Abstract
H-type thioredoxins (Trxs) constitute a particularly large Trx sub-group in higher plants. Here, the crystal structures are determined for the two barley Trx h isoforms, HvTrxh1 and HvTrxh2, in the partially radiation-reduced state to resolutions of 1.7 Å, and for HvTrxh2 in the oxidized state to 2.0 Å. The two Trxs have a sequence identity of 51% and highly similar fold and active-site architecture. Interestingly, the four independent molecules in the crystals of HvTrxh1 form two relatively large and essentially identical protein–protein interfaces. In each interface, a loop segment of one HvTrxh1 molecule is positioned along a shallow hydrophobic groove at the primary nucleophile Cys40 of another HvTrxh1 molecule. The association mode can serve as a model for the target protein recognition by Trx, as it brings the Met82 Cγ atom (γ position as a disulfide sulfur) of the bound loop segment in the proximity of the Cys40 thiol. The interaction involves three characteristic backbone–backbone hydrogen bonds in an antiparallel β-sheet-like arrangement, similar to the arrangement observed in the structure of an engineered, covalently bound complex between Trx and a substrate protein, as reported by Maeda et al. in an earlier paper. The occurrence of an intermolecular salt bridge between Glu80 of the bound loop segment and Arg101 near the hydrophobic groove suggests that charge complementarity plays a role in the specificity of Trx. In HvTrxh2, isoleucine corresponds to this arginine, which emphasizes the potential for specificity differences between the coexisting barley Trx isoforms.
Keywords: thioredoxin h, cysteine oxidoreduction, protein recognition, redox regulation
Thioredoxin (Trx) is a ubiquitous protein disulfide reductase of ∼12 kDa (Holmgren 1985). Trx employs two redox-active cysteines in the conserved active-site motif, Trp-CysN-Gly/Pro-Pro-CysC, to transfer electrons via dithiol/disulfide exchange reactions. For electron donation, the CysN thiol of reduced Trx exerts a nucleophilic attack on a protein disulfide and transiently forms an intermolecular disulfide bond (Kallis and Holmgren 1980). The subsequent attack from CysC releases reduced target protein and oxidized disulfide-bonded Trx. The oxidized Trx is converted to the reduced, dithiol form by accepting electrons from either NADPH via NADPH-dependent Trx reductase (NTR), or, in plant chloroplasts, from ferredoxin via ferredoxin-dependent Trx reductase.
Trx donates electrons to redox enzymes including ribonucleotide reductase, methionine sulfoxide reductase, and peroxiredoxin (Arnér and Holmgren 2000) and is moreover involved in the activity regulation of mammalian transcription factors and plant photosynthetic enzymes in a redox-dependent way (Schenk et al. 1994; Buchanan and Balmer 2005). Recent progress in proteomics has led to the identification of a large number of proteins possibly regulated by Trx (Verdoucq et al. 1999; Yano et al. 2001; Maeda et al. 2004, 2005). The coexistence of multiple Trx isoforms, other oxidoreductases, and low-molecular weight redox-active compounds in physiological environments makes it difficult to identify specific interaction partners of individual Trx isoforms. Undoubtedly, the most complex array of Trx isoforms is found in higher plants. For example, at least 19 Trx genes and numerous closely related genes have been identified in the genome of Arabidopsis thaliana (Meyer et al. 2002). Based on sequence similarities, plant Trxs are categorized into chloroplastic f-, m-, x-, and y-types; mitochondrial o-type; and the h-type, which has been identified in cytosol, nucleus, and mitochondria (Buchanan and Balmer 2005). The physiological importance of the nine Trx h isoforms of the A. thaliana genome (Meyer et al. 2002) has not been established, but complementation studies in Saccharomyces cerevisiae demonstrated functional differentiation among Trx h isoforms from A. thaliana, the green alga Chlamydomonas reinhardtii, and Pisum sativum (Mouaheb et al. 1998; Sarkar et al. 2005; Traverso et al. 2007). Moreover, genes encoding A. thaliana Trx h isoforms are differentially regulated in response to pathogens and under conditions of oxidative stress (Reichheld et al. 2002; Laloi et al. 2004).
Although Trx has a broad specificity, 3D structures of disulfide-linked Trx–substrate reaction intermediate mimics have provided evidence for recognition of structural elements in target protein motifs by Trxs (Qin et al. 1995, 1996; Maeda et al. 2006; Chartron et al. 2007). Moreover, mutational studies on Trxs have suggested that surface charge distribution patterns influence the reactivity of Trxs toward chloroplastic fructose-1,6-bisphosphatase (de Lamotte-Guery et al. 1991; Geck et al. 1996; Mora-García et al. 1998). Previously, NMR and X-ray structures were determined for Trx h from C. reinhardtii (Mittard et al. 1997; Menchise et al. 2001). Among higher plants, NMR structures have been reported for Trx h from A. thaliana (Peterson et al. 2005) and Populus tremula (Coudevylle et al. 2005). We recently reported the crystal structure of a barley Trx h isoform 2 (HvTrxh2) in an engineered, mixed disulfide-linked complex with a substrate protein, α-amylase/subtilisin inhibitor (BASI) (Maeda et al. 2006).
The crystal structures of the two barley Trx h isoforms, HvTrxh1 and HvTrxh2, determined in the present study allow the first 3D structure comparison of two Trx h isoforms from the same plant species. HvTrxh1 and HvTrxh2 share 51% sequence identity and coexist in barley seeds but vary in temporal and spatial distribution (Maeda et al. 2003). The two isoforms exhibit distinct kinetic properties in the insulin reduction assay and are reduced by A. thaliana NTR with a threefold difference in apparent K m values (Maeda et al. 2003).
