Abstract
IgA nephropathy (IgAN) is a complex trait determined by genetic and environmental factors. Most IgAN patients exhibit a characteristic undergalactosylation of the O-glycans of the IgA1 hinge region, which promotes formation and glomerular deposition of immune complexes. It is not known whether this aberrant glycosylation is the result of an acquired or inherited defect, or whether the presence of aberrant IgA1 glycoforms alone can produce IgAN. A newly validated lectin enzyme-linked immunosorbent assay (ELISA) was used to determine the serum level of galactose-deficient IgA1 (Gd-IgA1) in a cohort of 89 IgAN patients and 266 of their relatives. High Gd-IgA1 levels (≥95th percentile for controls) were observed in all 5 available patients with familial IgAN, in 21 of 45 (47%) of their at-risk relatives (assuming autosomal dominant inheritance), and in only 1 of 19 (5%) of unrelated individuals who married into the family. This provides evidence that abnormal IgA1 glycosylation is an inherited rather than acquired trait. Similarly, Gd-IgA1 levels were high in 65 of 84 (78%) patients with sporadic IgAN and in 50 of 202 (25%) blood relatives. Heritability of Gd-IgA1 was estimated at 0.54 (P = 0.0001), and segregation analysis suggested the presence of a major dominant gene on a polygenic background. Because most relatives with abnormal IgA1 glycoforms were asymptomatic, additional cofactors must be required for IgAN to develop. The fact that abnormal IgA1 glycosylation clusters in most but not all families suggests that measuring Gd-IgA1 may help distinguish patients with different pathogenic mechanisms of disease.
IgA nephropathy (IgAN) is one of the most common forms of primary glomerulonephritis.1 The diagnosis is based on examination of a renal biopsy specimen, demonstrating mesangial cellular proliferation with predominant or codominant deposition of IgA1 with C3 and variable presence of IgG and/or IgM. Because of the requirement for an invasive procedure for diagnosis, the population prevalence of IgAN trait is not known. Autopsy studies and transplant-donor biopsy series document a prevalence up to 4%, suggesting that this trait is very common but clinically silent in the majority of affected individuals.2,3
IgAN has complex determination, with contributions from genetic and environmental factors.4 For example, viral infections are well recognized as an environmental factor accentuating the clinical expression of this trait. Familial aggregation of IgAN, most compatible with autosomal dominant transmission with incomplete penetrance, has also been well documented, with some relatives displaying abnormalities in the synthesis of immunoglobulins and production of cytokines.4,5 Linkage studies have identified several IgAN susceptibility loci for familial forms of the disease, substantiating the role of genetic factors in this subpopulation.6–8 The contribution of genes to the development of the sporadic forms of IgAN has been less clearly defined. Many candidate genes for sporadic IgAN have been suggested, but these genetic associations have not been well replicated.4
Serum IgA levels are variably elevated in IgAN and are not diagnostic of disease.1,9 However, studies suggest that the majority of IgAN patients exhibit abnormalities in the glycosylation of the IgA1 heavy chain.9–15 In healthy individuals, O-linked glycans in the hinge region are composed of N-acetylgalactosamine with β1,3 linked galactose (Gal); one or both sugars may be sialylated. In IgAN patients, a significant portion of the circulating IgA1 is galactose-deficient, a configuration that predisposes to immune complex formation and mesangial deposition.16,17 It is not known whether abnormalities in IgA1 glycosylation are attributable to acquired or inherited defects, or whether defective IgA1 glycosylation is sufficient to produce IgAN.
We have recently developed an enzyme-linked immunosorbent assay (ELISA) using a lectin derived from Helix aspersa (HAA) to detect Gal-deficient IgA1 glycoforms in serum (Gd-IgA1).9 Studying a large population of IgAN cases and controls from the southeastern United States, we showed that 76% of IgAN patients had a significantly elevated Gd-IgA1 level, suggesting that this assay can be used as a screening tool for IgAN. To investigate whether this abnormality is antecedent to or a consequence of IgAN and to determine whether this trait is inherited or acquired, we have now investigated the distribution of this Gal-deficient glycoform among family members in kindreds with IgAN.
