Abstract
Leukemia-associated Rho guanine nucleotide exchange factor (LARG) is a RhoA-specific guanine nucleotide exchange factor (GEF) that can activate RhoA. The PDZ (PSD-95/Disc-large/ZO-1 homology) domain of LARG interacts with membrane receptors, which can relay extracellular signals to RhoA signal transduction pathways. Until now there is no structural and dynamic information about these interactions. Here we report the NMR structures of the LARG PDZ in the apo form and in complex with the plexin-B1 C-terminal octapeptide. Unobservable resonances of the residues in βB/βC and βE/αB loops in apo state were observed in the complex state. A distinct region of the binding groove in the LARG PDZ was found to undergo conformational change compared with other PDZs. Analysis of the 15N relaxation data using reduced spectral density mapping shows that the apo LARG PDZ (especially its ligand-binding groove) is flexible and exhibits internal motions on both picosecond to nanosecond and microsecond to millisecond timescales. Mutagenesis and thermodynamic studies indicate that the conformation of the βB/βC and βE/αB loops affects the PDZ–peptide interaction. It is suggested that the conformational flexibility could facilitate the change of structures upon ligand binding.
Keywords: LARG PDZ, NMR, complex structure, conformational change, internal motions, protein–peptide interaction
Rho GTPases play important roles in the cytoskeleton-involved cell processes such as cell migration, axon guidance, cell cycle events, and membrane transport (Hall 1994; Kaibuchi et al. 1999). They function as the molecular switches cycling between active GTP-bound and inactive GDP-bound states and are highly regulated (Schmidt and Hall 2002). The conversion from the GDP-bound form to the GTP-bound form is catalyzed by guanine nucleotide exchange factors (GEFs), which serve as critical mediators of Rho GTPases activation. Leukemia-associated RhoGEF (LARG) is a RhoA-specific RhoGEF and consists of four domains: PDZ, RGS, DH, and PH (Kourlas et al. 2000; Reuther et al. 2001). The DH and PH domains directly contribute to the substrate catalysis and RhoA activation (Kristelly et al. 2004).
The LARG PDZ domain can interact with several membrane proteins, including plexin-B1 (Aurandt et al. 2002; Swiercz et al. 2002), IGF-1 receptor (Taya et al. 2001), and CD44 (Bourguignon et al. 2006). Through these interactions, extracellular signals can stimulate the activation of RhoA. Plexin-B1 is a Semaphorin 4D receptor that governs cell adhesion and migration and is crucial for cell–cell signaling events in multiple tissues (Tamagnone et al. 1999; Oinuma et al. 2004). The interaction of the LARG PDZ with the carboxyl-terminal sequence of plexin-B1 promotes RhoA activation and thereby regulates cytoskeleton rearrangements during axonal and dendritic guidance, resulting in axonal growth cone collapse and cell migration (Artigiani et al. 1999; Tamagnone et al. 1999). The interaction with the carboxyl-terminal sequence of IGF-1 receptor also regulates cytoskeleton rearrangements (Taya et al. 2001). These interactions are essential in the activation of RhoA in response to extracellular stimulants.
Several reports suggested that the interactions of the PDZ domain in RhoGEFs with diverse target receptors initiate conformational changes leading to the activation of RhoA GTPase (Longhurst et al. 2006; Paduch et al. 2007), but until now there is no information about the structural aspect of these interactions. The important issues related to the trigger of structural changes remain unclear. Most PDZs recognize the extreme C termini of the target proteins and consequently have been classified according to their specificity for C-terminal peptides (Songyang et al. 1997). However, accumulating evidence shows that several PDZs recognize more than one class of C-terminal motifs (Kang et al. 2003; Basdevant et al. 2006). And, also, the dynamical properties of these recognitions have not been clearly delineated.
Here, using NMR spectroscopy, we have determined the solution structures of the LARG PDZ in the apo form and in complex with plexin-B1 C-terminal octapeptide; conformational differences between the two states are observed. 15N relaxation data and reduced spectral density mapping analysis reveal the dynamic properties of the LARG PDZ. The binding characteristics and the internal motions of the LARG PDZ are explored.
Results
Structure of the apo LARG PDZ
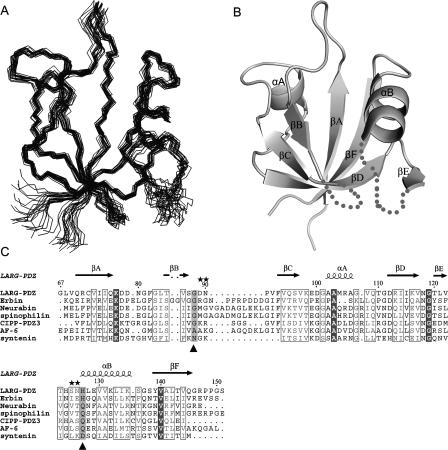
The ensemble of the 20 lowest-energy structures of the apo LARG PDZ is shown in Figure 1A and the structural statistics are listed in Table 1. The secondary structural elements include the following residues: Q70–Q76 (βA); T85–S87 (βB); F93–S96 (βC); A102–A106 (αA); G111–V117 (βD); T120–L121 (βE); H127–K135 (αB); and Y139–Q145 (βF). All of the β-strands are antiparallel (Fig. 1B).
Figure 1.
Structure ensemble and sequence alignment of the apo LARG PDZ. (A) Backbone superimposition of the 20 lowest-energy structures using residues 67–88, 93–121, and 128–147. (B) Ribbon representation of the average, energy-minimized structure. The βB/βC loop (89–92) and βE/αB loop (122–126) with unobservable resonances are shown in dashed lines. (C) Sequence alignment of the class V PDZ domains. Two critical positions, the first of which immediately follows the βB, and the second of which is at the beginning of the αB, are colored gray and marked below with a triangle (▲). Positions of the mutations are marked above with stars (★).
Table 1.
Structural statistics for the LARG PDZ domain in the apo state and the complex with plexin-B1 C-terminal peptidea
An interesting phenomenon is observed for the residues 89–92 and 122–127. For these two groups the resonance signals of the backbone amide groups could not be observed in 1H–15N HSQC spectra at temperatures ranging from 278K to 313K and pH ranging from 6.5 in phosphate buffer to 4.4 in acetate buffer. Carbon-13 resonances corresponding to the residues in βB/βC loop (89–91) and βE/αB loop (122–126) were not observed in 1H–13C HSQC either (data not shown). Consequently, the conformation of the βB/βC and βE/αB loops with missing resonances could not be defined.