Results and Discussion
Protein crystallization, data collection, and structure determination
Diffraction data were collected for HvTrxh1 crystals obtained from two different reservoir solutions: (1) 0.1 M MES pH 6.0, 2.4 M ammonium sulfate (HvTrxh1AS) and (2) 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% (w/v) polyethylene glycol (PEG) 4000 (HvTrxh1PEG). For HvTrxh2, crystals were obtained in nonbuffered 30% (w/v) PEG 1500 (HvTrxh2). The HvTrxh1PEG structure was solved by molecular replacement (MR) using the crystal structure of C. reinhardtii Trx h as a search model (PDB code 1EP7) (Menchise et al. 2001). The HvTrxh1PEG structure was subsequently used for MR of HvTrxh1AS and HvTrxh2. HvTrxh1PEG and HvTrxh1AS are both in the C2 space group, having nearly identical unit cell dimensions and crystal packing with four HvTrxh1 molecules (A–D) per asymmetric unit, while HvTrxh2 crystallizes in space group P21 with two HvTrxh2 molecules (A and B) per asymmetric unit.
The redox–cysteine pairs were parameterized as disulfides for the initial refinement, as none of the Trx samples had been treated with disulfide reductant. They were thus presumed to be in the oxidized state. However, the resultant F o–F c electron density maps had significant negative electron density at the center of the disulfide bonds. A revised model refinement without restraining the cysteine pairs to disulfide bonds resulted in disappearance of the negative electron density in F o–F c maps, giving Trx structures displaying various Sγ–Sγ distances of up to ∼3 Å, which greatly exceed the ideal disulfide bonded Sγ–Sγ distance of 2.03 Å. Disulfide bonds are in general highly radiation-sensitive (Ravelli and McSweeney 2000), and radiation disruption of redox-active disulfide bonds in Trxs and related proteins during X-ray data collection has often been observed (Friemann et al. 2003; Stirnimann et al. 2006). To confirm that the exposure of crystals to X-ray radiation was the cause of the enlarged Sγ–Sγ distances, the HvTrxh2 structure was independently refined against the data set partitioned into the initial half (HvTrxh2RED1) and the final half (HvTrxh2RED2) of data collection, each containing an equal number of images. The two time- and space-averaged HvTrxh2 structures confirmed the progressive disruption of the disulfide during data collection, as the averaged unrestrained Sγ–Sγ distances for the two independent HvTrxh2 molecules shifted from 3.1 Å in HvTrxh2RED1 with ∼50% of the molecules in the reduced state toward 3.5 Å in HvTrxh2RED2 with ∼80% of the molecules in the reduced state after prolonged X-ray exposure.
The structure of oxidized HvTrxh2 (HvTrxh2OX) was determined to study structural consequences of Trx oxidoreduction. Partially overlapping fractional data sets were collected from three isomorphous HvTrxh2 crystals in order to diminish the radiation-damaged portion of the diffraction data. The HvTrxh2 structure was refined against the merged fractional data sets with the redox–cysteine pairs fixed in disulfide bonds. The absence of significant negative electron density at the center of disulfide bonds in the resultant 1σ F o–F c electron density map confirmed that radiation damage was limited. The data collection and refinement statistics are listed in Table 1. The statistics for HvTrxh1AS are superior to those for HvTrxh1PEG, and unless otherwise specified, the following characterization is based on the HvTrxh1AS structure. Likewise, HvTrxh2RED1 is used to describe the HvTrxh2 structure unless otherwise specified.
Table 1.
Data collection and refinement statistics
Fold of HvTrxh1 and HvTrxh2
The primary structures of Trxs vary greatly both between species and between different isoforms from the same species. Large variations are indeed observed also in sequences of plant Trxs categorized as h-types based on the sequence similarity (Fig. 1A). The 3D structures of various Trxs nevertheless show little deviation in the overall fold. HvTrxh1 and HvTrxh2 thus have the typical fold of Trx consisting of a five-stranded central β-sheet surrounded by four α-helices in a βαβαβαββα topology (Fig. 1B,C). While the vast majority of residues in the folded regions of Trxs were modeled, the N-terminal regions preceding β1 in HvTrxh1 and HvTrxh2 displayed poorly interpretable electron densities for approximately five and 11 residues, respectively, and were not modeled. The same region was also distorted in the previously reported crystal structure of HvTrxh2 in complex with BASI (Maeda et al. 2006). The present HvTrxh2 structure and the structure of HvTrxh2 in complex with BASI show no major difference in the active-site architecture or in the overall fold and can be superimposed to a root mean square deviation (RMSD) of 0.5 Å using 97 Cα atoms (molecule A of HvTrxh2RED1).
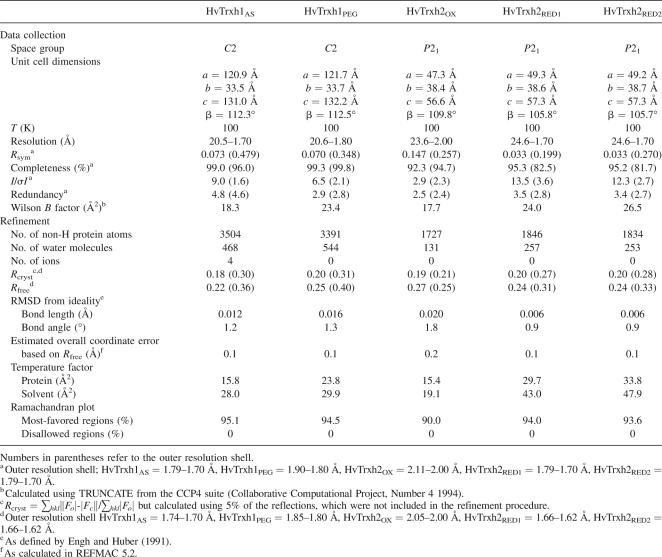
Figure 1.
