Abstract
Alanine racemase (Alr) is an important enzyme that catalyzes the interconversion of L-alanine and D-alanine, an essential building block in the peptidoglycan biosynthesis. For the small size of the Alr active site, its conserved substrate entryway has been proposed as a potential choice for drug design. In this work, we fully analyzed the crystal structures of the native, the D-cycloserine-bound, and four mutants (P219A, E221A, E221K, and E221P) of biosynthetic Alr from Escherichia coli (EcAlr) and studied the potential roles in substrate orientation for the key residues involved in the substrate entryway in conjunction with the enzymatic assays. Structurally, it was discovered that EcAlr is similar to the Pseudomonas aeruginosa catabolic Alr in both overall and active site geometries. Mutation of the conserved negatively charged residue aspartate 164 or glutamate 165 at the substrate entryway could obviously reduce the binding affinity of enzyme against the substrate and decrease the turnover numbers in both D- to L-Ala and L- to D-Ala directions, especially when mutated to lysine with the opposite charge. However, mutation of Pro219 or Glu221 had only negligible or a small influence on the enzymatic activity. Together with the enzymatic and structural investigation results, we thus proposed that the negatively charged residues Asp164 and Glu165 around the substrate entryway play an important role in substrate orientation with cooperation of the positively charged Arg280 and Arg300 on the opposite monomer. Our findings are expected to provide some useful structural information for inhibitor design targeting the substrate entryway of Alr.
Keywords: alanine racemase, substrate entryway, crystal structure, Escherichia coli, pyridoxal 5′-phosphate, charged residues
Alanine racemase (Alr, EC 5.1.1.1) is a pyridoxal 5′-phosphate (PLP)-dependent enzyme that catalyzes the interconversion of L-alanine and D-alanine, an essential precursor in the biosynthesis of the peptidoglycan layer of cell walls. Since Alr is unique to prokaryotes and a few of its counterparts were discovered in the D-Ala metabolism of yeast (Uo et al. 2001) and the D-alanine-containing peptides biosynthesis in fungi (Hoffmann et al. 1994; Cheng and Walton 2000), it has become an attractive target for antimicrobial development (Lambert and Neuhaus 1972), although the reported Alr inhibitors are almost structural analogs of alanine (Neuhaus and Hammes 1981; Copie et al. 1988), which usually lack specificity and act on some other PLP-containing enzymes. One of the representative inhibitors is D-cycloserine (DCS), a restricted drug against tuberculosis with serious side effects on the nervous system (Newton 1975).
For some organisms such as Lactobacillus plantarum (Hols et al. 1997), Mycobacteria (Strych et al. 2001), Corynebacterium glutamicum (Tauch et al. 2002), and Lactobacillus lactis (Steen et al. 2005), they each have only one Alr, while for others, including Salmonella typhimurium (Wasserman et al. 1983), Escherichia coli (Lobocka et al. 1994), and Pseudomonas aeruginosa (Strych et al. 2000), they each have two isozymes encoded by alr and dadX genes. The biosynthetic alr gene is expressed constitutively to provide D-Ala necessary for cell wall biosynthesis, while the expression of catabolic dadX gene is induced by L-alanine to a higher level than that of alr (Lobocka et al. 1994).
Structural studies on Alrs from Geobacillus stearothermophilus (Shaw et al. 1997), P. aeruginosa (LeMagueres et al. 2003), Streptomyces lavendulae (Noda et al. 2004), and Mycobacterium tuberculosis (LeMagueres et al. 2005) have revealed that Alr is a homodimer, with each monomer consisting of two different domains: an eight-stranded α/β barrel at the N terminus, and a C-terminal domain that is formed primarily with β-strands. Each of the two Alr active sites generated by dimer formation is composed of PLP, the PLP-binding residue Lys39 (residue number in G. stearothermophilus Alr), and the residues in the immediate environment of the pyridoxal cofactor (Noda et al. 2004). The complex structures of Alr with substrate analogs (Stamper et al. 1998; Morollo et al. 1999; Watanabe et al. 2002; Fenn et al. 2003) and related enzymatic features of the mutants (Sun and Toney 1999; Watanabe et al. 1999a,b) have revealed a two-base catalytic mechanism for Alr in which Lys39 is the catalytic base for the conversion of D-Ala to L-Ala and Tyr265′ from the opposing monomer catalyzes the reverse reaction.
Due to the small size of the active site for Alr, its conserved substrate entryway has been proposed as another choice for drug design (LeMagueres et al. 2003, 2005). This entryway corridor is formed within the N- and C-terminal domains of one monomer and the C-terminal domain of the other (Noda et al. 2004), and roughly conical with its base oriented toward the outside of the enzyme (LeMagueres et al. 2005). The conserved residues of this corridor are charged near the entrance, while dominantly hydrophobic near the active site (Noda et al. 2004). One of these hydrophobic residues is Tyr354, whose role in controlling substrate specificity has been reported (Patrick et al. 2002). However, the functions of the charged residues have not been yet identified.
In this work, by analyzing the crystal structures of the native, the DCS-bound, and four mutants of biosynthetic alr gene encoded Alr from E. coli (EcAlr) in conjunction with related a site-directed mutagenesis-based enzymatic assay, the potential roles for the key charged residues involved in the substrate entryway have been investigated. The results indicated that the negatively charged residues Asp164 and Glu165 around the substrate entryway play an important role in substrate orientation for EcAlr.