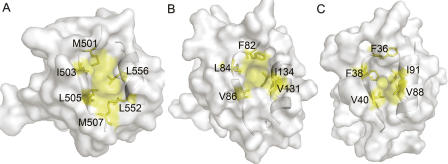
Using the DALI server (Holm and Sander 1993), structure of the LARG PDZ was identified to be most similar to that of GRIP1 PDZ7 (1M5Z), with a root mean square deviation (RMSD) of 1.4 Å for backbone Cα atoms in the secondary structure elements. Both of them have smaller βB/αB binding pockets compared with other PDZs (Fig. 2), which in GRIP1 PDZ7 is not capable of binding carboxyl peptide ligands (Feng et al. 2002). However, in spite of the shallow-binding groove occupied by bulky hydrophobic residues F82, V84, V86, V131, and I134 (Fig. 2B), the LARG PDZ can recognize classical carboxyl-terminal motifs, such as plexin-B1 C-terminal sequence.
Figure 2.
Comparison of the βB/αB binding pockets. Transparent solvent accessible surface models of PDZ domains, with ribbon representation of βB and αB. Side chains of the hydrophobic residues forming βB/αB binding pockets are labeled and colored yellow; other residues are colored white. (A) Human CASK/LIN-2 PDZ (1KWA). (B) LARG PDZ (2OMJ). (C) GRIP1 PDZ7 domain (1M5Z).
Structure of the LARG PDZ in complex with plexin-B1 C-terminal peptide
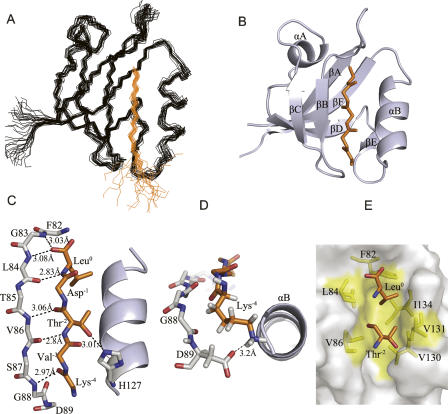
The solution structure of the LARG PDZ/plexin-B1 C-terminal peptide complex (Fig. 3A,B) was calculated using a total of 1306 experimental restraints including 1155 intramolecular NOE and hydrogen bond restraints, 90 dihedral angle restraints, and 61 intermolecular NOE restraints. The statistics for the complex structure are presented in Table 1. The structure of the plexin-B1 C-terminal peptide (VENKVTDL) within the complex is well defined for the last five C-terminal residues that contact with the PDZ domain. Standard PDZ ligand nomenclature is used to describe the peptide residues; the C terminus is designated as residue0 and the remaining residues are numbered with negative integers whose absolute value increases toward the N terminus (Aasland et al. 2002).
Figure 3.
Structure of the LARG PDZ/plexin-B1 peptide complex indicating the hydrogen bonding, electrostatic, and hydrophobic interactions. (A) Backbone overlay of the 20 structures with the lowest energy. The LARG PDZ is shown in black, and plexin-B1 peptide is shown in orange. (B) Ribbon representation of the energy-minimized average structure. For clarity, only the last five C-terminal residues of peptide are displayed. (C) Hydrogen bonds (dashed lines) between protein and peptide, with the bond lengths labeled. (D) Electrostatic interaction between the side chains of D89 and Lys−4. Rotated (x = −90°) from panel C. (E) Stick diagram depicting the hydrophobic interactions. Large hydrophobic pocket (yellow) composed of residues F82, L84, V86, V130, V131, and I134 in LARG PDZ adopts the side chains of Leu0 and Thr−2 (orange).
The plexin-B1 C-terminal peptide binds into the βB/αB groove of the LARG PDZ in a classical antiparallel fashion. Hydrogen bonds, electrostatic, and hydrophobic interactions between the protein and peptide are shown in detail in Figure 3C–E. Using the Protein–Protein Interaction Server, seven intermolecular hydrogen bonds were deduced from the geometry of the structure, including pairs L84–Leu0 and V86–Thr−2, which were also determined from H/D exchange experiments. The carbonyl oxygen of Leu0 also forms a hydrogen bond with the backbone amide of G83. The Nε atom of H127 in the helix αB bonds with the hydroxyl group of Thr−2. A hydrogen bond is also present between the backbone NH of G88 and the carbonyl oxygen of Lys−4. The bond lengths are shown in Figure 3C. The positively charged Lys−4, with its side chain stretching out of the binding groove, gives six weak intermolecular NOEs with residues G88 and D89, suggesting its proximity to the negatively charged D89 to be sufficient to form electrostatic interaction (Fig. 3D). Hydrophobic interactions also contribute to the binding of plexin-B1 peptide. The side chain of Leu0 is embedded tightly in a hydrophobic pocket formed by residues F82, L84, and I134. The methyl group of Thr−2 inserts into another hydrophobic pocket surrounded by V86, V130, and V131 (Fig. 3E). These hydrogen bonds, electrostatic interactions, and hydrophobic interactions stabilize the LARG PDZ/plexin-B1 peptide complex.
Conformational differences between the apo and complex states
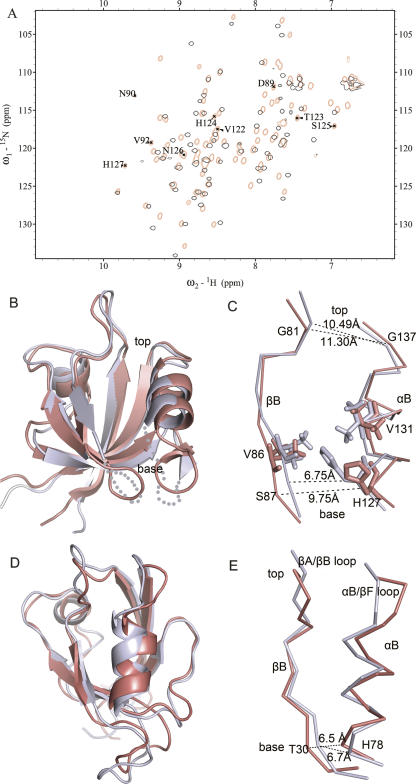
A remarkable difference between the apo and complex structures is that the unstructured βB/βC and βE/αB loops get stabilized conformation upon binding of the plexin-B1 peptide. Observable amide resonances for residues in the two loops are labeled in Figure 4A. Superimpositions of the 1H–15N HSQC spectra (Fig. 4A) and the structures in the two states (Fig. 4B) elucidate the binding-dependent conformational changes. RMSD reaches 1.7 Å for the 80 equivalent Cα atoms (residues 68–147); this is relatively large among PDZ domains that usually experience slight structural changes upon ligand binding with RMSDs lower than 1.2 Å for backbone atoms (Fuentes et al. 2004; von Ossowski et al. 2006). The major differences are found in the strands βB, βE, helix αB, and regions around the binding groove. Other secondary structure elements, including the strands βA, βC, and βF, undergo very small changes. Compared with the apo structure, peptide-bound LARG PDZ tilts the βB and αB apart to become parallel with each other; the rotation angle is ∼10° for βB and ∼15° for αB. These conformational changes open the base portion of the hydrophobic pocket which seems to be closed in the apo state and increase the accessible surface area of the pocket to provide better binding of the peptides (Fig. 5). The side chains of H127 and V131 (Supplemental Fig. S1) that extend into the binding groove in the apo form shift their positions away from the binding groove in the structure of complex (Fig. 4C).