Amino acid sequences and crystal structures of HvTrxh1 and HvTrxh2. (A) Multiple sequence alignment of HvTrxh1, HvTrxh2, and selected h-type Trxs. HvTrxh1 (Q7XZK3), HvTrxh2 (Q7XZK2), AtTrxh1 (A. thaliana Trx h-type 1, P29448), AtTrxh2 (A. thaliana Trx H-type 2, Q38879), AtTrxh3 (A. thaliana Trx h-type 3, Q42403), AtTrxh4 (A. thaliana Trx h-type 4, Q39239) AtTrxh5 (A. thaliana Trx H-type 5, Q39241), CrTrxh (C. reinhardtii Trx-h, P80028), and PtTrxh (P. tremula Trx-h, Q8S3L3) were aligned using ClustalW at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/). The redox-active–cysteine pair is indicated with bold red letters. Positions of the HvTrxh1 loop segments, Trp39-Pro42, Ala81-Pro83, and Val98-Gly100, are indicated with green boxes. The position of the catalytic aspartate (Asp34 in HvTrxh1) is indicated with a black box. Amino acids identical to those in HvTrxh1 are indicated with a gray background. The indication of secondary-structure elements and residue numbering are based on the structure of HvTrxh1 molecule A. (B,C) Cartoon display of the crystal structures of HvTrxh1 (B) and HvTrxh2 (C) colored magenta and green, respectively. (D) Structural alignment of HvTrxh1 (magenta) and HvTrxh2 (green) shown as Cα traces.
HvTrxh1 molecule A can be superimposed on the three other HvTrxh1 molecules in the asymmetric unit with RMSDs of up to 0.4 Å using 93 Cα atoms. The two HvTrxh2 molecules in the asymmetric unit superimpose on one another with an RMSD of 0.2 Å using 103 Cα atoms. This excellent superimposition of independent molecules shows that the structures of HvTrxh1 and HvTrxh2 are rigid. Despite having only 56% sequence identity in the folded regions, HvTrxh1 molecule A aligns with HvTrxh2 molecule A with an RMSD of 0.6 Å using 89 Cα atoms (Fig. 1D). Significant deviation is nevertheless seen at loop α3–β4, which is most likely caused by its involvement in the crystal packing of HvTrxh1, as will be discussed later.
The structure of HvTrxh1 (molecule A) superimposes on the crystal structure of C. reinhardtii Trx h (PDB code 1EP7, molecule A) (Menchise et al. 2001) with an RMSD of 0.7 Å using 91 Cα atoms, emphasizing the conservation of fold among h-type Trxs from different species. In contrast, HvTrxh1 (molecule A) only superimposes on Trxs from Escherichia coli (PDB code 2TRX, molecule A) (Katti et al. 1990) and human (PDB code 1ERU) (Weichsel et al. 1996) with RMSDs of 1.0 and 0.5 Å over a region of 68 and 74 Cα atoms, respectively. The most N-terminal ∼30 residues including β1 and α1 (HvTrxh1 sequence) with poor sequence conservation (Fig. 1A) show, as expected, the largest variation in 3D structure. While 14 residues thus constitute α1 in HvTrxh1, HvTrxh2, and C. reinhardtii Trx h (PDB code 1EP7) (Menchise et al. 2001), 13 in A. thaliana Trx h1 (PDB code 1XFL) (Peterson et al. 2005), and 11 in P. tremula Trx h (PDB code 1TI3) (Coudevylle et al. 2005), only four residues form α1 in E. coli Trx (PDB code 2TRX) (Katti et al. 1990).
Catalytic mechanism of HvTrxh1 and HvTrxh2
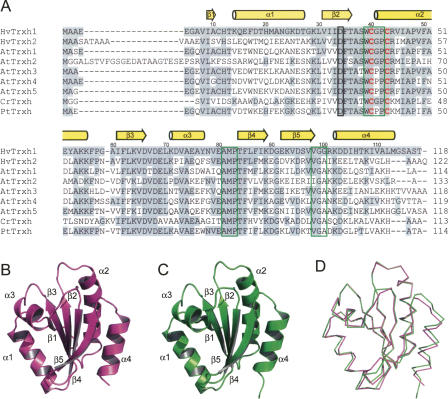
An aspartate (Asp26) in the hydrophobic interior of E. coli Trx acts as a general acid/base catalyst for the protonation/deprotonation of the buried CysC thiol group during the oxidoreduction (Chivers and Raines 1997). In HvTrxh1 and HvTrxh2, the corresponding residues are Asp34 and Asp40 (Fig. 1A), respectively. The carboxyl group is surrounded by hydrophobic residues but partly exposed to solvent in the narrow internal cavity separating it from the CysC Sγ atom (Fig. 2), as typically seen for Trx structures (Katti et al. 1990). The aspartate carboxyl group appears to have distinct environments in HvTrxh1 and HvTrxh2. The side chain of Ile46 occupies space in the internal cavity of HvTrxh1 with one of the Cγ atoms only 4.9 Å away from the aspartate carboxyl group on average, while the side chain of Met52 at the equivalent position in HvTrxh2 is at a distance of 5.3 Å (Cβ atom) (Fig. 2).
Figure 2.
Active-site architecture of HvTrxh1 (A) and HvTrxh2 (B). Carbon, nitrogen, oxygen, and sulfur atoms are colored green, blue, red, and yellow, respectively. The 2F o–F c electron density maps are presented as a gray isosurface mesh at the 1.0σ level. The water molecule buried in the internal cavity is shown as a sphere and is modeled in two alternative positions in HvTrxh1. For the HvTrxh2 structure, Cys46 and Cys49 are shown only in the reduced conformations, although they are also modeled in the oxidized conformations.