Results and Discussion
Overall structure of EcAlr
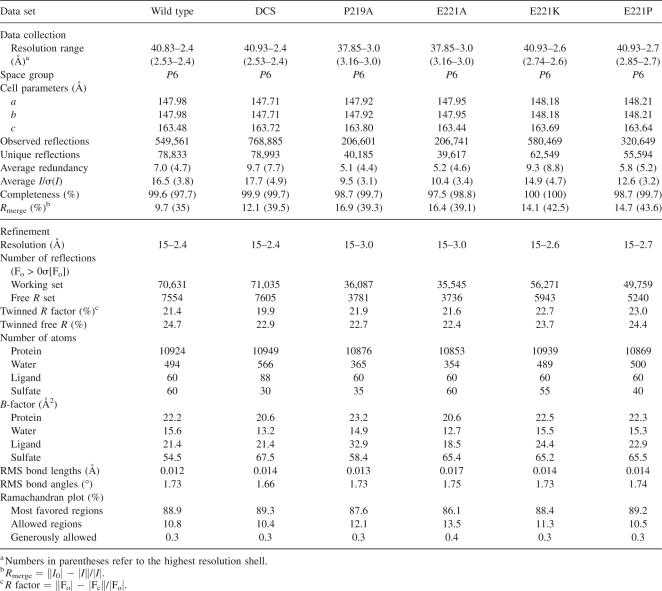
Since the protein sequence identity between EcAlr and P. aeruginosa catabolic dadX gene encoded alanine racemase (PaDadX) is high by 46.8% (Fig. 1A), the structures of these two enzymes are generally similar to each other, as expected. There are four EcAlr monomers forming two identical dimers in every crystallographic asymmetric unit. Except for a few disordered residues at the terminals of monomers C and D (residues 1–2 and 335–338), the native EcAlr structure is well defined, with the electron density maps continuous in all the monomers. As in PaDadX (LeMagueres et al. 2003), two EcAlr monomers keep a head-to-tail association with a low RMS difference of 0.29 Å obtained for their Cα atoms after least-square superimposition. Each monomer consists of two domains (colored differently in Fig. 1B): an N-terminal domain (residues 1–231) of the eight-stranded α/β-barrel, and a C-terminal domain (residues 232–359) containing mainly a β-structure. The composition of secondary structures in EcAlr is labeled above the sequences as shown in Figure 1A. In addition, there are three sulfate ions from the reservoir solution crystallized in each monomer: one in the substrate entryway (detailed below), and the other two on the protein surface and immobilized by arginines.
Figure 1.
Comparison of EcAlr and homologs. (A) Structure-based sequence alignment of alanine racemases from E. coli (EcAlr), P. aeruginosa (PaDadX), M. tuberculosis (MtAlr), G. stearothermophilus (GsAlr), and S. lavendulae (SlAlr). The secondary structures are labeled (“B” for strand and “H” for helix) and numbered on the top. Residues involved in the hydrogen-bond network of active site are labeled with an asterisk at the bottom, and the residues chosen for mutation are labeled by “#.” The alignment was calculated by the CE algorithm (Shindyalov and Bourne 1998) and produced by the program STRAP (Gille et al. 2003). (B) Ribbon diagram of the dimer structure of EcAlr. The N- and C-terminal domains are colored as orange and yellow in monomer A, and as red and pink in monomer B, respectively. The Lys34, PLP cofactor, and sulfate ions are shown in sticks. (C) Superposition of the monomers of EcAlr (cyan), PaDadX (green), and MtAlr (magenta) obtained after a least-squares fit of the Cα atoms from the N-terminal domains exclusively. The three regions showing major structure difference between EcAlr and PaDadX are labeled as a–c. The structures were all generated with PyMOL (Delano Scientific).
As shown in Figure 1C, the superimposition of the N-terminal domain from EcAlr with that from M. tuberculosis Alr (MtAlr) indicated that their C-terminal domains were significantly displaced from each other, while the superimposition between the N-terminal domains from EcAlr and PaDadX suggested their similar main-chain direction in the entirety of the monomers except for three regions (labeled a–c in Fig. 1C). Region a (residues Arg223 to Phe229) of EcAlr includes the helix H13 (Fig. 1A), whose direction is nearly vertical to that of its counterpart helix in PaDadX (LeMagueres et al. 2003). Region b contains residues Pro312 to Gln315, part of the loop between the two β-strands B16 and B17 (Fig. 1A). The difference of the main-chain directions between this region and its counterpart in PaDadX might arise from the unconserved residues. Region c (residues Glu332 to Val340) includes the α-helix H15, whose C terminus shifts a little toward the C-terminal domain. Since the loop near this helix composes part of the substrate entryway, this shift would make the entrance a little wider compared with that of PaDadX (LeMagueres et al. 2003).
Similar to the case for G. stearothermophilus Alr (GsAlr), binding of DCS induced no obvious conformation changes in the enzyme structure (Fenn et al. 2003). The RMS deviation between the native and DCS-bound EcAlr is 0.18 Å, calculated with 359 Cα atoms after least-square superimposition.
Active site
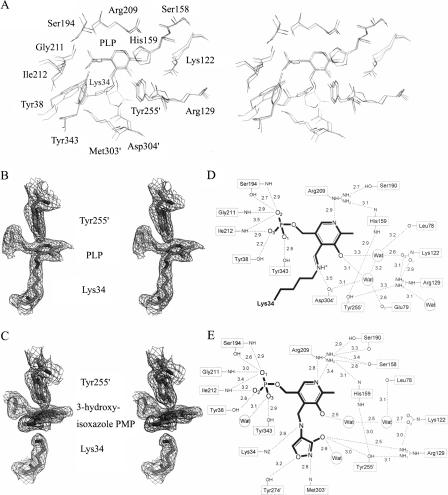
Generally, the geometries of the active sites of EcAlr and PaDadX are similar to each other. As shown in Figures 1A and 2A, the residues involved in the hydrogen bonds (H-bond) near PLP are conserved for these two racemases. Even after the binding of DCS to PLP forming the 3-hydroxyisoxazole PMP derivative, the active site residues keep their positions as in the native EcAlr, except Lys34, whose side chain lost the covalent aldimine linkage with the C4′ atom of PLP during the binding of DCS (Fig. 2B,C). In the meantime, the plane of the PLP pyridine ring turned toward Tyr255′ (255′ from the second monomer), making the linkage between hydroxyimine and pyridine just in the middle of the catalytic residues Lys34 and Tyr255′, with a distance around 3 Å, as in the case for GsAlr (Fenn et al. 2003).