Figure 4.
Distinct conformational change of the LARG PDZ compared with other PDZs. (A) Overlay of 1H, 15N-HSQC spectra of LARG PDZ in the apo form (black) and in complex with superfluous plexin-B1 peptide (red). The observable peaks for the residues in the βB/βC loop and βE/αB loop are assigned and labeled in the HSQC spectrum of the complex state. (B) Structure comparison of the LARG PDZ in the apo- (light blue) and ligand-bound (red) states. (C) Conformational changes of the binding groove and the side chains of H127 and V131, accompanied by the opening of the binding pocket at the base portion. The top and base widths of the binding groove are shown in dashed lines and measured. (D) Superposition of the free (PDB code 1GM1, light blue) and the complex (PDB code 1VJ6, red) structures of PTP-BL PDZ2. (E) The base widths of the binding groove of the PTP-BL PDZ2 in two forms are displayed.
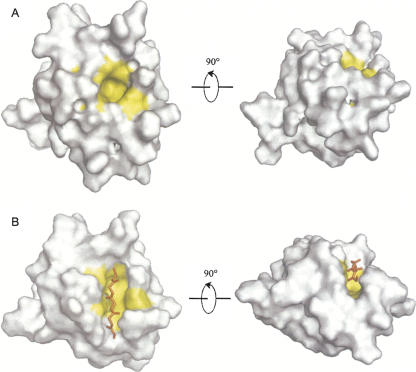
Figure 5.
Surface models of the LARG PDZ in two states highlighting the βB/αB binding pocket. (A) Apo state. (B) Plexin-B1 peptide-bound state. Residues F82, L84, V86, V130, V131, and I134 composing the binding pocket are colored yellow. Shown in the right panels are models rotated (x = −90°) from the left panels for a clear view.
Distinct conformational change as compared with other PDZs
Conformational change of the LARG PDZ observed at the base of the binding groove differs from the conformational changes taking place in other PDZs. Upon binding of the plexin-B1 peptide, the N-terminal part of the helix αB is tilted counterclockwise; the C-terminal part of the strand βB is tilted clockwise; and the βB/βC and βE/αB loops are rearranged, resulting in opening of the binding groove at the bottom. The bottom width increases from 6.75 ± 0.27 Å in the apo state to 9.75 ± 0.55 Å in the bound state, calculated from the S87–H127 Cα–Cα distance. The top width calculated from the G81–G137 Cα–Cα distance exhibits a minor change from 10.49 ± 0.55 Å to 11.30 ± 0.51 Å (Fig. 4C). In contrast, other PDZs, such as SAP97 PDZ2 and PTP-BL PDZ2, undergo conformational changes at the top instead of the base part with the movement of the βA/βB loop and αB/βF loop. Superposition of free (PDB code 1GM1) and peptide-bound (PDB code 1VJ6) structures of PTP-BL PDZ2 (Fig. 4D) reveals that the C-terminal portion of αB reorients away from the hydrophobic core by ∼10° relative to the βB, coupling with overall position shifting of the αB/βF loop and βA/βB loop (Gianni et al. 2006). The bottom width of the hydrophobic groove, calculated from the corresponding T30–H78 Cα–Cα distance, shows small change (from 6.50 ± 0.40 Å to 6.70 ± 0.22 Å, Fig. 4E). In conclusion, different parts of the binding groove undergo rearrangements in LARG PDZ as compared with other PDZs.
Internal motions indicated by 15N relaxation data
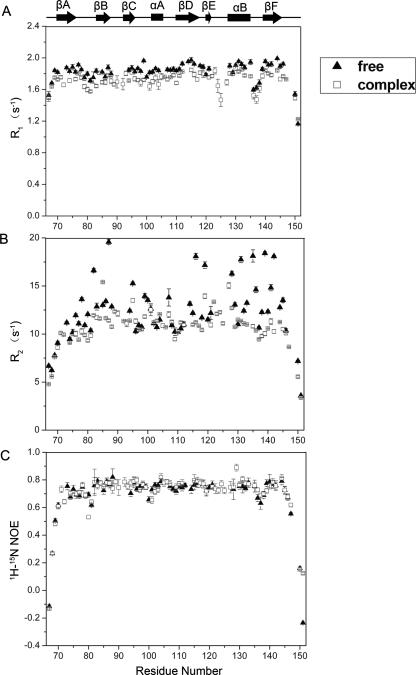
Backbone relaxation data R 1, R 2, and 15N–1H NOE (Fig. 6) revealed the dynamic properties of the LARG PDZ; 61 and 72 of the 85 residues were used for analysis of the apo and complex state, respectively. Unanalyzed residues were prolines and the ones that were not assigned, overlapped, or too weak to be accurately quantified. The R 2 values in the apo state are variable and heterogeneous, exhibiting large fluctuations ranging from 7.78 to 19.58 s−1, with the mean value of 12.17 (±2.70) s−1. In contrast, the R 2 parameters in the complex state are relatively stable with the mean value of 10.70 (±1.35) s−1. The protein was finally purified using gel filtration under reducing conditions and was present as the monomer in the NMR sample. This was also confirmed by chemical cross-linking experiments (Supplemental Fig. S2) and is consistent with Paduch and colleagues’ report that the LARG PDZ exists as monomers under reducing conditions (Paduch et al. 2007). Residues with R 2/R 1 values that differ more than one standard deviation from the mean value are considered undergoing conformational exchange processes (Clore et al. 1990). The much higher R 2 values for some residues in βB, βD, and αB may result from the contribution of internal motions. The 1H–15N NOE, with average values of 0.69 ± 0.11 and 0.72 ± 0.16 for the apo and the complex state, respectively, is lower than +0.82 expected for NH groups of a rigid globular protein tumbling isotropically (Kay et al. 1989). The relaxation data imply internal motions in the apo LARG PDZ.
Figure 6.
15N relaxation parameters for the LARG PDZ in free (▲) and peptide-bound (□) states. Only those residues for which 1H–15N cross-peaks are good enough to permit accurate measurements of their intensities are analyzed. The secondary structure elements are indicated at the top. (A) R 1. (B) R 2. (C) 15N–1H NOE.