RESULTS
Familial IgAN
We examined the distribution of Gd-IgA1 in two large white kindreds comprising 7 individuals with biopsy-documented IgAN (K1904 and K3041, Table 1; Figure 1, A and B). As reported for other IgAN pedigrees, the disease transmission pattern was consistent with dominant inheritance with incomplete penetrance. Members of K1904 lived in northeastern Alabama, whereas members of K3041 were in one county in eastern Kentucky. We selected these kindreds because their members resided in the same region and the index case in each family had Gd-IgA1 above the 95th percentile of serum levels derived from 141 normal white controls (≥1145 U/ml). In total, 64 family members were recruited and studied. We next measured serum Gd-IgA1 levels in all available individuals and compared levels between IgAN patients, their relatives, and controls.
Table 1.
Characteristics of patients with IgAN, their relatives, and controls
| Familial IgAN Patients | Relatives of Familial IgAN Patients | Sporadic IgAN Patients | Relatives of Sporadic IgAN Patients | Controls | |
|---|---|---|---|---|---|
| N | 5 | 64 | 84 | 202 | 141 |
| Age, yr | 44 ± 5 | 42 ± 18 | 39 ± 13 | 43 ± 16 | 37 ± 13 |
| Ethnicity, white/black | 5/0 | 63/0 | 81/3 | 192/10 | 141/0 |
| Gender, male/female | 5/0 | 28/35 | 48/37 | 67/125 | 73/68 |
| BMI, kg/m2 | 32 ± 11 | 27.5 ± 6 | 31.5 ± 8 | 29.5 ± 7 | 26.5 ± 5 |
| GFR, ml/min/1.73 m2 | 43 ± 15 | 76 ± 17 | 61.5 ± 37 | 83 ± 21 | 88 ± 17 |
| GFR <30, N | 1 | 0 | 19 | 0 | 0 |
| Systolic BP, mm Hg | 125 ± 5 | 129 ± 16 | 131 ± 20 | 130 ± 20 | 123 ± 13 |
| Diastolic BP, mm Hg | 82 ± 9 | 77 ± 9 | 77 ± 10 | 75 ± 10 | 73 ± 7 |
| Hypertension, N | 5 | 17 | 56 | 67 | 21 |
| Urine P/C | 0.3 ± 0.1 | 0.1 ± 0.1 | 1.0 ± 1.4 | 0.2 ± 1.3 | — |
| Urine P/C > 3, N | 0 | 0 | 5 | 1 | — |
| Hematuria, N (%) | 5 (100) | 6 (9) | 33 (40) | 7 (3.5) | 0 |
Values are mean ± SD. BMI, body mass index; GFR, glomerular filtration rate; BP, blood pressure; P/C, protein to creatinine ratio; —, not applicable.
Figure 1.
Structure of K1904 (A) K3041 (B). Symbols are described in the figure. Arrows identify the patients with biopsy-proven IgAN (2 deceased). (C) Distribution of Gd-IgA1 levels among IgAN patients, relatives at risk under an autosomal dominant model, marry-in relatives, and unrelated controls. Mean values for each group is indicated by solid black bars. The dashed horizontal lines indicate the 95th and 99th percentile cutoffs derived from controls. Gd-IgA1 values above the 95th percentile are shown in red. The P values for comparisons between the different groups are shown.