Figure 2.
Active sites of native and DCS-bound EcAlr. (A) Stereoview of the superposition of the active site residues of PaDadX, native, and DCS-bound EcAlr obtained after a least-squares fit of Cα atoms in the N-terminal domains. The residue numbers are those of EcAlr and residues from the second monomer are identified with primed numbers. (B) A stereoview of the catalytic resides and PLP of native EcAlr embedded in 2Fo−Fc type electron density map contoured at 1.0 σ. (C) A stereoview of the catalytic residues and 3-hydroxyisoxazole–PMP of DCS-bound EcAlr embedded in 2Fo−Fc type electron density map contoured at 1.0 σ. (D–E) Schematic diagrams of the hydrogen-bond network in the active sites of native (D) and DCS-bound EcAlr (E).
Unlike PaDadX, which contains an external aldimine involving PLP and a guest substrate D-lysine (LeMagueres et al. 2003), EcAlr has only the internal aldimine linkage between PLP and Lys34. Figure 2D and E shows the H-bond networks in the active sites of native and DCS-bound EcAlr, respectively. Apart from the conserved residues and PLP, water molecules also participate in the networks.
As shown in Figure 2D, within the active site of native EcAlr, two water molecules form not only the internal H-bond (3.2 Å), but also two H-bonds with the oxygen atom of PLP (3.1 and 3.3 Å) and four additional H-bonds with the oxygen atoms from the main chains or side chains of residues Leu78, Glu79, Asp304′, and the carbamylated Lys122. It is noticeable that the side chain of Arg129 has no direct interaction with PLP, different from the cases for its counterparts Arg129 of PaDadX (LeMagueres et al. 2003) and Arg136 of GsAlr (Shaw et al. 1997; Fig. 1A), which form H-bonds with the oxygen atom of PLP directly. For EcAlr, Arg129 joins the network by forming H-bonds with Tyr255′, the carbamylated Lys122, and one water molecule (Fig. 2D). Such diversity might probably arise from the different ligand occupations near the substrate binding sites of these three racemases as revealed by the crystal structure analyses. For EcAlr, there is nothing but water for this occupation, whereas for PaDadX the guest D-lysine occupies the room (LeMagueres et al. 2003), and for GsAlr the acetate molecular exists in the room (Shaw et al. 1997).
As for the DCS-bound EcAlr, most of the residues involved in the H-bond network of 3-hydroxyisoxazole PMP derivatives are generally similar to those in the network of PLP for native EcAlr, except Tyr274′ and Met303′, which interact with the hydroxyimine (Fig. 2E) like their counterparts Tyr284′ and Met312′ of GsAlr (Fenn et al. 2003; Fig. 1A). In addition, the hydroxyl moiety derived from DCS forms H-bonds with the side chains of Arg129, Tyr255′, and a water molecule. As shown in Figure 2E, there are three other water molecules participating in the network. One water molecule forms an H-bond with the O3 atom at the phosphate tail of PLP, and the other two interact with the oxygen atom of PLP one by one.
Characterization of substrate entryway
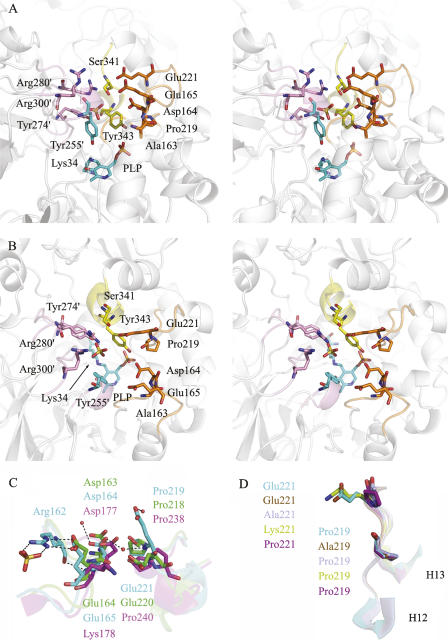
The substrate entryway is a key feature of Alr, and has been proposed to be a potential target for drug design since it consists of highly conserved residues (LeMagueres et al. 2005). For EcAlr, as indicated in Figure 3A and B, this entryway is composed of three parts: (1) two loops from the N-terminal domain (colored in orange); (2) one loop with part of the neighbor helix from the C-terminal domain (colored in yellow), and (3) two loops and one short helix from the C-terminal domain of the other monomer (colored in pink). The residues for directly shaping the entryway are Ala163, Asp164, Glu165, Pro219, Glu221, Ser341, Tyr343, and Tyr255′, Tyr274′, Arg280′, Arg300′ from the other monomer.
Figure 3.
Substrate entryway of EcAlr. Stereo pictures of the conserved residues at the substrate entryway in both side view (A) and top view (B). Residues forming the entryway from the N-terminal domain, C-terminal domain, and C-terminal domain of the second monomer are colored in orange, yellow, and pink the same as in Figure 1B, respectively. The catalytic residues Lys34, Tyr255′, and PLP are colored in cyan. The rest of the part of the EcAlr dimer is shown as a ribbon diagram and colored in white. (C) Superposition of the residues chosen for mutation (plus nearby secondary structures) in EcAlr (cyan) with the counterparts in PaDadX (green) and MtAlr (magenta). The residues are labeled in the corresponding colors. (D) Superposition of the loop containing mutations from wild-type EcAlr (cyan), P219A (brown), E221A (sky blue), E221K (yellow), and E221P (purple). The residues after mutation are labeled in the corresponding colors.