Internal motions derived from reduced spectral density mapping
The ratios of the principle components of inertia tensors for the apo and complex forms of LARG PDZ are 1:0.80:0.62 and 1:0.78:0.57, respectively, suggesting motional anisotropy. Fitting relaxation data to axially symmetric and fully asymmetric diffusion tensor models using the program TENSOR2 (Dosset et al. 2000) resulted in rejected models. Hence, we could not analyze the data using the conventional model-free approach with acceptable confidence.
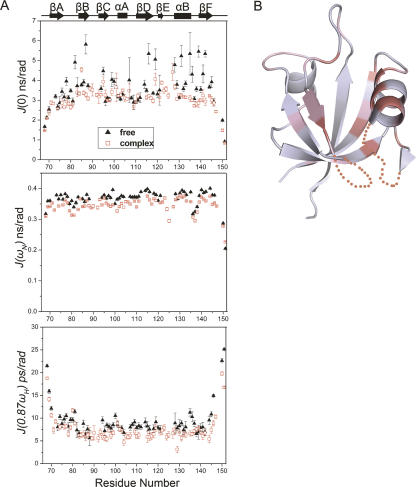
Reduced spectral density mapping (Peng and Wagner 1992; Farrow et al. 1995; Palmer III 2004; Eldho and Dayie 2007) describing the internal motions is used to analyze the relaxation data. The spectral density functions at three frequencies, J(0), J(ω N), and J(0.87ω H), correspond to relaxation contributions from the motions on different timescales (Maguire et al. 2005). High J(0) value may be indicative of slow motions on the millisecond to microsecond timescale, whereas J(0.87ω H) is sensitive only to fast internal motions on the picosecond to nanosecond timescale (Dyson and Wright 2001). The results are summarized in Figure 7A. For the apo LARG PDZ, almost all of the residues in the secondary structural elements exhibited J(0.87ω H) > 7.5 ps/rad, with an average value of 9.4 ps/rad, indicating fast internal motions (Kuang et al. 2006). The J(0) varied dramatically for different parts of the protein. Residues F82, S87, Q95, K116, G119, L128, V131, K135, Y139, and L142 showed J(0) values >4.51 ns/rad (one standard deviation above the mean value) and high J(0.87ω H) values, strongly implying slow internal motions on a microsecond to millisecond timescale in and around the binding groove (Fig. 7B). Since the J(0) were considerably heterogeneous, some residues with J(0) larger than the mean value of 3.59 ns/rad but smaller than 4.51 ns/rad might undergo slow internal motions. For the PDZ/peptide complex, most of the residues in the secondary structure elements showed J(0.87ω H) values <7.5 ps/rad and relatively uniform J(0) values (3.08 ns/rad on the average). As a result, the apo LARG PDZ exhibits fast and slow internal motions; these internal motions are considerably reduced upon the binding of plexin-B1 peptide.
Figure 7.
Reduced spectral density function analysis and mapping of the residues with slow internal motions onto the structure of the apo LARG PDZ. (A) Spectral density functions J(0), J(ω N), and J(0.87ω H) and their uncertainties were calculated for the apo (▲) and peptide-bound (□) LARG PDZ. (B) Structure of the apo LARG PDZ. Residues showing J(0) > 4.51 ns/rad (the mean value plus one standard deviation) are colored deep pink; residues with J(0) values between 3.59 nsc/rad (mean value) and 4.51 ns/rad are colored light pink.
Contributions of the βB/βC and βE/αB loops to binding affinity
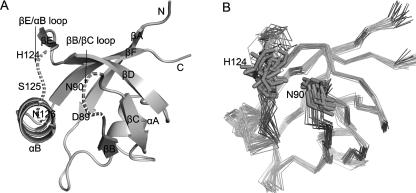
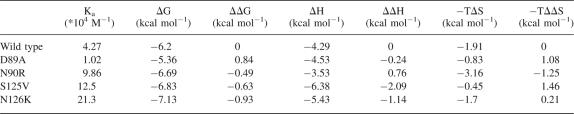
A set of mutants, including D89A, N90R in the βB/βC loop, and S125V, N126K in the βE/αB loop (Fig. 8A), was constructed in order to determine the contribution of the two loops to the interaction between PDZ and plexin-B1 peptide by isothermal titration calorimetry (ITC). The structural integrity of mutants was assessed by 1H–15N HSQC spectra (Supplemental Fig. S3). Thermodynamic parameters calculated from the titration curves are presented in Table 2. In the D89A mutant, disruption of the electrostatic interaction between D89 and Lys−4 and alteration of the packing of nearby residues resulted in a threefold lower binding constant of ∼1.02 × 104 M−1. The entropy loss (−TΔS) due to binding increased by 1.08 kcal mol−1 while the enthalpy slightly decreased, indicating the deletion of the electrostatic interaction is entropically unfavorable. N90R is a charge-introducing mutation, creating additional steric bulk and electrostatic interaction with residue H124, which is situated opposite to N90 (Fig. 8B). These may drive the two loops apart from each other and stabilize them. Superimposing the 1H, 15N-HSQC spectra of mutant N90R and wild type revealed that there were several new cross-peaks in that of N90R, which might be assigned to residues in the βB/βC and βE/αB loops (Supplemental Fig. S3A). Mutant N90R showed a onefold increase in the binding affinity, completely compensating the unfavorable enthalpy increase by a decrease in the negative entropy change (–TΔΔS). In two other mutants, S125V and N126K, the decreases of enthalpy change (ΔΔH) were about −2.09 and −1.14 kcal mol−1, respectively, outweighing the unfavorable entropy change (–TΔΔS) of ∼1.46 and 0.21 kcal mol−1 and resulting in enhanced binding affinities.
Figure 8.
Locations of the mutations. (A) Dashed lines denote the positions of mutated residues: D89A, N90R in the βB/βC loop and S125V, N126K in the βE/αB loop. (B) The backbone atoms of the 20 lowest-energy structures are shown in the same orientation as A, highlighting the opposite positions of N90 and H124. Although the two loops are unstructured in the apo state, they were geometrically constrained in a limited district, especially the short βB/βC loop.
Table 2.
Thermodynamic parameters of interactions between mutant PDZ and plexin-B1 peptide
Binding characteristics of the LARG PDZ
Chemical shift perturbation experiments on the LARG PDZ showed different affinities and exchange rates upon binding of different ligands. The dissociation constants (K d) for plexin-B1 peptide (VENKVTDL) and the IGF-1 receptor peptide (LPLPQSSTC) were ∼23.4 μM and 40 μM, respectively. Upon titration of each peptide, the interactions were in the intermediate-fast-exchange regime. Most of the residues were greatly perturbed and the backbone amide cross-peaks for the residues in the βB/βC and βE/αB loops became observable in the 1H–15N HSQC spectrum (Fig. 4A; Supplemental Fig. S4A). Titration with the CD44 C-terminal peptide (QNVDMKIGV) or cytoplasmic domain of CD44 implied that the interaction between PDZ and CD44 was in the fast-exchange regime with a minor change in chemical shifts. Resonances for the residues in the βB/βC and βE/αB loops were still unobservable in the 1H–15N HSQC spectrum (Supplemental Fig. S4B).