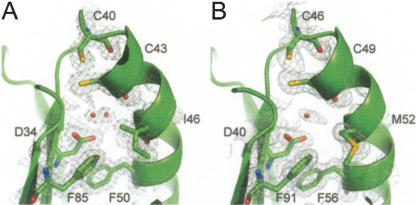
The HvTrxh2 crystal structures in oxidized (HvTrxh2OX) and the partially disulfide-disrupted states (HvTrxh2RED1/RED2) enable a detailed description of structural changes following the Trx oxidoreduction. The average CysN Sγ to CysC Sγ distance increases from HvTrxh2OX to HvTrxh2RED2, in association with shifts in the average CysN χ1 angle from 160° toward −174° and in the average CysC χ1 angle from −61° toward −72° (Fig. 3A,B). Disulfide reduction seems to only slightly alter the main-chain conformation at this resolution (Fig. 3A,B), in accordance with studies on Trxs from E. coli (Jeng et al. 1994), human (Weichsel et al. 1996), and spinach (Capitani et al. 2000). Neither is any major change observed for the catalytic aspartate nor for the water molecules buried in the cavity. Moreover, by comparing the structure amplitudes of HvTrxh2RED1 and HvTrxh2RED2, a time-dependent difference electron density map [1/V∑ | F(h)RED1 − F(h)RED2 | ei ϕRED2(h) e −2πi(h − r)] is generated and shown with the HvTrxh2RED1 structure in Figure 3C. The map reveals an area of intensive positive density (less electron density in HvTrxh2RED2) (green) that covers the positions of the disulfide-bonded Sγ atoms of CysN and CysC, and two areas of negative density (more electron density in HvTrxh2RED2) (red) flanking the disulfide bond. This feature confirms that the oxidoreduction of HvTrxh2 is primarily associated with side-chain conformational changes of the two redox-active cysteines.
Figure 3.
Radiation disruption of the active-site disulfide in HvTrxh2. Stereo images of HvTrxh2OX (A) and HvTrxh2RED2, 80% reduced (B), showing a close-up view of segment Cys46−Cys49. The 2F o–F c electron density maps are shown as gray isosurface mesh at the 1.0σ level. (C) HvTrxh2RED1 with the 1/V∑ | F(h)RED1 − F(h)RED2 | ei ϕRED2(h) e −2πi(h − r) difference electron density contoured at the 7σ level. Significant negative (red) density represents areas where the HvTrxh2RED2 structure has a higher level of electron density than the HvTrxh2RED1 structure, and positive (green) electron density, the opposite. For HvTrxh2RED1 and HvTrxh2RED2 structures, Cys46 and Cys49 are modeled and shown in two alternative conformations (for oxidized and reduced states).
Hydrophobic groove on HvTrxh1 implicated in protein recognition is buried in a protein–protein interface
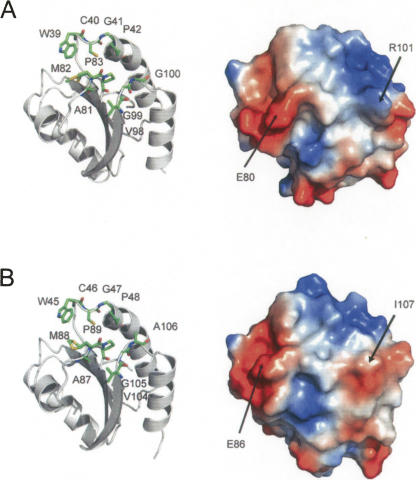
Solvent-exposed hydrophobic and uncharged residues form an elongated shallow groove next to the redox-active cysteines of HvTrxh1 and HvTrxh2. We refer to this conserved feature in Trx structures as the substrate-binding loop motif, since it is proposed to play a central role in the recognition of substrate proteins (Qin et al. 1995; 1996; Maeda et al. 2006; Chartron et al. 2007). This motif involves three loop segments, Trp39-Cys40(CysN)-Gly41-Pro42, Ala81-Met82-Pro83, and Val98-Gly99-Gly100 of HvTrxh1 (Fig. 4A). The Gly100 corresponds to an alanine in HvTrxh2 that is more commonly present at this position in Trxs (Figs. 1A, 4B).
Figure 4.
Substrate-binding loop motif in HvTrxh1 (A) and HvTrxh2 (B). Cartoon display of HvTrxh1 with sticks showing the loop segments Trp39-Pro42, Ala81-Pro83, and Val98-Gly100 (A, left). The vacuum electrostatic potential surface of HvTrxh1 (from the same angle as the left image) with the positions for Glu80 and Arg101 indicated (A, right). HvTrxh2 is presented accordingly (B). Met82 of HvTrxh1 and Met88 of HvTrxh2 are modeled and shown in two alternative conformations. For the HvTrxh2 structure, Cys46 is only shown in the reduced conformation, although it was also modeled in the oxidized conformation.
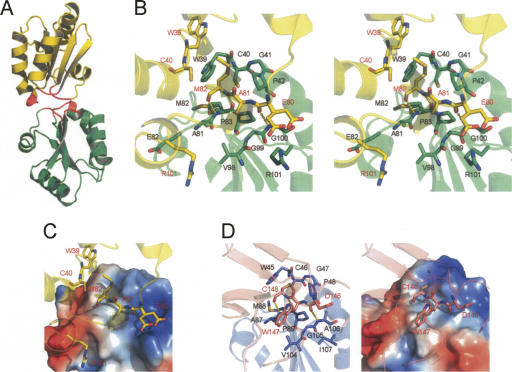
In the HvTrxh1 structure, the substrate-binding loop motif is largely buried in the two protein–protein interfaces formed between the two pairs of molecules in the asymmetric unit, molecules A and D and molecules B and C, respectively (Fig. 5A). The residues in the three loop segments account for >85% of the total buried accessible surface area (ASA) (902 Å2 [A:D] and 989 Å2 [B:C]). The two interfaces display essentially identical protein–protein interactions, as the two HvTrxh1 molecular pairs can be aligned with an RMSD of 1.1 Å using 199 Cα atoms. The HvTrxh1 crystal dimers do not lie on crystallographic twofold axes as often seen in protein homodimers, and there is no evidence that the dimer represents the in vivo quaternary structure of HvTrxh1. The interface areas in the HvTrxh1 crystals are in accordance with those found in crystals of monomeric proteins, typically burying 200–1200 Å2 ASA (Janin and Rodier 1995), and are smaller than protein dimer interfaces that generally bury >2000 Å2 ASA (Bahadur et al. 2003). Nevertheless, the HvTrxh1 crystal dimers must either be formed in solution under crystallization conditions or be strongly favored to form during crystal growth. The residues in the AD interface have well-defined electron density both in the HvTrxh1AS and HvTrxh1PEG crystals. A few residues around the BC interface in the HvTrxh1PEG crystals have high B factors or are not modeled. In both crystal forms, molecule C seems to be forced into a slightly constrained conformation by the crystal packing, as the helical structure is deformed at the most N-terminal part of α2, which is in contact with molecule B.