Except residues Ser341 and Glu221, most of the residues within the substrate entryway are conserved in EcAlr compared with other Alrs (Fig. 1A). The counterpart of Ser341 is the hydrophobic isoleucine among all the other homologs, while the counterpart of Glu221 is also glutamate in PaDadX but proline in the other three Alrs. In addition, Glu165 is substituted by lysine only in MtAlr (LeMagueres et al. 2005), as indicated in Figure 1A.
By investigating the charges of the residues around the entryway, we discovered that the three residues Asp164, Glu165, and Glu221 with negative charges take nearly opposite positions against the two positively charged residues Arg280′ and Arg300′ from the other monomer (Fig. 3A,B). In the crystal structure of EcAlr, these three negatively charged residues join a small H-bond network around them as shown in Figure 3C. Glu165 forms three H-bonds (two by side chain and one by main chain) with Arg162 in the same loop, whose side chain forms two additional H-bonds with a sulfate ion. Asp164 keeps H-bonds with two water molecules, one of which also interacts with the main-chain nitrogen of Glu221 from the nearby loop. Moreover, Glu221 forms the second H-bond with another water molecule by its side-chain oxygen (Fig. 3C).
In addition to the conserved residues, there is a sulfate ion located approximately in the middle of the above-mentioned entryway corridor of EcAlr (Fig. 3A,B). It forms two H-bonds with the NE and NH2 of Arg280′ via two oxygen atoms, one of which also forms another H-bond with the hydroxyl of Tyr274′. The average B-factor of this sulfate is about 45.3 Å2, higher than that of the protein (22.2 Å2) while lower than that of all the sulfate ions in the crystal structure (54.5 Å2) (Table 1). The position of this sulfate may provide a clue for the substrate's entry, in which the substrate might probably get the help of Tyr274′ and Arg280′ to break through the inner gate (LeMagueres et al. 2005) of the active site formed by Tyr255′ and Tyr343 in certain orientation.
Table 1.
Summary of diffraction data and structure refinement statistics
To further explore the possible roles of the negatively charged residues for substrate entry and catalysis, we constructed eight single-point mutants (D164A, D164K, E165A, E165K, P219A, E221A, E221K, and E221P), and carried out the related enzymatic and structural characterization against these mutants.
Enzymatic characterization of the mutants
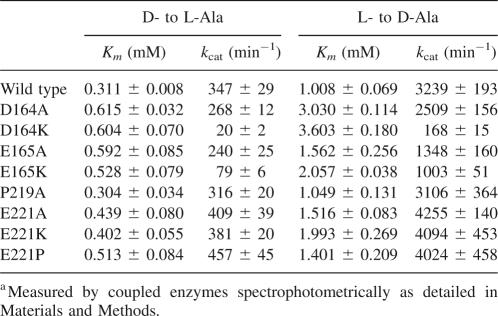
The kinetic parameters of EcAlr and its mutants were spectrophotometrically inspected according to the published approach (Esaki and Walsh 1986), and the results were listed in Table 2. Similar to the discovered results for S. lavendulae Alr by Noda et al. (2005), we also found that the equilibrium constant value (K eq), calculated from a Haldane equation defined by (k cat/Km for D-Ala)/(k cat/Km for L-Ala), was quite different from the theoretical value of 1.0, which might be caused by the incompatibility that the change in the amount of NADH was not consistent with the amount of Ala racemized in both D- to L-Ala and L- to D-Ala directions (Noda et al. 2005). However, the results might also facilitate us in comparing the parameters of the wild type and mutants in the same direction.
Table 2.
Kinetic parameters of the wild-type EcAlr and its mutantsa
As shown in Table 2, in comparison with the wild-type EcAlr, mutant D164A exhibited about one- or twofold increases in the Km values of D-Ala and L-Ala, and its two k cat values were found to be obviously lower than those of the wild type in the two directions. However, for mutant D164K, its Km values were relatively close to those of D164A, and its two k cat values were dramatically decreased to <6% compared with those of the wild type. As for the mutants E165A and E165K, their kinetic parameters changed in a similar manner compared with the Asp164 mutants (Table 2). Especially, the turnover numbers of E165K were only 20%∼30% of those of the wild type, while three- to approximately fivefold higher than those of D164K. These results thus indicated that residues Asp164 and Glu165 functioned potently for EcAlr catalysis. Since these two neighbor residues share the same charge, it is thereby proposed that these two residues might function similarly in controlling the substrate entry and/or product exit, and their negative charges possibly play an important role. However, it seems that residue Asp164 is more crucial as investigated by its more severe influence on the mutant k cat compared with Glu165 (Table 2). This might follow the fact that Asp164 is a little closer to the active site and fully conserved across all the Alrs (LeMagueres et al. 2005). Interestingly, our charge-involved substrate entry-controlling proposal might be further verified by the MtAlr enzyme research (Strych et al. 2001). In comparison with EcAlr, MtAlr has a much lower maximum velocity with the positively charged Lys178 as the counterpart for Glu165 of EcAlr (Figs. 1A, 3C).
Subsequently, we turned to the conserved residue Pro219 and partly conserved residue Glu221 of EcAlr on the other loop that forms the substrate entryway (Fig. 3A,B). As shown in Figure 1A, Pro219 is conserved in all the five Alr homologs. However, as indicated in Table 2, P219A mutant exhibited very close catalytic activity compared with the wild type, which implies that residue Pro219 might not directly participate in the substrate entry. On the other hand, it is discovered that the Km and k cat values of mutants E221A, E221K, and E221P were all slightly increased, suggesting that the negatively charged residue Glu221 possibly exhibits a small influence on the enzymatic activity and substrate entry (Table 2), which is not comparable with the cases for residues Asp164 and Glu165.