Discussion
According to the classification rule described by Songyang et al. (1997), C-terminal peptide of plexin-B1 is a class I PDZ recognition sequence; C termini of CD44 is a class II ligand and the IGF-1 receptor peptide belongs to another class. LARG PDZ is classified to be a class V PDZ domain, which can bind both class I and II motifs (Kelker et al. 2007). There are about six other known PDZs belonging to this class, namely Erbin PDZ (Birrane et al. 2003), CIPP PDZ3 (Kurschner et al. 1998; Bezprozvanny and Maximov 2001), Syntenin PDZ2 (Kang et al. 2003), AF-6 PDZ (Zhou et al. 2005), neurabin PDZ, and spinophilin PDZ (Kelker et al. 2007). Sequence alignment of class V PDZs revealed that LARG PDZ and Erbin PDZ differ from others in two positions critical for binding selectivity: The first immediately follows the strand βB, and the second one is at the beginning of the helix αB (Bezprozvanny and Maximov 2001). LARG PDZ and Erbin PDZ are of (G, H) group and canonical to bind class I motif, whereas the other class V PDZs have Gln or Asp instead of His at the second critical position (Fig. 1C). Another noticeable feature of the LARG PDZ is its short βB/βC loop, instead of characteristic long loops in other PDZs, especially in Erbin PDZ. Structural comparison of the LARG PDZ and Erbin PDZ reveals that their major differences are located in the helix αA, αB, and loop regions. Erbin PDZ shows no structural change in the binding pocket and slight changes in the βB/βC and βE/αB loops upon binding of the class I or class II ligands, indicating less structural variability (Skelton et al. 2003). In contrast, the LARG PDZ shows substantial conformational changes in the hydrophobic pocket, the βB/βC loop, and the βE/αB loop upon the class I ligand binding, suggesting mobility of these parts of the protein. Moreover, major conformational changes in the LARG PDZ take place at the base of the binding groove, rather than the top portion as in the case of other PDZs.
The apo form of LARG PDZ undergoes internal motions on both fast and slow timescales, especially the residues in and around the ligand-binding pocket which are surface exposed, relatively free from steric hindrance, and thereby with more motional degrees of freedom. Considering that signals for residues in the βB/βC loop (D89–V91) and βE/αB loop (V122–N126) could not be observed, we focused on the adjacent residues, including S87, G88, and L128. J(0) values for residues S87 and E128 exceeded the mean value by more than one standard deviation, indicating conformational exchange contributions; G88 showed very weak NH cross-peak. Consequently, we interpret the absence of the resonances in 1H, 13C-HSQC and 1H, 15N-HSQC spectra as the result of intermediate timescale conformational exchange between two or more states, with signals too broad to be detected. Mapping of the residues with slow internal motions onto the solution structure of the apo PDZ revealed that these residues are clustered in the ligand-binding side (Fig. 7B). Fast and slow internal motions in the binding groove, especially in the βB/βC loop and βE/αB loop, indicate the possibility of reorientation of the binding pocket. Steric interactions, hydrogen bonds, and electrostatic interactions dramatically reduced the internal motions upon binding of the plexin-B1 peptide.
Mutagenesis analysis and thermodynamic data demonstrated that the βB/βC and βE/αB loops affect protein–peptide interactions. The intramolecular electrostatic repulsion in the mutant N90R and steric hindrances with bulkier side chains in mutants S125V and N126K were introduced; all mutations may contribute to widening the base of the binding groove and thereby to increasing the binding constants. These facts suggest that the conformation of the βB/βC loop and βE/αB loop affects the binding characteristics of the LARG PDZ. These two loops with fast and slow internal motions are apt to work in a cooperative way to drive the reorientation of βB and αB to widen the binding groove and accommodate the ligands. In the LARG PDZ domain, the loop mobility may contribute to the conformational change of the binding groove. The conformational flexibility could facilitate the changes in structures upon the binding of different ligands.
Materials and Methods
Protein expression and purification
The DNA fragment encoding residues 67–151 of human LARG PDZ (GenBank number AF180681) was amplified and cloned into plasmid pET15b (+) (Novagen) using the NdeI and XhoI cloning sites. Sequencing verified the construct. The recombinant plasmid was transformed into Escherichia coli BL21 (DE3) and cells were grown in M9 medium supplemented with uniformly 15N-labeled 15NH4Cl (0.5 g/L) and 13C6-glucose (2.5 g/L) as stable isotope sources for protein production. Recombinant protein was purified using Hi-trap chelating column (Pharmacia) chromatography and completely digested with thrombin protease (Amersham Biosciences) at 4°C to remove the N-terminal His-tag. At last the protein was purified by means of gel-filtration chromatography on a Superdex G75 column (Amersham Pharmacia Biotech) under reducing conditions (buffer with 5 mM DTT). Purity of the recombinant protein was confirmed by SDS-PAGE, and the concentration was estimated using BCA kits (Pierce). The CD44 cytoplasmic domain (amino acids 296–361, code in GenBank M59040) was cloned, expressed, and purified as described above.
The C-terminal peptides of VENKVTDL (residues 2128–2135 of plexin-B1, GenBank accession number NM_002673), LPLPQSSTC (residues 4096–4104 of IGF-1 receptor, GenBank NM_000875), and QNVDMKIGV (residue 353–361 of CD44, GenBank M59040) were chemically synthesized using a standard Fmoc method at GL Biochem (Shanghai) Ltd. The synthetic peptides were purified on a reverse-phase HPLC C18 column, eluted with an acetonitrile gradient of 15%–30%. The final product was lyophilized and verified by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry and NMR signal assignments.
Site-directed mutagenesis
Mutants D89A, N90R, S125V, and N126K represent mutations of Asp89 to Ala, Asn90 to Arg, Ser125 to Val, and Asn126 to Lys, respectively. Site-directed mutations were generated by the conventional PCR method using the LARG PDZ-containing pET15b (+) plasmid as a template; the plasmid template was digested by the endonuclease after the PCR reaction. Correctness of the mutations was confirmed by sequencing. The integrity of the mutants was assessed by the 1H, 15N-HSQC spectra.
Isothermal titration calorimetry
ITC experiments were performed at 293K using a VP-ITC titration microcalorimeter (MicroCal). Protein and peptide samples in the same buffer as that used for NMR experiments were centrifuged and thoroughly degassed before the experiments. The 2.72 mM solution of the plexin-B1 C-terminal peptide was titrated into the sample cell containing the LARG PDZ or its mutants (50–100 μM). Control experiments and dilution experiments were performed by making identical injection of peptide into the sample cell containing only buffer to correct the heat effects not directly related to the binding reaction. Data were analyzed with Microcal's Origin software. The enthalpy change (ΔH), entropy change (TΔS = −ΔG + ΔH), and the binding constant (K a) of the interactions were obtained from fitting of the experimental titration curve.