The Gd-IgA1 levels were normally distributed in controls (Figures 1C and S2). The five available IgAN patients all had Gd-IgA1 levels above the 95th percentile of controls (Figure 1C). Among all relatives, Gd-IgA1 levels were also significantly elevated compared with controls (Figure 1C, P = 0.002). The relatives with high Gd-IgA1 levels displayed values that were similar to those observed in IgAN patients. When we dichotomized Gd-IgA1 levels based on 95th percentile reference value for normal controls, 22 of 64 (35%) of all relatives had Gd-IgA1 levels above this level. The pattern of transmission of Gd-IgA1 levels was most compatible with an autosomal dominant component: high Gd-IgA1 levels were observed across multiple generations and in different branches of the families, were evenly distributed between sexes, and were present among 9 of 23 (39%) first-degree relatives of IgAN patients. This pattern suggested that high Gd-IgA1 levels co-segregate with IgAN susceptibility alleles. To test this hypothesis, we examined the distribution of Gd-IgA1 among the subset of relatives who would be at risk for IgAN under an autosomal dominant model (n = 45). Gd-IgA1 levels were significantly higher among at-risk relatives compared with controls (P = 1.2 × 10−4, Figure 1C), with 21 of 45 (47%) individuals demonstrating levels above the 95th percentile. In contrast, individuals who married into the family (marry-ins) and were not at risk for IgAN under autosomal dominant transmission showed Gd-IgA1 levels that were not significantly different from controls (P = 0.4) but were significantly different from at-risk relatives (P = 3 × 10−3, Figure 1C). Only 1 of 19 (5%) marry-ins displayed a level above the 95th percentile. This difference in the distribution of Gd-IgA1 levels between at-risk relatives versus marry-ins or controls was highly significant (P = 0.001 and P < 0.0001, respectively). This unilineal segregation pattern of high Gd-IgA1 levels in these families strongly supports genetic determination.
In contrast to the strong clustering Gd-IgA1 levels within families, total serum IgA levels did not cluster (Figure S1). There was no difference in mean serum IgA levels between IgAN patients, at risk relatives, and marry-ins (P = 0.5). The portion of cases displaying serum IgA levels above the 95th percentile was also similar among IgAN patients (20%), at-risk relatives (26%), and marry-in relatives (21%). These data are consistent with the low sensitivity of serum IgA levels as a screening test for IgAN.1,9 As predicted by these data, the ratio of Gd-IgA1 to total serum IgA performed intermediately compared with absolute Gd-IgA1 and total serum IgA levels. Finally, urinalysis screening also proved less specific than Gd-IgA1 measurements: although both at-risk (n = 4) and marry-in individuals (n = 2) demonstrated abnormal urinalyses, only the at-risk relatives displayed abnormal Gd-IgA1 levels.
Sporadic IgAN
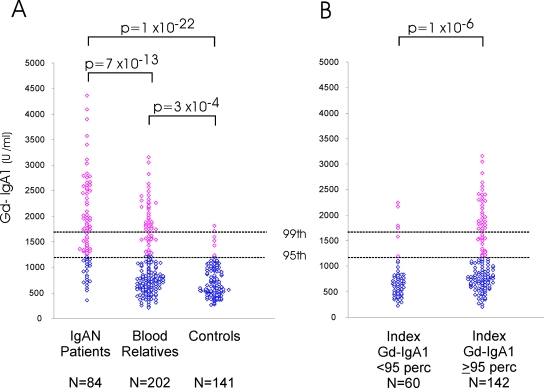
We next asked whether IgA1 glycosylation defects are also present in family members of patients with sporadic forms of IgAN. We studied 84 IgAN patients and 202 blood relatives recruited from the southeastern United States (Table 1). IgAN patients had significantly elevated Gd-IgA1 values compared with controls (P = 1 × 10−22, Figure 2A), with 65 (78%) individuals displaying Gd-IgA1 levels above the 95th percentile. As observed in our previous study, there were no differences in clinical parameters (such as age, blood pressure [BP], glomerular filtration rate [GFR] or proteinuria, hematuria) between IgAN patients with high or low Gd-IgA1 levels.9
Figure 2.
(A) Distribution of Gd-IgA1 levels in sporadic IgAN patients (n = 84), their blood relatives (n = 202), and unrelated controls (n = 141). The dashed horizontal lines indicate the 95th and 99th percentile cutoffs for controls. Values above the 95th percentile are shown in red. (B) Distribution of Gd-IgA1 levels in relatives, after stratification based on Gd-IgA1 levels in the index case. The P values for comparisons between the different groups are shown.
Similar to the situation in familial IgAN, Gd-IgA1 levels were significantly higher in blood relatives, with 50 of 202 (25%) individuals displaying Gd-IgA1 levels above the 95th percentile (P = 3 × 10−4, Figure 2A; Table S1). As expected for a heritable trait, the proportion of relatives with high Gd-IgA1 was larger in first-degree relatives (28%) versus second-degree relatives (17%). We further examined the inheritance pattern of this trait in a subset of 22 families in which both parents of the IgAN patient were available. In 15 families, high Gd-IgA1 was present in only one parent, a pattern compatible with unilineal transmission; in 2 families, high Gd-IgA1 was found in both parents (bilineal transmission); and in 5 families, Gd-IgA1 was normal in both parents. Therefore, familial aggregation of high Gd-IgA1 levels and unilineal transmission is a common feature of sporadic as well as familial forms of IgAN.