Crystal structure-based analysis of the key residues involved in the substrate entryway
To further explore the potential roles in substrate entry for the negatively charged residues at atomic level, we carried out the crystal structure-based characterization against the related EcAlr mutants. Although here the four mutants D164A, D164K, E165A, and E165K could not be crystallized for X-ray crystallographic use, the solved crystal structures of mutants P219A, E221A, E221K, and E221P have largely facilitated us in understanding the major functions of the negative charges for the key residues involved in the substrate entryway.
As expected, the structures of P219A and the wild type are quite similar with an RMS difference of only 0.05 Å obtained for 359 Cα atoms after least-square superimposition, since their catalytic activities were very close to each other (Table 2). As shown in Figure 3D, the structures of the loop between helices H12 and H13 (Fig. 1A) are nearly identical in both P219A and the wild type, except for the mutated side chain. As for the mutants E221A, E221K, and E221P, the RMS differences between them and the wild type are 0.21, 0.09, and 0.09 Å, respectively, suggesting that mutation of Glu221 did not change the overall structure. Moreover, the RMS differences of this loop between the mutants and wild type are 0.63, 0.22, and 0.34 Å, respectively. All the results thereby indicate that the mutations caused relatively larger variation on the loop than the whole protein, especially on the residues flanking Glu221 (Fig. 3D).
As have been already indicated in Table 2, unlike the mutations of Asp164 and Glu165, which significantly decreased the EcAlr catalytic efficiency (especially D164K and E165K), mutations of Glu221 only slightly influenced the enzymatic activity, even mutated to lysine with the reverse charge. To figure out the reason why these three negatively charged residues exhibited different effects on the substrate entry, we thus compared the electrostatic surface of wild-type EcAlr, mutants E221K and E221P (Fig. 4A–C). As indicated, the mutation decreased the negative charges around the substrate entrance and caused negative charges concentrated toward residue Asp164 that is just opposite to the positively charged residues Arg280′ and Arg300′. Interestingly, as shown in Figure 4B, besides the sulfate ion in the middle of the corridor, another sulfate could be modeled at the edge of the entrance. It locates at the center of the positive charges by forming H-bonds to Lys221, Arg280′, and Arg300′ with an average B-factor of 62.7 Å2, slightly lower than that of the middle sulfate (69.8 Å2) and all the sulfate ions in the asymmetric unit (65.2 Å2). This new sulfate has provided another indirect but convictive evidence for the charge redistribution around the entryway. Therefore, in Glu221 mutants, the weakened and concentrated negative charges, together with positive charges on the opposite side, might offer a little lower affinity to the amphoteric substrate alanine but slightly higher efficiency in the substrate orientation before it arrives at the active site. This could partly explain the somewhat increased Km and k cat values of Glu221 mutants compared with those of the wild type.
Figure 4.
Electrostatic surfaces around the substrate entryways of EcAlr wild type (A), E221K (B), E221P (C), PaDadX (D), GsAlr (E), MtAlr (F), PLP–L-Ala-bound (G), and PLP–D-Ala-bound GsAlr (H). Red colored to blue suggests the charges from negative to positive. The charged residues around the entrance and PLP or PLP–Ala in the active sites are shown in sticks and labeled. The sulfate ions in EcAlr structures are also shown in sticks.
Taking all these results together, we thus presume that the negatively charged residues Asp164 and Glu165 at the substrate entryway might play essential roles in substrate binding, and more importantly, in substrate orientation. Hence, the mutations of these two residues caused lower binding affinities to the substrate and lower turnover numbers (Table 2), especially when mutated to lysines with positive charge, which might remarkably influence the charge distribution. By comparing the electrostatic surface around the substrate entryways of EcAlr (Fig. 4A) and its homologs PaDadX (Fig. 4D), GsAlr (Fig. 4E), and MtAlr (Fig. 4F), we found that their charges distributed similarly except for MtAlr, whose Lys178 protrudes its side chain over neighbor Asp177 (Fig. 3C) and dramatically diminishes the negative charges around the entrance. This could be the partial reason for the 20- to ∼40-fold lower turnover number of MtAlr than that of EcAlr (Strych et al. 2001), since the substrate orientation facilitated by the certain charges around the entrance seems crucial for the reaction efficiency of Alr, which catalyzes the substrate at the small active site one by one (Shaw et al. 1997).
Further evidence of this above-mentioned presumption can be also found from the crystal structures of GsAlr bound with reaction intermediate analogs N-(5′-phosphopyridoxyl)-L-alanine (PLP-L-Ala) and N-(5′-phosphopyridoxyl)-D-alanine (PLP-D-Ala) (Watanabe et al. 2002). As shown in Figure 4G and H, when studying the PLP-L-Ala or PLP-D-Ala at the active site from the outside of the substrate entryway, it is discovered that the orientation of L-Ala or D-Ala in the reaction intermediate analogs is probably consistent with what we supposed in the substrate entry process. The nitrogen atom of alanine is toward the negatively charged residues Asp171 and Glu172, and the carboxyl is close to the positively charged Arg290′ and Arg309′. This consistency suggests that the substrate after preorientation at the entrance would be catalyzed near the PLP efficiently since no more energy or time for rotation is needed. A similar effect might happen to the product in the release process, as for racemases substrate and product can replace each other in the reverse reactions.