Figure 5.
Features of the HvTrxh1 crystal dimer. (A) Cartoon display shows the dimerization of HvTrxh1 molecules A (green) and D (yellow). Residues Trp39−Pro42, Ala81−Pro83, and Val98−G100 that constitute the substrate-binding loop motif are colored red. (B) Stereoview of the interface between HvTrxh1 molecules A and D. Cartoon display is shown transparently and colored green (molecule A) and yellow (molecule D). Key residues are shown in stick representation and labeled in black (molecule A) and red (molecule D), respectively. Hydrogen bonds are shown as dashed yellow lines. Glu80 (molecule D) is modeled in two alternative conformations. (C) The image is identical to the left image of Figure 5B, except that the vacuum electrostatic potential surface is shown for HvTrxh1 molecule A. (D) Cartoon display shows the interface between HvTrxh2 (blue) and BASI (pink) in the structure of HvTrxh2–BASI complex (Maeda et al. 2006). Key residues are shown in stick representations and labeled with black (HvTrxh2) and red (BASI) letters. The vacuum electrostatic potential surface is shown for HvTrxh2 in the right image.
HvTrxh1 crystal dimer interfaces suggest a mechanism for substrate recognition by Trx
The following descriptions of the dimer interfaces are based on the AD interface unless specified. The indole group of Trp39 in molecule A is positioned close to the CysN thiol group of the same molecule, as seen in most other Trx structures, and greatly contributes to the buried ASA (Fig. 5B). In contrast, the indole group in molecule D is “flipped out” and located distantly from CysN (Fig. 5B). The equivalent tryptophan residue is also observed in both conformations in crystal structures of Trx from spinach (Capitani et al. 2000) and Trypanosoma brucei brucei (Friemann et al. 2003). The α3–β4 loop segment of molecule D, Glu80-Ala81-Met82 (partially overlapping with the substrate-binding loop motif), has extended main chain conformation and is bound along the hydrophobic groove formed by the substrate-binding loop motif on the surface of molecule A (Fig. 5B,C). Of particular importance, this mode of binding brings the Met82 side chain Cγ of the bound molecule D α3–β4 loop to a distance of 4.1 Å from the CysN thiol group Sγ of molecule A (Fig. 5B,C). The dithiol/disulfide exchange reaction between Trx and a substrate protein requires the approximation of the CysN thiol group and a cysteine Sγ atom in the disulfide form (positionally analogous to the Met Cγ atom) of the substrate protein. Therefore, the protein–protein interaction features of the HvTrxh1 dimer interfaces can represent a model for the Trx–substrate interaction. The α3–β4 loop segment in molecule D is bound on the surface of molecule A by several van der Waals interactions and three intermolecular backbone–backbone hydrogen bonds, resembling antiparallel β-sheet formation. This pattern of intermolecular hydrogen bonds was previously seen in structures of engineered, disulfide-linked Trx–substrate complexes mimicking reaction intermediates. These include the NMR structure of human Trx in complex with a synthetic peptide from Ref-1 (Qin et al. 1996) and the crystal structure of HvTrxh2 in complex with BASI, representing the outcome of the initial nucleophilic attack from HvTrxh2 CysN on BASI disulfide Cys144–Cys148 (Fig. 5D; Maeda et al. 2006). An essentially identical pattern of intermolecular hydrogen bond formation was also observed in the crystal structure of the N-terminal Trx-like domain of E. coli DsbD in a disulfide-linked complex with DsbD C-terminal domain (Rozhkova et al. 2004). The crystal structure of 3′-phosphoadenosine-5′-phosphosulfate reductase in a disulfide-linked complex with E. coli Trx showed a similar mode of binding except that one of the hydrogen bonds is absent (Chartron et al. 2007). Based on the HvTrxh2-BASI complex structure, we previously proposed that this mode of intermolecular-hydrogen-bond formation plays an important role in substrate recognition and selectivity of Trx. The present study provides remarkable structural evidence for such selective binding of a protein motif with extended backbone conformation to the active site of Trx also in the absence of an engineered disulfide linkage. Moreover, this study emphasizes the possible involvement of the substrate-binding loop motif in protein–protein association not involving dithiol/disulfide exchanging, as seen in the binding of the reduced E. coli Trx with bacteriophage T7 DNA polymerase (PDB code 1T7P) (Doublié et al. 1998).
Involvement of electrostatic interactions in HvTrxh1 crystal dimer formations
The protein–protein interactions found in HvTrxh1 are absent in the HvTrxh2 crystal. Inspection of the existing PDB entries of Trx structures neither revealed such interaction patterns although twofold symmetry-related dimerization is reported in crystals of a C. reinhardtii Trx h mutant (Menchise et al. 2001) and for Drosophila melanogaster Trx (Wahl et al. 2005). Hence, the HvTrxh1 crystal complex formation must signify its unique structural self-complementarity. While the fold and hydrophobicity of the substrate-binding loop motif are well conserved among Trxs, surrounding solvent-exposed residues are divergent. In HvTrxh1, the substrate-binding loop motif is flanked by solvent-exposed Glu80 and Arg101 (Figs. 4, 5B). In the crystal dimer AD, the Arg101 guanidinium cation from molecule A forms an electrostatic interaction with the Glu80 carboxylate anion of the bound molecule D loop segment with distances of 2.7 and 3.9 Å (Fig. 5B,C), while the same groups are 6.7 and 10.2 Å apart in the BC interface (Glu80 modeled in two alternative confirmations in both molecules C and D). The other pair of Glu80 carboxylate anion (of molecule A) and Arg101 guanidinium cation (of molecule D) in HvTrxh1 crystal dimers are separated by 7.2 Å, and the HvTrxh1 interfaces involve no obvious conflicting contacts of charged groups. Similar structural self-complementarity is not present in the HvTrxh2 structure, as the HvTrxh1 Arg101 is not conserved among h-type Trxs (Fig. 1A). This residue is substituted with Ile107 in HvTrxh2.