The finding of normal Gd-IgA1 levels in 22% of IgAN patients may be attributable to temporal fluctuation of this trait or assay variability. Alternatively, the skewed, almost bimodal distribution of Gd-IgA1 levels among patients and relatives suggests that Gd-IgA1 measurements correctly differentiate between two subpopulations of IgAN (Figure S2). One can discriminate between these two possibilities by examining Gd-IgA1 levels in relatives after stratification for Gd-IgA1 levels in the index case. If there is random fluctuation of this trait or significant assay variability, one would expect high or low Gd-IgA1 levels to be similarly distributed among all relatives, independent of the Gd-IgA1 level in the index patient. Alternatively, if Gd-IgA1 is a reliable and highly heritable biomarker, high Gd-IgA1 levels should be found predominantly among relatives of IgAN patients with high levels. Among relatives of IgAN patients with high Gd-IgA1 values, 47 of 142 (33%) individuals had levels above the 95th percentile (Table S1; Figure 2B). In contrast, relatives of IgAN patients with normal Gd-IgA1 levels had levels that were indistinguishable from controls (P = 0.3), with only 6 of 60 (10%) relatives displaying values above the 95th percentile. The difference in distribution of the Gd-IgA1 levels between the two groups of relatives was highly significant, whether considered as a continuous (P = 1 × 10−6) or dichotomous trait (P = 0.0004). These data strongly argue in favor of Gd-IgA1 values as a reliable biomarker that identifies distinct subpopulations among IgAN patients.
Heritability and Segregation Analysis
To better quantify the genetic effect on Gd-IgA1 levels, we calculated heritability values and performed segregation analysis in the entire cohort of familial and sporadic IgAN. The heritability of Gd-IgA1 was significant (h2 = 0.54, P = 1 × 10−4) with IgAN affection status and IgA levels emerging as the only significant independent covariates. Estimations of heritability for other traits, such as height, body mass index (BMI), BP, GFR, or serum IgA levels, demonstrated values consistent with published data in other cohorts, demonstrating a lack of bias in our calculations (Table S2). As expected from the observed distribution of the trait, heritability was significant in the subset of families in which the index case had a Gd-IgA1 level above the 95th percentile (h2 = 0.62, P = 1 × 10−5); on the other hand, we found no evidence for heritability in the 19 families in which the index case displayed an Gd-IgA1 level below the 95th percentile (h2 = 0.12, P = 0.4). Segregation analysis of Gd-IgA1 as a quantitative trait rejected environmental, sporadic, as well as pure Mendelian models (P < 0.05). Altogether, a mixed model suggesting inheritance of major dominant gene with an additional polygenic component provided the best fit; a mixed polygenic and environmental model could not be excluded (Tables 2 and S3). High Gd-IgA1 was equally present among parents (39%), siblings (28%), and children (30%) of index cases, providing further support for a major dominant component.
Table 2.
Results of segregation analysis of Gd-IgA1 in study cohort
| Model | No. of Parameters | χ2 | df | P versus Free Model |
|---|---|---|---|---|
| Free | 9 | — | — | — |
| Mendelian dominant | 4 | 11 | 5 | 0.05 |
| Mendelian recessive | 4 | 13 | 5 | 0.02 |
| Additive | 4 | 13 | 5 | 0.02 |
| Environmental | 6 | 17 | 3 | 0.0008 |
| Sporadic | 2 | 25,808 | 7 | 0 |
| Polygenic | 3 | 25,713 | 6 | 0 |
| Major recessive + polygenes | 5 | 2 | 4 | 0.73 |
| Major dominant + polygenes | 5 | 0 | 4 | 0.99 |
| Environmental + polygenes | 2 | 1 | 7 | 0.99 |
—, not applicable. The significance of each model was estimated by computing the difference in natural log likelihoods between the general (free) model and the model of interest. This difference approximates a χ 2 statistic, with degrees of freedom (df) equal to the difference in the number of parameters used in the two models. The best fitting models are the ones that are not significantly different compared with the free model. A detailed table is included in supplemental materials online.