NMR spectroscopy and data processing
The NMR sample of the apo LARG PDZ contained ∼0.8 mM of protein dissolved in 50 mM phosphate buffer with 50 mM NaCl, 5 mM EDTA, 5 mM DTT in 90% H2O, 10% D2O (pH 6.0). For the PDZ/peptide complex sample, the 15N, 13C-labeled LARG PDZ (∼0.8 mM) mixed with plexin-B1 C-terminal peptide (∼7.6 mM) was dissolved in the same buffer as described above. NMR experiments were carried out at 293K on Bruker DMX500 and DMX600 spectrometers with self-shielded Z-axis gradients. The following spectra were recorded to obtain backbone and side chain resonance assignments: 2D 1H, 15N-HSQC, 2D 1H, 13C-HSQC, 3D CBCANH, CBCA(CO)NH, HNCO, HN(CA)CO and HBHA(CBCACO)NH, C(CO)NH-TOCSY, H(CCO)NH-TOCSY, HCCH-TOCSY, and HCCH-COSY. 3D 15N-separated and 13C-separated NOESY were acquired with a mixing time of 110 ms. For the peptide in apo state, the resonances were assigned to individual protons by conventional 2D TOCSY and NOESY experiments. Peptide resonance assignments in complex state were obtained from heteronuclear X-Filter 1H PFG double-quantum experiments (Dalvit et al. 1998) both in H2O and D2O. 2D 13C-filtered (F1), 13C-edited (F2) NOESY spectrum was used to detect the intermolecular interactions. Data were processed using the program NMRPipe software (Delaglio et al. 1995). Spectra were analyzed and assigned using the program SPARKY 3 (T.D. Goddard and D.G. Kneller, University of California, San Francisco).
NMR structure calculation
NOEs were classified into four distance ranges according to their intensities: strong (1.8–3.0 Å), medium (1.8–4.0 Å), weak (1.8–5.0 Å), and very weak (1.8–6.0 Å). The 1.8 Å lower limits were imposed only implicitly by the van der Waals repulsion force. Phi and psi dihedral angle restraints were obtained based on analysis of 13Cα, 13Cβ, 13CO, 15N, and 1Hα chemical shifts using the program TALOS (Cornilescu et al. 1999). Structures were calculated using the program CNS v1.1 (Brunger et al. 1998). For the initial round of calculation, only unambiguous NOEs of sequential, and medium-range, and dihedral angle restraints were used; other long-range NOEs and hydrogen bonds were then introduced into calculation steps. For the final stage, 200 structures were calculated and the 20 conformers with the lowest energy were selected to form the final ensemble. The calculated structures were analyzed and visualized by the programs PROCHECK (Laskowski et al. 1996), MOLMOL (Koradi et al. 1996), and PyMOL (Delano Scientific).
Chemical cross-linking of the LARG PDZ
Proteins at concentrations of 5 mg/mL and 8 mg/mL under reducing condition (5 mM DTT) and another sample of 12 mg/mL under nonreducing condition (without DTT) were centrifuged at 12,000g for 5min. The supernatants were then treated with DMSO alone or disuccinimidyl suberate (DSS) in DMSO from a 10-fold stock solution to the final concentration of 10 mM. After 30-min incubation at 37°C, the cross-linking reaction was stopped and samples were solubilized with the sample buffer (Bio-Rad), boiled for 15 min, and centrifuged at 12,000g for 10 min. The proteins were loaded on 15% SDS-PAGE gel, followed by Coomassie brilliant blue staining.
15N relaxation measurements
15N relaxation experiments were carried out on a Bruker DMX 500 NMR spectrometer at 293K using the published methods (Farrow et al. 1994). The samples of the apo PDZ and PDZ/plexin-B1 peptide complex were dissolved in the same buffer as described above. With a 1-s recycle delay, the T 1 and T 2 were measured with eight relaxation delays (11.15, 61.3, 141.54, 241.84, 362.2, 522.68, 753.37, and 1144.54 ms) and seven relaxation delays (0, 17.6, 35.2, 52.8, 70.4, 105.6, and 140.8 ms), respectively. The spectra measuring 1H–15N NOE were acquired with a 2-s relaxation delay, followed by a 3-s period of proton saturation. In the absence of proton saturation, the spectra were recorded by a relaxation delay of 5 s. The exponential curve fitting and data analysis were carried out with the program Sparky (T.D. Goddard and D.G. Kneller, University of California, San Francisco).
Reduced spectral density mapping
Reduced spectral density mapping with the assumption that the variation in J(ω) is relatively constant between J(ωH + ωN) and J(ωH − ωN) (Peng and Wagner 1992; Farrow et al. 1995) was used to analyze the relaxation data. The magnitudes of J(ω) are sensitive to motions at the corresponding frequencies. The J(0), J(ω N), and J(0.87ω H) values were obtained according to the Equations 1–4:
where d = (μ 0 hγNγH/(8π 2))/(r 3 NH), c = ω NΔσ/√3, μ 0 is the permeability of free space, h is the Planck's constant; γH and γN are the gyromagnetic ratios of 1H and 15N, respectively; r NH = 1.02 Å is the average amide bond length; Δσ = −160 ppm is the chemical shift anisotropy for the 15N nuclei. The residues in the secondary structural elements were used to calculate the mean values.
Data deposition
The atomic coordinates for the LARG PDZ have been deposited in the Protein Data Bank with accession codes 2OMJ (apo state) and 2OS6 (in complex with plexin-B1 C-terminal peptide). The 1H, 15N, and 13C assignments have been deposited in BioMagResBank under the accession numbers BMRB-15168 and BMRB-15163, respectively.
Electronic supplemental material
The Supplemental material includes the structure ensembles of residues H127 and V131 in two states (Supplemental Fig. S1); the evidence for the monomeric state of the LARG PDZ from the chemical cross-linking experiment (Supplemental Fig. S2); the structural integrity of mutants and overlay of 1H, 15N-HSQC spectra of the wild-type and mutated LARG PDZ (Supplemental Fig. S3); and the structural integrity analysis of the LARG PDZ titrated with different ligands (Supplemental Fig. S4).
Acknowledgments
We thank Professor Changlin Tian for help with this manuscript. This work was supported by the Chinese National Fundamental Research Project (grants 2006CB806507, 2006CB910201, 2002CB713806, and 2006AA02A315) and the Chinese National Natural Science Foundation (grants 30121001 and 30570361).