In general, electrostatic complementarity is considered to be a major driving force for the rapid association of electron donors and acceptor proteins into complexes (Crowley and Carrondo 2004). The present study has indicated that HvTrxh1 Arg101 can play a particularly crucial role in the association of HvTrxh1 with target proteins by forming electrostatic interactions with a protein motif bound in the substrate-binding loop motif. Indeed, a charge mutation of E. coli Trx at this particular position is shown to affect its reactivity toward fructose-1,6-bisphosphatase (Mora-García et al. 1998). The presence of Arg101 in HvTrxh1 and the uncharged Ile107 in HvTrxh2 may therefore give rise to differential isoform interaction with some redox partners.
Materials and Methods
X-ray crystallography
Recombinant HvTrxh1 and HvTrxh2 were expressed in E. coli and purified as described previously (Maeda et al. 2003). Purified Trxs were dialyzed against water and concentrated using Centricon-10 filters (Millipore). Crystals of HvTrxh1 and HvTrxh2 were obtained using the hanging-drop vapor-diffusion method at room temperature with 2 μL each of reservoir and protein solutions. HvTrxh1 solutions of 10 and 20 mg/mL were used for crystallization in 0.1 M MES pH 6.0, 2.4 M ammonium sulfate and in 0.2 M ammonium acetate, 0.1 M sodium acetate trihydrate pH 4.6, 30% (w/v) PEG 4000, respectively. A HvTrxh2 solution of 20 mg/mL was used for crystallization in nonbuffered 30% (w/v) PEG 1500. All diffraction data were collected on a Rigaku RU-H3RHB rotating Cu anode X-ray generator equipped with an R-AXIS IV++ imaging plate detector and a 700 series Cryostream cooler (Oxford Cryosystems) at –160°C. To generate a structural model for oxidized HvTrxh2, diffraction data were collected from three isomorphous HvTrxh2 crystals by 40° rotation (80 diffraction images) each. Reflections were indexed, integrated, and scaled with MOSFLM (Leslie 1992) and SCALA (Collaborative Computational Project, Number 4 1994). MR was performed in Phaser (Collaborative Computational Project, Number 4 1994; Storoni et al. 2004). The structure of C. reinhardtii Trx h was used as a search model, because the MR was performed before the previously published crystal structure of HvTrxh2 in complex with BASI was available (PDB code 2IWT). The refinement was carried out with the programs CNS (Brünger et al. 1998) and Refmac 5 (Collaborative Computational Project, Number 4 1994; Murshudov et al. 1997). The electron density maps of HvTrxh1 and HvTrxh2 allowed manual building of the models in O (Jones et al. 1991) and Coot (Emsley and Cowtan 2004). Final refinements were performed in Refmac 5 and CNS. Two alternate conformations of Cys46/Cys49 were included for the refinement of the two HvTrxh2RED structures, one with the distance between sulfur atoms restrained by a disulfide bond and one with no bond restraint. The occupancy of each conformation was optimized by evaluation of the resultant F o–F c difference Fourier keeping the total occupancy of each residue = 1 and the occupancy of the cysteines in a Cys46/Cys49 pair equal. The geometries of the refined structures were checked with PROCHECK (Laskowski et al. 1993). Secondary structures were assigned in Stride (http://webclu.bio.wzw.tum.de/cgi-bin/stride/stridecgi.py/) (Frishman and Argos 1995). The protein–protein interfaces were analyzed in the Protein–Protein Interaction Server (V1.5) (http://www.biochem.ucl.ac.uk/bsm/PP/server/) (Jones and Thornton 1996). Figures of protein structures are made in PyMOL (Delano Scientific).
Data deposition
The coordinates of the present crystal structures have been deposited to the RCSB Protein Data Bank with the codes 2VM1 (HvTrxh1AS), 2VM2 (HvTrxh1PEG), 2VLT (HvTrxh2OX), 2VLU (HvTrxh2RED1), and 2VLV (HvTrxh2RED2).
Acknowledgments
Mette Therese Christensen, Karina Rasmussen (the Technical University of Denmark), and Annette Kure Andreassen (Carlsberg Laboratory) are acknowledged for excellent technical assistance. Jakob R. Winther (University of Copenhagen) is gratefully thanked for helpful discussions. The Danish Technical Research Council (STVF, grant 26-03-0194) and the Carlsberg Foundation are acknowledged for financial support (to B.S.). C.F. received a Young Research Leader grant (23-03-0073) from the Danish Agricultural and Veterinary Research Council (SJVF). K.M. received a Ph.D. scholarship from the Technical University of Denmark.
Footnotes
Reprint requests to: Birte Svensson, Enzyme and Protein Chemistry, Department of Systems Biology, Søltofts Plads, Building 224, Technical University of Denmark, DK-2800 Kongens Lyngby, Denmark; e-mail: bis@bio.dtu.dk; fax: 45-4588-6307.
Abbreviations: ASA, accessible surface area; BASI, barley α-amylase/subtilisin inhibitor; HvTrxh1, barley thioredoxin h isoform 1; HvTrxh2, barley thioredoxin h isoform 2; MR, molecular replacement; NTR, NADPH-dependent thioredoxin reductase; PEG, polyethylene glycol; RMSD, root mean square deviation; Trx, thioredoxin.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.083460308.