DISCUSSION
In the present study, we used a newly developed and validated assay9 to systematically study the prevalence of an IgA1 glycosylation defect among IgAN patients, their relatives, and controls. As previously observed, an elevated serum level of Gd-IgA1 was highly associated with IgAN.9 Moreover, elevated Gd-IgA1 levels, similar to those found in IgAN patients, were found among a significant proportion of relatives who did not have IgAN. Examination of familial forms of IgAN clearly demonstrated that Gd-IgA1 levels were elevated among some, but not all, at-risk relatives and were essentially normal among individuals who married into the family. These findings provide strong evidence that elevated Gd-IgA1 levels are not merely a consequence of IgAN, and implicate genetic transmission in the production of Gal-deficient IgA1. Gd-IgA1 levels displayed heritability values that were comparable to values for height and BMI, some of the most heritable traits in humans, and the distribution of values within kindreds with both familial and sporadic IgAN suggests an autosomal dominant component contributing to variation in this trait. In contrast, total serum IgA levels provided much less discrimination between cases and controls; these values were equally high among at-risk relatives and marry-ins and showed significantly lower heritability.
Defects in the O-linked glycans of the hinge region in serum IgA1 have been documented in multiple cohorts and appear to be specific to IgAN.9,10,12,13 Abnormal IgA1 glycoforms may directly participate in the disease pathogenesis by promoting interaction with naturally occurring anti-glycan IgG and/or IgA1 antibodies, resulting in formation of circulating immune complexes and mesangial deposition.10,11,14 The finding of high Gd-IgA1 levels in a significant proportion of relatives suggests that aberrant IgA1 glycosylation may constitute an initial, inherited risk factor for the development of disease. However, because most relatives who displayed an abnormal IgA1 glycosylation were asymptomatic, one can conclude that elevated Gd-IgA1 levels alone are not sufficient to produce clinical IgAN but likely require additional genetic or environmental cofactors for full expression of disease. This conclusion is in agreement with our observation that the proliferation of cultured mesangial cells is not stimulated with Gal-deficient IgA1 alone but only after it formed immune complexes.17 This may be a situation analogous to the Tn syndrome (OMIM 300622), a rare autoimmune disorder in which a Gal-deficient membrane glycoprotein in blood cells of all lineages is recognized by naturally occurring complement-activating antibodies.18 Although the prevalence of this biochemical abnormality is about 1:100,000 in the general population, fewer than 30 patients with the clinical syndrome have been reported.19 Recently, it was found that some Tn syndrome patients display somatic mutations in the gene encoding the chaperone protein Cosmc, which is required for proper folding of glycosyltransferases.20 Defective glycosyltransferase enzyme activity resulting from somatic mutations in COSMC produces the autoimmune Tn antigen.20 In IgAN, a decisive factor in formation of nephritogenic immune complex may be the proportion of antigen (Gd-IgA1) and antiglycan antibody10,11,14,21–23 Therefore, one can hypothesize that clinical manifestations of IgAN occur when abnormal IgA1 glycosylation is coupled to additional defects (such as somatic mutations in genes in the IgA1 glycosylation pathway) that further perturb the balance of Gd-IgA1 and antiglycan antibody toward production of pathogenic immune complexes.21–23
The present findings raise several questions that can be formally addressed in clinical and genetic studies of IgAN. Prospective follow-up of individuals with elevated Gd-IgA1 levels should clarify whether abnormal IgA1 glycosylation imparts a significant risk for the future development of IgAN. The aggregation of high Gd-IgA1 levels among subsets of IgAN families also suggests that this trait identifies patients with distinct pathogenic mechanisms of disease. The notion of disease subtypes is supported by associations of IgAN with other diseases, such as thin basement membrane nephropathy or evidence of linkage of familial IgAN to distinct genetic loci.6–8,24,25 Further evidence is provided by a previous study showing that abnormal IgA1 glycosylation is more frequent in IgAN patients with glomerular basement membranes of normal thickness, compared with those with thin glomerular basement membranes.26 Genetic association studies can test whether distinct genetic loci contribute to disease among IgAN populations with contrasting IgA1 glycosylation profiles. Alternatively, Gd-IgA1 levels may be used as an endophenotype to identify asymptomatic individuals carrying IgAN susceptibility alleles, thereby augmenting the power of linkage studies of familial IgAN. Genetic studies of Gd-IgA1 levels are under way to further clarify the relationship between IgAN and abnormal IgA1 glycosylation.
