. In: Manual of Clinical Microbiology, 6th Ed., edited by Murray PR, Washington, DC, American Society for Microbiology (ASM) Press, 1995, pp 308–314
- 15.Clinical and Laboratory Standards Institute: Performance Standards for Antimicrobial Susceptibility Testing. Supplement M100–S16, Wayne, PA, Clinical and Laboratory Standards Institute, 2006
- 16.D’Agata EMC, Green K, Schulman G, Li H, Tang YW, Schaffner W: Vancomycin-resistant enterococci among chronic hemodialysis patients. Clin Infect Dis 32: 23–29, 2001 [DOI] [PubMed] [Google Scholar]
- 17.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B: Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol 33: 2223–2229, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donskey CJ: The role of the intestinal tract as a reservoir and source for transmission of nosocomial pathogens. Clin Infect Dis 39: 219–226, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Tancrede CH, Andremont AO: Bacterial translocation and gram-negative bacteremia in patients with hematological malignancies. J Infect Dis 152: 99–103, 1985 [DOI] [PubMed] [Google Scholar]
- 20.Ben-Ami R, Schwaber MJ, Navon-Venezia S, Schwartz D, Giladi M, Chimelnistky I, Leavitt A, Carmeli Y: Influx of extended-spectrum beta-lactamase-producing Enterobacteriaceae into the hospital. Clin Infect Dis 42: 925–934, 2006 [DOI] [PubMed] [Google Scholar]
- 21.Flamm RK, Weaver MK, Thornsberry C, Jones ME, Karlowsky JA, Sahm DF: Factors associated with relative rates of antibiotic resistance in Pseudomonas aeruginosa isolates tested in clinical laboratories in the United States form 1999 to 2002. Antimicrob Agents Chemother 48: 2431–2436, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muder RR, Breene C, Drenning SD, Stout JE, Wagener MM: Multiply antibiotic-resistant gram-negative bacilli in a long-term-care facility: A case-control study of patient risk factors and prior antibiotic use. Infect Control Hosp Epidemiol 18: 809–813, 1997 [PubMed] [Google Scholar]
- 23.Wiener J, Quinn JP, Bradford PA, Goering RV, Nathan C, Bush K, Weinstein RA: Multiple antibiotic-resistant Klebsiella and Escherichia coli in nursing homes. JAMA 281: 517–523, 1999 [DOI] [PubMed] [Google Scholar]
- 24.Kurella M, Covinsky KE, Collins J, Chertow GM: Octogenarians and nonagenarians starting dialysis in the United States. Ann Intern Med 146: 177–183, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Roghmann MC, Fink JC, Polish L, Maker T, Brewrink J, Morris JG, Light PD: Colonization with vancomycin-resistant enterococci in chronic hemodialysis patients. Am J Kidney Dis 32: 254–257, 1998 [DOI] [PubMed] [Google Scholar]
- 26.Paterson DL: The epidemiological profile of infections with multidrug-resistant Pseudomonas aeruginosa and Acinetobacter species. Clin Infect Dis 43: S43–S48, 2006 [DOI] [PubMed] [Google Scholar]
- 27.Harris AD, McGregor JC, Furuno JP: What infection control interventions should be undertaken to control multidrug-resistant gram-negative bacteria? Clin Infect Dis 43 [Suppl 2]: S57–S61, 2006 [DOI] [PubMed] [Google Scholar]