References
- Arnér, E.S.J., Holmgren, A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Bahadur, R.P., Chakrabarti, P., Rodier, F., Janin, J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buchanan, B.B., Balmer, Y. Redox regulation: A broadening horizon. Annu. Rev. Plant Biol. 2005;56:187–220. doi: 10.1146/annurev.arplant.56.032604.144246. [DOI] [PubMed] [Google Scholar]
- Capitani, G., Markovic-Housley, Z., DelVal, G., Morris, M., Jansonius, J.N., Schurmann, P. Crystal structures of two functionally different thioredoxins in spinach chloroplasts. J. Mol. Biol. 2000;302:135–154. doi: 10.1006/jmbi.2000.4006. [DOI] [PubMed] [Google Scholar]
- Chartron, J., Shiau, C., Stout, C.D., Carroll, K.S. 3′-Phosphoadenosine-5′-phosphosulfate reductase in complex with thioredoxin: A structural snapshot in the catalytic cycle. Biochemistry. 2007;46:3942–3951. doi: 10.1021/bi700130e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivers, P.T., Raines, R.T. General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry. 1997;36:15810–15816. doi: 10.1021/bi971504l. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Coudevylle, N., Thureau, A., Hemmerlin, C., Gelhaye, E., Jacquot, J.P., Cung, M.T. Solution structure of a natural CPPC active site variant, the reduced form of thioredoxin h1 from poplar. Biochemistry. 2005;44:2001–2008. doi: 10.1021/bi047816n. [DOI] [PubMed] [Google Scholar]
- Crowley, P.B., Carrondo, M.A. The architecture of the binding site in redox protein complexes: Implications for fast dissociation. Proteins. 2004;55:603–612. doi: 10.1002/prot.20043. [DOI] [PubMed] [Google Scholar]
- de Lamotte-Guery, F., Miginiac-Maslow, M., Decottignies, P., Stein, M., Minard, P., Jacquot, J.P. Mutation of a negatively charged amino acid in thioredoxin modifies its reactivity with chloroplastic enzymes. Eur. J. Biochem. 1991;196:287–294. doi: 10.1111/j.1432-1033.1991.tb15816.x. [DOI] [PubMed] [Google Scholar]
- Doublié, S., Tabor, S., Long, A.M., Richardson, C.C., Ellenberger, T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engh, R.A., Huber, R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- Friemann, R., Schmidt, H., Ramaswamy, S., Forstner, M., Krauth-Siegel, R.L., Eklund, H. Structure of thioredoxin from Trypanosoma brucei brucei . FEBS Lett. 2003;554:301–305. doi: 10.1016/s0014-5793(03)01173-6. [DOI] [PubMed] [Google Scholar]
- Frishman, D., Argos, P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- Geck, M.K., Larimer, F.K., Hartman, F.C. Identification of residues of spinach thioredoxin f that influence interactions with target enzymes. J. Biol. Chem. 1996;271:24736–24740. doi: 10.1074/jbc.271.40.24736. [DOI] [PubMed] [Google Scholar]
- Holmgren, A. Thioredoxin. Annu. Rev. Biochem. 1985;54:237–271. doi: 10.1146/annurev.bi.54.070185.001321. [DOI] [PubMed] [Google Scholar]
- Janin, J., Rodier, F. Protein–protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Jeng, M.F., Campbell, A.P., Begley, T., Holmgren, A., Case, D.A., Wright, P.E., Dyson, H.J. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- Jones, S., Thornton, J.M. Principles of protein–protein interactions. Proc. Natl. Acad. Sci. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Kallis, G.B., Holmgren, A. Differential reactivity of the functional sulfhydryl groups of cysteine-32 and cysteine-35 present in the reduced form of thioredoxin from Escherichia coli . J. Biol. Chem. 1980;255:10261–10265. [PubMed] [Google Scholar]
- Katti, S.K., LeMaster, D.M., Eklund, H. Crystal structure of thioredoxin from Escherichia coli at 1.68 Å resolution. J. Mol. Biol. 1990;212:167–184. doi: 10.1016/0022-2836(90)90313-B. [DOI] [PubMed] [Google Scholar]
- Laloi, C., Mestres-Ortega, D., Marco, Y., Meyer, Y., Reichheld, J.P. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004;134:1006–1016. doi: 10.1104/pp.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., McArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Leslie, A.G. Daresbury Laboratory; Warrington, UK: 1992. MOSFLM Joint CCP4+ ESF-EAMCB Newsletter on Protein Crystallography, No. 26. [Google Scholar]
- Maeda, K., Finnie, C., Østergaard, O., Svensson, B. Identification, cloning and characterization of two thioredoxin h isoforms, HvTrxh1 and HvTrxh2, from the barley seed proteome. Eur. J. Biochem. 2003;270:2633–2643. doi: 10.1046/j.1432-1033.2003.03637.x. [DOI] [PubMed] [Google Scholar]
- Maeda, K., Finnie, C., Svensson, B. Cy5 maleimide labelling for sensitive detection of free thiols in native protein extracts: Identification of seed proteins targeted by barley thioredoxin h isoforms. Biochem. J. 2004;378:497–507. doi: 10.1042/BJ20031634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda, K., Finnie, C., Svensson, B. Identification of thioredoxin h−reducible disulphides in proteomes by differential labelling of cysteines: Insight into recognition and regulation of proteins in barley seeds by thioredoxin h. Proteomics. 2005;5:1634–1644. doi: 10.1002/pmic.200401050. [DOI] [PubMed] [Google Scholar]
- Maeda, K., Hägglund, P., Finnie, C., Svensson, B., Henriksen, A. Structural basis for target protein recognition by the protein disulfide reductase thioredoxin. Structure. 2006;14:1701–1710. doi: 10.1016/j.str.2006.09.012. [DOI] [PubMed] [Google Scholar]