CONCISE METHODS
Study Subjects
Adult patients residing in Alabama and eastern Kentucky were ascertained based on the diagnosis of IgAN documented by renal biopsy and recruited. Familial IgAN was defined by the presence of at least 2 family members (first cousins or more closely related) with biopsy-documented IgAN. Relatives were invited to participate in the study. In total, we recruited 192 first-degree relatives (parents, offspring, and siblings), 43 second-degree relatives (aunt/uncle/niece/nephew/first cousin), 11 second-cousins, and one third-cousin. Nineteen individuals who married into two kindreds with familial IgAN (K1904 and K3041) were also recruited. A blood sample was obtained for measurement of clinical laboratory chemistries and total serum IgA and Gd-IgA1 levels. Urine was obtained for dipstick analysis and quantitation of protein/creatinine ratio. Abnormal urinalyses were defined based on the presence of hematuria (≥2+ blood) and/or proteinuria (urine protein/creatinine ratio ≥0.5). A group of residents of Alabama and Kentucky with normal renal function and no family history of kidney disease were recruited as healthy controls. The study protocol adhered to the Declaration of Helsinki. All study subjects provided signed, informed consent, and the protocol was approved by the local Institutional Review Board at each participating institution.
Measurement of Serum Total IgA and Gal-deficient IgA1 (Gd-IgA1)
Total IgA levels were measured by ELISA.9 Levels of Gd-IgA1 were quantitated using a previously validated ELISA that uses biotinylated HAA (Sigma-Aldrich, St. Louis, MO).9 This lectin binds specifically to terminal GalNAc, but not to GalNAc with attached Gal or sialic acid. The assay uses neuraminidase treatment and thus measures total Gal-deficient IgA1. Because only IgA1, but not IgA2, has the hinge region of the heavy chain with O-linked glycans, this assay measures only Gal-deficient IgA1. The standard for Gd-IgA1 consisted of a polymeric naturally Gal-deficient IgA1 protein isolated from plasma of a patient with IgA1 myeloma.9 The results are expressed as units per milliliter, with 1 unit of Gd-IgA1 defined as 1 μg of the standard protein.
Statistical Analysis
All analyses were performed using the SPSS 15.0 package. Gd-IgA1 were compared between the different study groups (IgAN patients, relatives, and controls) using analysis of covariance with the SPSS General Linear Model command. Covariates tested in the model included age, sex, patient group (IgAN versus relative versus control), chronic kidney disease stage, BMI, total serum IgA levels, urine protein/creatinine ratio, and mean BP. Only patient group status and total IgA levels emerged as independent predictors of Gd-IgA1 levels. All P values were corrected for significant covariates and multiple comparisons using the Bonferroni method. High serum Gd-IgA1 levels were defined as above the 95th percentile value for normal controls and compared as a categorical trait with a Fisher exact test. Heritability calculations were performed using SOLAR.27 Traits deviating from a normal distribution (such as serum Gd-IgA1 and total IgA levels) were log10-transformed to approximate a normal distribution. Covariates tested in the heritability analysis of Gd-IgA1 included serum IgA levels, affection status, age, sex, GFR, BP, and proteinuria. Segregation analysis of Gd-IgA1 levels was performed using the Pedigree Analysis Package.28 Gd-IgA1 levels were analyzed as a continuous trait (log10-transformed) and adjusted for significant covariates (affection status and total serum IgA levels). Inheritance models tested included a general free model, environmental, sporadic, polygenic, single-gene dominant/recessive, and major gene dominant/recessive with polygenic residual transmission.
DISCLOSURES
None.