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Yunyu Shi, School of Life Science, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China; e-mail: yyshi@ustc.edu.cn; fax: 86-551-3601443; or Jihui Wu, School of Life Science, University of Science and Technology of China, Hefei, Anhui 230026, People's Republic of China; e-mail: wujihui@ustc.edu.cn; fax: 86-551-3601443.
Abbreviations: LARG, leukemia-associated Rho guanine nucleotide exchange factor; GEF, guanine nucleotide exchange factor; PDZ, PSD-95/Disc-large/ZO-1 homology; RMSD, root mean square deviation; ITC, isothermal titration calorimetry.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073416508.
References
- Aasland, R., Abrams, C., Ampe, C., Ball, L.J., Bedford, M.T., Cesareni, G., Gimona, M., Hurley, J.H., Jarchau, T., Lehto, V.P., et al. Normalization of nomenclature for peptide motifs as ligands of modular protein domains. FEBS Lett. 2002;513:141–144. doi: 10.1016/s0014-5793(01)03295-1. [DOI] [PubMed] [Google Scholar]
- Artigiani, S., Comoglio, P.M., Tamagnone, L. Plexins, semaphorins, and scatter factor receptors: A common root for cell guidance signals? IUBMB Life. 1999;48:477–482. doi: 10.1080/713803563. [DOI] [PubMed] [Google Scholar]
- Aurandt, J., Vikis, H.G., Gutkind, J.S., Ahn, N., Guan, K.L. The semaphorin receptor plexin-B1 signals through a direct interaction with the Rho-specific nucleotide exchange factor, LARG. Proc. Natl. Acad. Sci. 2002;99:12085–12090. doi: 10.1073/pnas.142433199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basdevant, N., Weinstein, H., Ceruso, M. Thermodynamic basis for promiscuity and selectivity in protein–protein interactions: PDZ domains, a case study. J. Am. Chem. Soc. 2006;128:12766–12777. doi: 10.1021/ja060830y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezprozvanny, I., Maximov, A. Classification of PDZ domains. FEBS Lett. 2001;509:457–462. doi: 10.1016/s0014-5793(01)03214-8. [DOI] [PubMed] [Google Scholar]
- Birrane, G., Chung, J., Ladias, J.A. Novel mode of ligand recognition by the Erbin PDZ domain. J. Biol. Chem. 2003;278:1399–1402. doi: 10.1074/jbc.C200571200. [DOI] [PubMed] [Google Scholar]
- Bourguignon, L.Y., Gilad, E., Brightman, A., Diedrich, F., Singleton, P. Hyaluronan-CD44 interaction with leukemia-associated RhoGEF and epidermal growth factor receptor promotes Rho/Ras co-activation, phospholipase Cε-Ca2+ signaling, and cytoskeleton modification in head and neck squamous cell carcinoma cells. J. Biol. Chem. 2006;281:14026–14040. doi: 10.1074/jbc.M507734200. [DOI] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Clore, G.M., Driscoll, P.C., Wingfield, P.T., Gronenborn, A.M. Analysis of the backbone dynamics of interleukin-1β using two-dimensional inverse detected heteronuclear 15N-1H NMR spectroscopy. Biochemistry. 1990;29:7387–7401. doi: 10.1021/bi00484a006. [DOI] [PubMed] [Google Scholar]
- Cornilescu, G., Delaglio, F., Bax, A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- Dalvit, C., Ramage, P., Hommel, U. Heteronuclear X-filter 1H PFG double-quantum experiments for the proton resonance assignment of a ligand bound to a protein. J. Magn. Reson. 1998;131:148–153. doi: 10.1006/jmre.1997.1342. [DOI] [PubMed] [Google Scholar]
- Delaglio, F., Grzesiek, S., Vuister, G.W., Zhu, G., Pfeifer, J., Bax, A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Dosset, P., Hus, J.C., Blackledge, M., Marion, D. Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR. 2000;16:23–28. doi: 10.1023/a:1008305808620. [DOI] [PubMed] [Google Scholar]
- Dyson, H.J., Wright, P.E. Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol. 2001;339:258–270. doi: 10.1016/s0076-6879(01)39317-5. [DOI] [PubMed] [Google Scholar]
- Eldho, N.V., Dayie, K.T. Internal bulge and tetraloop of the catalytic domain 5 of a group II intron ribozyme are flexible: Implications for catalysis. J. Mol. Biol. 2007;365:930–944. doi: 10.1016/j.jmb.2006.10.037. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Muhandiram, R., Singer, A.U., Pascal, S.M., Kay, C.M., Gish, G., Shoelson, S.E., Pawson, T., Forman-Kay, J.D., Kay, L.E. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Farrow, N.A., Zhang, O., Szabo, A., Torchia, D.A., Kay, L.E. Spectral density function mapping using 15N relaxation data exclusively. J. Biomol. NMR. 1995;6:153–162. doi: 10.1007/BF00211779. [DOI] [PubMed] [Google Scholar]
- Feng, W., Fan, J.S., Jiang, M., Shi, Y.W., Zhang, M. PDZ7 of glutamate receptor interacting protein binds to its target via a novel hydrophobic surface area. J. Biol. Chem. 2002;277:41140–41146. doi: 10.1074/jbc.M207206200. [DOI] [PubMed] [Google Scholar]
- Fuentes, E.J., Der, C.J., Lee, A.L. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Gianni, S., Walma, T., Arcovito, A., Calosci, N., Bellelli, A., Engstrom, A., Travaglini-Allocatelli, C., Brunori, M., Jemth, P., Vuister, G.W. Demonstration of long-range interactions in a PDZ domain by NMR, kinetics, and protein engineering. Structure. 2006;14:1801–1809. doi: 10.1016/j.str.2006.10.010. [DOI] [PubMed] [Google Scholar]
- Hall, A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu. Rev. Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Holm, L., Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Kaibuchi, K., Kuroda, S., Amano, M. Regulation of the cytoskeleton and cell adhesion by the Rho family GTPases in mammalian cells. Annu. Rev. Biochem. 1999;68:459–486. doi: 10.1146/annurev.biochem.68.1.459. [DOI] [PubMed] [Google Scholar]
- Kang, B.S., Cooper, D.R., Devedjiev, Y., Derewenda, U., Derewenda, Z.S. Molecular roots of degenerate specificity in syntenin's PDZ2 domain: Reassessment of the PDZ recognition paradigm. Structure. 2003;11:845–853. doi: 10.1016/s0969-2126(03)00125-4. [DOI] [PubMed] [Google Scholar]