Conclusion
We report here the crystal structures of EcAlr in native, DCS-bound, and mutated forms. Together with kinetic parameters of the wild-type and mutants, we presume that the negatively charged residues around the substrate entryway, Asp164 and Glu165, play an important role in substrate orientation with the cooperation of the positively charged Arg280′ and Arg300′ from the other monomer. What is more, Asp164, which is fully conserved across Alr homologs (LeMagueres et al. 2005), may form a middle gate with Arg280′ on the opposite side (Fig. 3A,B), compared with the inner gate formed by Tyr255′ and Tyr343 near to the active site (LeMagueres et al. 2005). The middle gate is probably responsible for the final orientation of the substrate before it enters the inner one. Our work is expected to supply some useful information for drug-design targeting the substrate entryway of Alr.
Materials and Methods
Materials
E. coli strains BL21 (DE3) and JM109 were purchased from Stratagene. PLP, DCS, D-Ala, NAD+, NADH, L-Ala dehydrogenase, L-lactic dehydrogenase, and D-amino acid oxidase were from Sigma-Aldrich. Kanamycin, IPTG, and L-Ala were from Sangon.
Cloning of alr gene from E. coli
Based on the sequence of E. coli strain K12 (GenBank accession number NC_000913), the alr gene of E. coli was cloned from the genome of JM109 by genomic DNA extraction kit (Sangon). The DNA fragment containing alr was amplified by PCR with the primers (sense: 5′-AACACATATGCAAGCGGCAACTGTTGTGATTAAC-3′ and anti-sense: 5′-TACACTCGAGTTAATCCACGTATTTCATCGCGAC-3′), and inserted into pET28a (Novagen) to produce the recombinant protein with an N-terminal His-tag. The resulting plasmid pET28a-EcAlr was sequenced, and the deduced protein sequence was identical to that of alanine racemase 1 from K12 genome (GenBank accession number NP_418477).
Site-directed mutagenesis
Eight EcAlr single-point mutants D164A, D164K, E165A, E165K, P219A, E221A, E221K, and E221P were constructed with Quikchange Site-Directed Mutagenesis kit (Stratagene) following the instruction manual and verified by sequencing. The PCR primers containing the desired single mutations (in boldface) are as follows (sense only): 5′-CATTTTGCGCGCGCGGCTGAACCAAAATGTGG-3′ (D164A), 5′-CCATTTTGCGCGCGCGAAAGAACCAAAATGTGGCG-3′ (D164K), 5′-GCGCGCGCGGATGCACCAAAATGTGGCGC-3′ (E165A), 5′-GCGCGCGCGGATAAACCAAAATGTGGCGC-3′ (E165K), 5′-CTTTATGGCGTCTCGGCGCTGGAAGATCGC-3′ (P219A), 5′-GCGTCTCGCCGCTGGCAGATCGCTCCACCG-3′ (E221A), 5′-GCGTCTCGCCGCTGAAAGATCGCTCCACCG-3′ (E221K), and 5′-GCGTCTCGCCGCTGCCAGATCGCTCCACCG-3′ (E221P).
Expression and purification of EcAlr enzyme
The plasmid pET28a–EcAlr was transformed into E. coli strain BL21 (DE3), which grew in LB media supplemented with 100 μg/mL Kanamycin at 37°C. When the OD600 reached 1.0, the culture was induced by 0.2 mM IPTG and incubated at 22°C for an additional 4 h. After purification by His • Bind resin (Novagen), EcAlr was dialyzed against buffer A (20 mM Tris–HCl, pH 8.0, 100 μM PLP) and concentrated by ultrafiltration with an Amicon centrifugal filter device (Millipore). The expression and purification of the EcAlr mutants were handled in the same way as for the wild type. Protein concentration was determined by Bradford protein assay kit (Beyotime).
Crystallization and diffraction
EcAlr (15–30 mg/mL) was crystallized at 4°C in hanging-drops equilibrated with the reservoir solution (0.1 M HEPES, pH 7.0, 1.6 M ammonium sulfate). Yellow crystals were obtained with dimensions of 0.4 × 0.3 × 0.1 mm in about 2 wk. Colorless DCS-bound EcAlr crystals were grown in the similar way except that 2 mM DCS was added into the protein solution before mixed with the same reservoir solution. Similarly, four mutated proteins (P219A, E221A, E221K, and E221P) were also crystallized successfully.
Crystals were soaked in mother liquor containing a glycerol concentration of 30% as cryoprotectant before flash freezing. Diffraction data were collected at −180°C on an in-house R-Axis IV++ image-plate detector equipped with a Rigaku rotating-anode generator. The data sets were integrated with MOSFLM (Leslie 1999) and scaled with programs of the CCP4 suite (Collaborative Computational Project, Number 4 1994). Summary of the diffraction data statistics is given in Table 1.
Structure determination and refinement
The structure of the wild-type EcAlr was solved by molecular replacement (MR) with the programs in CCP4 (Collaborative Computational Project, Number 4 1994) using the coordinate of PaDadX (PDB code 1RCQ) as the search model. Structure refinement with hemi-hedral twinning (twinning fraction of 0.4) was carried out using CNS (Brunger et al. 1998) standard protocols (energy minimization, simulated annealing, water picking, and B-factor refinement), and model building was facilitated by using the program Coot (Emsley and Cowtan 2004). Anisotropic B-scaling, bulk-solvent correction, and non-crystallographic restraints were employed throughout the refinement. For the DCS-bound EcAlr and the four mutants, their structures were placed in the isomorphous unit cell and directly refined by rigid-body refinement and other standard procedures. The stereochemical quality of the structure models during the course of refinement and model building was evaluated with the program PROCHECK (Laskowski et al. 1993). The statistics of structure refinement are summarized in Table 1. The structures of native EcAlr, DCS-bound EcAlr, mutants P219A, E221A, E221K, and E221P have been deposited in the RCSB Protein Data Bank under accession codes 2RJG, 2RJH, 3B8T, 3B8U, 3B8V, and 3B8W, respectively.