Supplementary Material
Acknowledgments
This study was supported by grants DK78244, DK61525, DK71802, DK47322, and DK64400 from the National Institutes of Health and by General Clinical Research Centers of the University of Alabama at Birmingham (M01 RR00032) and the University of Tennessee Health Sciences Center (M01 RR00211). A.G.G. is supported by the Emerald Foundation and the Columbia Center for Glomerular Diseases.
The authors thank the patients, their family members, and control subjects for their participation in this study as well as Wen-Qiang Huang, Rhubell Brown, Stacy Hall, Claretha R. Nichols, and Candace Kirksey for technical assistance and Krzysztof Kiryluk, Simone Sanna-Cherchi, and Patricia Weng for helpful discussions.
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1.Barratt J, Feehally J: IgA nephropathy. J Am Soc Nephrol 16, 2088–2097, 2005 [DOI] [PubMed] [Google Scholar]
- 2.Varis J, Rantala I, Pasternack A, Oksa H, Jäntti M, Paunu ES, Pirhonen R: Immunoglobulin and complement deposition in glomeruli of 756 subjects who had committed suicide or met with a violent death. J Clin Pathol 46: 607–610, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzuki K, Honda K, Tanabe K, Toma H, Nihei H, Yamaguchi Y: Incidence of latent mesangial IgA deposition in renal allograft donors in Japan. Kidney Int 63: 2286–2294, 2003 [DOI] [PubMed] [Google Scholar]
- 4.Beerman I, Novak J, Wyatt RJ, Julian BA, Gharavi AG: The genetics of IgA nephropathy. Nat Clin Pract Nephrol 3: 325–338, 2007 [DOI] [PubMed] [Google Scholar]
- 5.Schena FP, Scivittaro V, Ranieri E, Sinico R, Benuzzi S, Di Cillo M, Aventaggiato L: Abnormalities of the IgA immune system in members of unrelated pedigrees from patients with IgA nephropathy. Clin Exp Immunol 92: 139–144, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gharavi AG, Yan Y, Scolari F, Schena FP, Frasca GM, Ghiggeri GM, Cooper K, Amoroso A, Viola BF, Battini G, Caridi G, Canova C, Farhi A, Subramanian V, Nelson-Williams C, Woodford S, Julian BA, Wyatt RJ, Lifton RP: IgA nephropathy, the most common cause of glomerulonephritis, is linked to 6q22–23. Nat Genet 26: 354–357, 2000 [DOI] [PubMed] [Google Scholar]
- 7.Bisceglia L, Cerullo G, Forabosco P, Torres DD, Scolari F, Di Perna M, Foramitti M, Amoroso A, Bertok S, Floege J, Mertens PR, Zerres K, Alexopoulos E, Kirmizis D, Ermelinda M, Zelante L, Schena FP; European IgAN Consortium: Genetic heterogeneity in Italian families with IgA nephropathy: Suggestive linkage for two novel IgA nephropathy loci. Am J Hum Genet 79: 1130–1134, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson AD, Liu XQ, Wang K, Magistroni R, Song X, Kappel J, Klassen J, Cattran D, St George-Hyslop P, Pei Y: Genome-wide linkage scan of a large family with IgA nephropathy localizes a novel susceptibility locus to chromosome 2q36. J Am Soc Nephrol 18: 2408–2415, 2007 [DOI] [PubMed] [Google Scholar]
- 9.Moldoveanu Z, Wyatt RJ, Lee JY, Tomana M, Julian BA, Mestecky J, Huang WQ, Anreddy SR, Hall S, Hastings MC, Lau KK, Cook WJ, Novak J: Patients with IgA nephropathy have increased serum galactose-deficient IgA1 levels. Kidney Int 71: 1148–1154, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J: Circulating immune complexes in IgA nephropathy consist of IgA1 with galactose-deficient hinge region and antiglycan antibodies. J Clin Invest 104: 73–81, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allen AC, Bailey EM, Brenchley PE, Buck KS, Barratt J, Feehally J: Mesangial IgA1 in IgA nephropathy exhibits aberrant O-glycosylation: Observations in three patients. Kidney Int 60: 969–973, 2001 [DOI] [PubMed] [Google Scholar]