- Kay, L.E., Torchia, D.A., Bax, A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: Application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kelker, M.S., Dancheck, B., Ju, T., Kessler, R.P., Hudak, J., Nairn, A.C., Peti, W. Structural basis for spinophilin-neurabin receptor interaction. Biochemistry. 2007;46:2333–2344. doi: 10.1021/bi602341c. [DOI] [PubMed] [Google Scholar]
- Koradi, R., Billeter, M., Wuthrich, K. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:29–32. 51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kourlas, P.J., Strout, M.P., Becknell, B., Veronese, M.L., Croce, C.M., Theil, K.S., Krahe, R., Ruutu, T., Knuutila, S., Bloomfield, C.D., et al. Identification of a gene at 11q23 encoding a guanine nucleotide exchange factor: Evidence for its fusion with MLL in acute myeloid leukemia. Proc. Natl. Acad. Sci. 2000;97:2145–2150. doi: 10.1073/pnas.040569197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristelly, R., Gao, G., Tesmer, J.J. Structural determinants of RhoA binding and nucleotide exchange in leukemia-associated Rho guanine-nucleotide exchange factor. J. Biol. Chem. 2004;279:47352–47362. doi: 10.1074/jbc.M406056200. [DOI] [PubMed] [Google Scholar]
- Kuang, Z., Yao, S., Keizer, D.W., Wang, C.C., Bach, L.A., Forbes, B.E., Wallace, J.C., Norton, R.S. Structure, dynamics and heparin binding of the C-terminal domain of insulin-like growth factor-binding protein-2 (IGFBP-2) J. Mol. Biol. 2006;364:690–704. doi: 10.1016/j.jmb.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Kurschner, C., Mermelstein, P.G., Holden, W.T., Surmeier, D.J. CIPP, a novel multivalent PDZ domain protein, selectively interacts with Kir4.0 family members, NMDA receptor subunits, neurexins, and neuroligins. Mol. Cell. Neurosci. 1998;11:161–172. doi: 10.1006/mcne.1998.0679. [DOI] [PubMed] [Google Scholar]
- Laskowski, R.A., Rullmannn, J.A., MacArthur, M.W., Kaptein, R., Thornton, J.M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Longhurst, D.M., Watanabe, M., Rothstein, J.D., Jackson, M. Interaction of PDZRhoGEF with microtubule-associated protein 1 light chains: Link between microtubules, actin cytoskeleton, and neuronal polarity. J. Biol. Chem. 2006;281:12030–12040. doi: 10.1074/jbc.M513756200. [DOI] [PubMed] [Google Scholar]
- Maguire, M.L., Guler-Gane, G., Nietlispach, D., Raine, A.R., Zorn, A.M., Standart, N., Broadhurst, R.W. Solution structure and backbone dynamics of the KH-QUA2 region of the Xenopus STAR/GSG quaking protein. J. Mol. Biol. 2005;348:265–279. doi: 10.1016/j.jmb.2005.02.058. [DOI] [PubMed] [Google Scholar]
- Oinuma, I., Ishikawa, Y., Katoh, H., Negishi, M. The Semaphorin 4D receptor Plexin-B1 is a GTPase activating protein for R-Ras. Science. 2004;305:862–865. doi: 10.1126/science.1097545. [DOI] [PubMed] [Google Scholar]
- Paduch, M., Biernat, M., Stefanowicz, P., Derewenda, Z.S., Szewczuk, Z., Otlewski, J. Bivalent peptides as models for multimeric targets of PDZ domains. Chembiochem. 2007;8:443–452. doi: 10.1002/cbic.200600389. [DOI] [PubMed] [Google Scholar]
- Palmer A.G., III NMR characterization of the dynamics of biomacromolecules. Chem. Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- Peng, J.W., Wagner, G. Mapping of the spectral densities of N–H bond motions in Eglin c using heteronuclear relaxation experiments. Biochemistry. 1992;31:8571–8586. doi: 10.1021/bi00151a027. [DOI] [PubMed] [Google Scholar]
- Reuther, G.W., Lambert, Q.T., Booden, M.A., Wennerberg, K., Becknell, B., Marcucci, G., Sondek, J., Caligiuri, M.A., Der, C.J. Leukemia-associated Rho guanine nucleotide exchange factor, a Dbl family protein found mutated in leukemia, causes transformation by activation of RhoA. J. Biol. Chem. 2001;276:27145–27151. doi: 10.1074/jbc.M103565200. [DOI] [PubMed] [Google Scholar]
- Schmidt, A., Hall, A. Guanine nucleotide exchange factors for Rho GTPases: Turning on the switch. Genes & Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Skelton, N.J., Koehler, M.F., Zobel, K., Wong, W.L., Yeh, S., Pisabarro, M.T., Yin, J.P., Lasky, L.A., Sidhu, S.S. Origins of PDZ domain ligand specificity. Structure determination and mutagenesis of the Erbin PDZ domain. J. Biol. Chem. 2003;278:7645–7654. doi: 10.1074/jbc.M209751200. [DOI] [PubMed] [Google Scholar]
- Songyang, Z., Fanning, A.S., Fu, C., Xu, J., Marfatia, S.M., Chishti, A.H., Crompton, A., Chan, A.C., Anderson, J.M., Cantley, L.C. Recognition of unique carboxyl-terminal motifs by distinct PDZ domains. Science. 1997;275:73–77. doi: 10.1126/science.275.5296.73. [DOI] [PubMed] [Google Scholar]
- Swiercz, J.M., Kuner, R., Behrens, J., Offermanns, S. Plexin-B1 directly interacts with PDZ-RhoGEF/LARG to regulate RhoA and growth cone morphology. Neuron. 2002;35:51–63. doi: 10.1016/s0896-6273(02)00750-x. [DOI] [PubMed] [Google Scholar]
- Tamagnone, L., Artigiani, S., Chen, H., He, Z., Ming, G.I., Song, H., Chedotal, A., Winberg, M.L., Goodman, C.S., Poo, M., et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell. 1999;99:71–80. doi: 10.1016/s0092-8674(00)80063-x. [DOI] [PubMed] [Google Scholar]
- Taya, S., Inagaki, N., Sengiku, H., Makino, H., Iwamatsu, A., Urakawa, I., Nagao, K., Kataoka, S., Kaibuchi, K. Direct interaction of insulin-like growth factor-1 receptor with leukemia-associated RhoGEF. J. Cell Biol. 2001;155:809–820. doi: 10.1083/jcb.200106139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Ossowski, I., Oksanen, E., von Ossowski, L., Cai, C., Sundberg, M., Goldman, A., Keinanen, K. Crystal structure of the second PDZ domain of SAP97 in complex with a GluR-A C-terminal peptide. FEBS J. 2006;273:5219–5229. doi: 10.1111/j.1742-4658.2006.05521.x. [DOI] [PubMed] [Google Scholar]
- Zhou, H., Xu, Y., Yang, Y., Huang, A., Wu, J., Shi, Y. Solution structure of AF-6 PDZ domain and its interaction with the C-terminal peptides from Neurexin and Bcr. J. Biol. Chem. 2005;280:13841–13847. doi: 10.1074/jbc.M411065200. [DOI] [PubMed] [Google Scholar]