- Menchise, V., Corbier, C., Didierjean, C., Saviano, M., Benedetti, E., Jacquot, J.P., Aubry, A. Crystal structure of the wild-type and D30A mutant thioredoxin h of Chlamydomonas reinhardtii and implications for the catalytic mechanism. Biochem. J. 2001;359:65–75. doi: 10.1042/0264-6021:3590065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, Y., Vignols, F., Reichheld, J.P. Classification of plant thioredoxins by sequence similarity and intron position. Methods Enzymol. 2002;347:394–402. doi: 10.1016/s0076-6879(02)47039-5. [DOI] [PubMed] [Google Scholar]
- Mittard, V., Blackledge, M.J., Stein, M., Jacquot, J.P., Marion, D., Lancelin, J.M. NMR solution structure of an oxidised thioredoxin h from the eukaryotic green alga Chlamydomonas reinhardtii . Eur. J. Biochem. 1997;243:374–383. doi: 10.1111/j.1432-1033.1997.0374a.x. [DOI] [PubMed] [Google Scholar]
- Mora-García, S., Rodríguez-Suárez, R., Wolosiuk, R.A. Role of electrostatic interactions on the affinity of thioredoxin for target proteins. Recognition of chloroplast fructose-1,6-bisphosphatase by mutant Escherichia coli thioredoxins. J. Biol. Chem. 1998;273:16273–16280. doi: 10.1074/jbc.273.26.16273. [DOI] [PubMed] [Google Scholar]
- Mouaheb, N., Thomas, D., Verdoucq, L., Monfort, P., Meyer, Y. In vivo functional discrimination between plant thioredoxins by heterologous expression in the yeast Saccharomyces cerevisiae . Proc. Natl. Acad. Sci. 1998;95:3312–3317. doi: 10.1073/pnas.95.6.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov, G.N., Vagin, A.A., Dodson, E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Peterson, F.C., Lytle, B.L., Sampath, S., Vinarov, D., Tyler, E., Shahan, M., Markley, J.L., Volkman, B.F. Solution structure of thioredoxin h1 from Arabidopsis thaliana . Protein Sci. 2005;14:2195–2200. doi: 10.1110/ps.051477905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin, J., Clore, G.M., Kennedy, W.M., Huth, J.R., Gronenborn, A.M. Solution structure of human thioredoxin in a mixed disulfide intermediate complex with its target peptide from the transcription factor NFκB. Structure. 1995;3:289–297. doi: 10.1016/s0969-2126(01)00159-9. [DOI] [PubMed] [Google Scholar]
- Qin, J., Clore, G.M., Kennedy, W.P., Kuszewski, J., Gronenborn, A.M. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure. 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- Ravelli, R.B., McSweeney, S.M. The “fingerprint” that X-rays can leave on structures. Structure. 2000;8:315–328. doi: 10.1016/s0969-2126(00)00109-x. [DOI] [PubMed] [Google Scholar]
- Reichheld, J.P., Mestres-Ortega, D., Laloi, C., Meyer, Y. The multigenic family of thioredoxin h in Arabidopsis thaliana: Specific expression and stress response. Plant Physiol. Biochem. 2002;40:685–690. [Google Scholar]
- Rozhkova, A., Stirnimann, C.U., Frei, P., Grauschopf, U., Brunisholz, R., Grutter, M.G., Capitani, G., Glockshuber, R. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, N., Lemaire, S., Wu-Scharf, D., Issakidis-Bourguet, E., Cerutti, H. Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage. Eukaryotic Cell. 2005;4:262–273. doi: 10.1128/EC.4.2.262-273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk, H., Klein, M., Erdbrügger, W., Dröge, W., Schulze-Osthoff, K. Distinct effects of thioredoxin and antioxidants on the activation of transcription factors NF-κB and AP-1. Proc. Natl. Acad. Sci. 1994;91:1672–1676. doi: 10.1073/pnas.91.5.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnimann, C.U., Rozhkova, A., Grauschopf, U., Bockmann, R.A., Glockshuber, R., Capitani, G., Grutter, M.G. High-resolution structures of Escherichia coli cDsbD in different redox states: A combined crystallographic, biochemical and computational study. J. Mol. Biol. 2006;358:829–845. doi: 10.1016/j.jmb.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Storoni, L.C., McCoy, A.J., Read, R.J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. 2004;D60:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- Traverso, J.A., Vignols, F., Cazalis, R., Pulido, A., Sahrawy, M., Cejudo, F.J., Meyer, Y., Chueca, A. PsTRXh1 and PsTRXh2, are both pea (Pisum sativum) h-type thioredoxins with antagonistic behaviour in redox imbalances. Plant Physiol. 2007;143:300–311. doi: 10.1104/pp.106.089524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoucq, L., Vignols, F., Jacquot, J.P., Chartier, Y., Meyer, Y. In vivo characterization of a thioredoxin h target protein defines a new peroxiredoxin family. J. Biol. Chem. 1999;274:19714–19722. doi: 10.1074/jbc.274.28.19714. [DOI] [PubMed] [Google Scholar]
- Wahl, M.C., Irmler, A., Hecker, B., Schirmer, R.H., Becker, K. Comparative structural analysis of oxidized and reduced thioredoxin from Drosophila melanogaster . J. Mol. Biol. 2005;345:1119–1130. doi: 10.1016/j.jmb.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Weichsel, A., Gasdaska, J.R., Powis, G., Montfort, W.R. Crystal structures of reduced, oxidized, and mutated human thioredoxins: Evidence for a regulatory homodimer. Structure. 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- Yano, H., Wong, J.H., Lee, Y.M., Cho, M.J., Buchanan, B.B. A strategy for the identification of proteins targeted by thioredoxin. Proc. Natl. Acad. Sci. 2001;98:4794–4799. doi: 10.1073/pnas.071041998. [DOI] [PMC free article] [PubMed] [Google Scholar]