Enzyme activity assay
The activities of EcAlr wild type and mutants were assayed spectrophotometrically as described previously (Esaki and Walsh 1986). All assays were conducted in a 96-well-plate system (Tecan GENios reader) at 30°C and monitored continuously at 340 nm. Reactions for the D- to L-Ala direction were performed in buffer containing 100 mM Tris (pH 8.0), 10 mM NAD+, 0.5 U/mL L-Ala dehydrogenase, and D-Ala at desired concentrations; while those for the reverse direction contained 100 mM Tris (pH 8.0), 0.5 mM NADH, 100 U/mL L-lactic dehydrogenase, 1 U/mL D-amino acid oxidase, and L-Ala at desired concentrations. The reactions were initiated by the addition of 25 nM EcAlr enzymes (except for 100 nM E165K and 250 nM D164K) into the assay solution and the kinetic parameters were determined by double-reciprocal plots.
Acknowledgments
This work was supported by the State Key Program of Basic Research of China (Grants 2004CB58905, 2007CB914304, and 2006AA09Z447). We also acknowledge the National Natural Science Foundation of China (Grants 30525024, 90713046, and 20721003), Shanghai Basic Research Project from the Shanghai Science and Technology Commission (Grants 06JC14080 and 03DZ19228), and Foundation of Chinese Academy of Sciences (Grant KSCX1-YW-R-18).
Footnotes
Reprint requests to: Hualiang Jiang, Drug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, People's Republic of China; e-mail: hljiang@mail.shcnc.ac.cn; or Xu Shen, Drug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, People's Republic of China; e-mail: xshen@mail.shcnc.ac.cn; fax: 86-21-50806918.
Abbreviations: Alr, alanine racemase; EcAlr, Escherichia coli Alr encoded by gene alr; PLP, pyridoxal 5′-phosphate; DCS, D-cycloserine; PaDadX, Pseudomonas aeruginosa Alr encoded by gene dadX; MtAlr, Mycobacterium tuberculosis Alr; GsAlr, Geobacillus stearothermophilus Alr; H-bond, hydrogen bond.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.083495908.
References
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cheng, Y.Q., Walton, J.D. A eukaryotic alanine racemase gene involved in cyclic peptide biosynthesis. J. Biol. Chem. 2000;275:4906–4911. doi: 10.1074/jbc.275.7.4906. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Copie, V., Faraci, W.S., Walsh, C.T., Griffin, R.G. Inhibition of alanine racemase by alanine phosphonate: Detection of an imine linkage to pyridoxal 5′-phosphate in the enzyme-inhibitor complex by solid-state 15N nuclear magnetic resonance. Biochemistry. 1988;27:4966–4970. doi: 10.1021/bi00414a002. [DOI] [PubMed] [Google Scholar]
- Emsley, P., Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Esaki, N., Walsh, C.T. Biosynthetic alanine racemase of Salmonella typhimurium: Purification and characterization of the enzyme encoded by the alr gene. Biochemistry. 1986;25:3261–3267. doi: 10.1021/bi00359a027. [DOI] [PubMed] [Google Scholar]
- Fenn, T.D., Stamper, G.F., Morollo, A.A., Ringe, D. A side reaction of alanine racemase: Transamination of cycloserine. Biochemistry. 2003;42:5775–5783. doi: 10.1021/bi027022d. [DOI] [PubMed] [Google Scholar]
- Gille, C., Lorenzen, S., Michalsky, E., Frommel, C. KISS for STRAP: User extensions for a protein alignment editor. Bioinformatics. 2003;19:2489–2491. doi: 10.1093/bioinformatics/btg354. [DOI] [PubMed] [Google Scholar]
- Hoffmann, K., Schneider-Scherzer, E., Kleinkauf, H., Zocher, R. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J. Biol. Chem. 1994;269:12710–12714. [PubMed] [Google Scholar]
- Hols, P., Defrenne, C., Ferain, T., Derzelle, S., Delplace, B., Delcour, J. The alanine racemase gene is essential for growth of Lactobacillus plantarum. J. Bacteriol. 1997;179:3804–3807. doi: 10.1128/jb.179.11.3804-3807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert, M.P., Neuhaus, F.C. Mechanism of D-cycloserine action: Alanine racemase from Escherichia coli W. J. Bacteriol. 1972;110:978–987. doi: 10.1128/jb.110.3.978-987.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- LeMagueres, P., Im, H., Dvorak, A., Strych, U., Benedik, M., Krause, K.L. Crystal structure at 1.45 Å resolution of alanine racemase from a pathogenic bacterium, Pseudomonas aeruginosa, contains both internal and external aldimine forms. Biochemistry. 2003;42:14752–14761. doi: 10.1021/bi030165v. [DOI] [PubMed] [Google Scholar]
- LeMagueres, P., Im, H., Ebalunode, J., Strych, U., Benedik, M.J., Briggs, J.M., Kohn, H., Krause, K.L. The 1.9 Å crystal structure of alanine racemase from Mycobacterium tuberculosis contains a conserved entryway into the active site. Biochemistry. 2005;44:1471–1481. doi: 10.1021/bi0486583. [DOI] [PubMed] [Google Scholar]
- Leslie, A.G. Integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- Lobocka, M., Hennig, J., Wild, J., Klopotowski, T. Organization and expression of the Escherichia coli K-12 dad operon encoding the smaller subunit of D-amino acid dehydrogenase and the catabolic alanine racemase. J. Bacteriol. 1994;176:1500–1510. doi: 10.1128/jb.176.5.1500-1510.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morollo, A.A., Petsko, G.A., Ringe, D. Structure of a Michaelis complex analogue: Propionate binds in the substrate carboxylate site of alanine racemase. Biochemistry. 1999;38:3293–3301. doi: 10.1021/bi9822729. [DOI] [PubMed] [Google Scholar]