- 12.Allen AC, Harper SJ, Feehally J: Galactosylation of N- and O-linked carbohydrate moieties of IgA1 and IgG in IgA nephropathy. Clin Exp Immunol 100: 470–474, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hiki Y, Horii A, Iwase H, Tanaka A, Toda Y, Hotta K, Kobayashi Y: O-linked oligosaccharide on IgA1 hinge region in IgA nephropathy: Fundamental study for precise structure and possible role. Contrib Nephrol 111: 73–84, 1995 [PubMed] [Google Scholar]
- 14.Hiki Y, Horii A, Iwase H, Tanaka A, Toda Y, Hotta K, Kobayashi Y: Mass spectrometry proves under-O-glycosylation of glomerular IgA1 in IgA nephropathy. Kidney Int 59: 1077–1085, 2001 [DOI] [PubMed] [Google Scholar]
- 15.Andre PM, Le Pogamp P, Chevet D: Impairment of jacalin binding to serum IgA in IgA nephropathy. J Clin Lab Anal 4: 115–119, 1990 [DOI] [PubMed] [Google Scholar]
- 16.Tomana M, Novak J, Julian BA, Matousovic K, Konecny K, Mestecky J: Galactose-deficient IgA1 in sera of IgA nephropathy patients is present in complexes with IgG. Kidney Int 52: 509–516, 1997 [DOI] [PubMed] [Google Scholar]
- 17.Novak J, Tomana M, Matousovic K, Brown R, Hall S, Novak L, Julian BA, Wyatt RJ, Mestecky J: IgA1-containing immune complexes in IgA nephropathy differentially affect proliferation of mesangial cells. Kidney Int 67: 504–513, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Dahr W, Uhlenbruck G, Gunson HH, van der Hart M: Molecular basis of Tn-polyagglutinability. Vox Sang 29: 36–50, 1975 [DOI] [PubMed] [Google Scholar]
- 19.Berger EG: Tn-syndrome. Biochim Biophys Acta 1455: 255–268, 1999 [DOI] [PubMed] [Google Scholar]
- 20.Ju T, Cummings RD: Protein glycosylation: chaperone mutation in Tn syndrome. Nature 437: 1252, 2005 [DOI] [PubMed] [Google Scholar]
- 21.Mestecky J, Suzuki H, Yanagihara T, Moldoveanu Z, Tomana M, Matousovic K, Julian BA, Novak J: IgA nephropathy: Current views of immune complex formation. Contrib Nephrol 157: 56–63, 2007 [DOI] [PubMed] [Google Scholar]
- 22.Novak J, Moldoveanu Z, Renfrow MB, Yanagihara T, Suzuki H, Raska M, Hall S, Brown R, Huang WQ, Goepfert A, Kilian M, Poulsen K, Tomana M, Wyatt RJ, Julian BA, Mestecky J: IgA nephropathy and Henoch-Schoenlein purpura nephritis: Aberrant glycosylation of IgA1, formation of IgA1-containing immune complexes, and activation of mesangial cells. Contrib Nephrol 157: 134–138, 2007 [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Moldoveanu Z, Hall S, Brown R, Julian BA, Wyatt RJ, Tomana M, Tomino Y, Novak J, Mestecky J: IgA nephropathy: Characterization of IgG antibodies specific for galactose-deficient IgA1. Contrib Nephrol 157: 129–133, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Levy M: Multiplex families in IgA nephropathy. Contrib Nephrol 104: 46–53, 1993 [DOI] [PubMed] [Google Scholar]
- 25.Frascá GM, Soverini L, Gharavi AG, Lifton RP, Canova C, Preda P, Vangelista A, Stefoni S: Thin basement membrane disease in patients with familial IgA nephropathy. J Nephrol 17: 778–785, 2004 [PubMed] [Google Scholar]
- 26.Linossier MT, Palle S, Berthoux F: Different glycosylation profile of serum IgA1 in IgA nephropathy according to the glomerular basement membrane thickness: Normal versus thin. Am J Kidney Dis 41: 558–564, 2003 [DOI] [PubMed] [Google Scholar]
- 27.Almasy L, Blangero J: Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 62: 1198–1211, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hasstedt SJ: jPAP: Document-driven software for genetic analysis. Genet Epidemiol 29: 255, 2005 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.