- Neuhaus, F.C., Hammes, W.P. Inhibition of cell wall biosynthesis by analogues and alanine. Pharmacol. Ther. 1981;14:265–319. doi: 10.1016/0163-7258(81)90030-9. [DOI] [PubMed] [Google Scholar]
- Newton, R.W. Side effects of drugs used to treat tuberculosis. Scott. Med. J. 1975;20:47–49. doi: 10.1177/003693307502000204. [DOI] [PubMed] [Google Scholar]
- Noda, M., Matoba, Y., Kumagai, T., Sugiyama, M. Structural evidence that alanine racemase from a D-cycloserine-producing microorganism exhibits resistance to its own product. J. Biol. Chem. 2004;279:46153–46161. doi: 10.1074/jbc.M404605200. [DOI] [PubMed] [Google Scholar]
- Noda, M., Matoba, Y., Kumagai, T., Sugiyama, M. A novel assay method for an amino acid racemase reaction based on circular dichroism. Biochem. J. 2005;389:491–496. doi: 10.1042/BJ20041649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick, W.M., Weisner, J., Blackburn, J.M. Site-directed mutagenesis of Tyr354 in Geobacillus stearothermophilus alanine racemase identifies a role in controlling substrate specificity and a possible role in the evolution of antibiotic resistance. ChemBioChem. 2002;3:789–792. doi: 10.1002/1439-7633(20020802)3:8<789::AID-CBIC789>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Shaw, J.P., Petsko, G.A., Ringe, D. Determination of the structure of alanine racemase from Bacillus stearothermophilus at 1.9 Å resolution. Biochemistry. 1997;36:1329–1342. doi: 10.1021/bi961856c. [DOI] [PubMed] [Google Scholar]
- Shindyalov, I.N., Bourne, P.E. Protein structure alignment by incremental combinatorial extension (CE) of the optimal path. Protein Eng. 1998;11:739–747. doi: 10.1093/protein/11.9.739. [DOI] [PubMed] [Google Scholar]
- Stamper, G.F., Morollo, A.A., Ringe, D. Reaction of alanine racemase with 1-aminoethylphosphonic acid forms a stable external aldimine. Biochemistry. 1998;37:10438–10445. doi: 10.1021/bi980692s. [DOI] [PubMed] [Google Scholar]
- Steen, A., Palumbo, E., Deghorain, M., Cocconcelli, P.S., Delcour, J., Kuipers, O.P., Kok, J., Buist, G., Hols, P. Autolysis of Lactococcus lactis is increased upon D-alanine depletion of peptidoglycan and lipoteichoic acids. J. Bacteriol. 2005;187:114–124. doi: 10.1128/JB.187.1.114-124.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strych, U., Huang, H.C., Krause, K.L., Benedik, M.J. Characterization of the alanine racemases from Pseudomonas aeruginosa PAO1. Curr. Microbiol. 2000;41:290–294. doi: 10.1007/s002840010136. [DOI] [PubMed] [Google Scholar]
- Strych, U., Penland, R.L., Jimenez, M., Krause, K.L., Benedik, M.J. Characterization of the alanine racemases from two mycobacteria. FEMS Microbiol. Lett. 2001;196:93–98. doi: 10.1111/j.1574-6968.2001.tb10547.x. [DOI] [PubMed] [Google Scholar]
- Sun, S., Toney, M.D. Evidence for a two-base mechanism involving tyrosine-265 from arginine-219 mutants of alanine racemase. Biochemistry. 1999;38:4058–4065. doi: 10.1021/bi982924t. [DOI] [PubMed] [Google Scholar]
- Tauch, A., Gotker, S., Puhler, A., Kalinowski, J., Thierbach, G. The alanine racemase gene alr is an alternative to antibiotic resistance genes in cloning systems for industrial Corynebacterium glutamicum strains. J. Biotechnol. 2002;99:79–91. doi: 10.1016/s0168-1656(02)00159-1. [DOI] [PubMed] [Google Scholar]
- Uo, T., Yoshimura, T., Tanaka, N., Takegawa, K., Esaki, N. Functional characterization of alanine racemase from Schizosaccharomyces pombe: A eucaryotic counterpart to bacterial alanine racemase. J. Bacteriol. 2001;183:2226–2233. doi: 10.1128/JB.183.7.2226-2233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman, S.A., Walsh, C.T., Botstein, D. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J. Bacteriol. 1983;153:1439–1450. doi: 10.1128/jb.153.3.1439-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, A., Kurokawa, Y., Yoshimura, T., Kurihara, T., Soda, K., Esaki, N. Role of lysine 39 of alanine racemase from Bacillus stearothermophilus that binds pyridoxal 5′-phosphate. Chemical rescue studies of Lys39 → Ala mutant. J. Biol. Chem. 1999a;274:4189–4194. doi: 10.1074/jbc.274.7.4189. [DOI] [PubMed] [Google Scholar]
- Watanabe, A., Yoshimura, T., Mikami, B., Esaki, N. Tyrosine 265 of alanine racemase serves as a base abstracting α-hydrogen from L-alanine: The counterpart residue to lysine 39 specific to D-alanine. J. Biochem. 1999b;126:781–786. doi: 10.1093/oxfordjournals.jbchem.a022517. [DOI] [PubMed] [Google Scholar]
- Watanabe, A., Yoshimura, T., Mikami, B., Hayashi, H., Kagamiyama, H., Esaki, N. Reaction mechanism of alanine racemase from Bacillus stearothermophilus: X-ray crystallographic studies of the enzyme bound with N-(5′-phosphopyridoxyl)alanine. J. Biol. Chem. 2002;277:19166–19172. doi: 10.1074/jbc.M201615200. [DOI] [PubMed] [Google Scholar]