Abstract
When researchers build high-quality models of protein structure from sequence homology, it is today common to use several alternative target-template alignments. Several methods can, at least in theory, utilize information from multiple templates, and many examples of improved model quality have been reported. However, to our knowledge, thus far no study has shown that automatic inclusion of multiple alignments is guaranteed to improve models without artifacts. Here, we have carried out a systematic investigation of the potential of multiple templates to improving homology model quality. We have used test sets consisting of targets from both recent CASP experiments and a larger reference set. In addition to Modeller and Nest, a new method (Pfrag) for multiple template-based modeling is used, based on the segment-matching algorithm from Levitt's SegMod program. Our results show that all programs can produce multi-template models better than any of the single-template models, but a large part of the improvement is simply due to extension of the models. Most of the remaining improved cases were produced by Modeller. The most important factor is the existence of high-quality single-sequence input alignments. Because of the existence of models that are worse than any of the top single-template models, the average model quality does not improve significantly. However, by ranking models with a model quality assessment program such as ProQ, the average quality is improved by ∼5% in the CASP7 test set.
Keywords: protein structure/folding, structure, protein structure prediction, homology modeling
The gap between the number of known protein sequences in genome databases and corresponding three-dimensional structures is rapidly increasing, and for the vast majority of proteins we will likely never determine experimental structures. One important tool to bridge this gap and deduce structural properties from sequence is theoretical modeling based on homology. Even if the quality of these models cannot yet compete with experimental structures, they are extremely cheap to produce and can be applied on a much larger scale. Homology modeling methods use the fact that evolutionarily related proteins frequently share a similar structure. Therefore, if the sequence identity is high enough a three-dimensional model of a protein with unknown structure (target) can be built using a sequence alignment to a protein of known structure (template). Improving these model-building algorithms is important not only for decreasing the structure–sequence gap, but also to achieve higher-quality individual models that, e.g., are accurate enough for drug design.
The accuracy of homology models is directly related to how similar the target is to the template sequence, and there is pretty solid consensus that the two most important factors are to (1) choose the best possible template and then (2) optimally align the target sequence onto this template (Moult 2005). When the sequence identity is >40%, the alignment is usually considered to be trivial and the main reason for model inaccuracies is due to structural divergence. However, if there are several different templates with similar sequence identity it is hard or impossible to choose the best. With more distance targets, neither the selection of the best template nor its alignment is trivial. Many studies have analyzed different ways to obtain better models, for instance to use profile–profile, or HMM–HMM methods, which appear to do best at identifying the template folds (Rychlewski et al. 2000; Ohlson et al. 2004). For the actual alignments, profile–profile methods seem to achieve better results than methods that do not use information from profiles for both the target and the query sequences (Honig 1999; Wang and Dunbrack 2004; Ohlson and Elofsson 2005). Finally, with finished models, many methods have been developed to attempt to identify the best model out of a set of predictions (Colovos and Yeates 1993; Sippl 1993; Eisenberg et al. 1997; Wallner and Elofsson 2003), and it has clearly been shown in the latest CASP experiments that consensus methods (Lundström et al. 2001) using input from several predictors excel in this context. In particular, these methods are excellent at resolving and predicting relative quality of different parts of a model (Wallner and Elofsson 2006).
Significantly less focus has been given to the final step in the homology modeling, i.e., the model building itself. In a recent study (Wallner and Elofsson 2006), we showed that three methods, Nest (Petrey et al. 2003), Modeller (Sali and Blundell 1993), and SegMod (Levitt 1992), all perform quite well for single-template homology modeling, while several other methods frequently failed to produce close-to-optimal models. In addition, the performance of some common modeling programs using alignments of low sequence identity has been tested recently (Dalton and Jackson 2007). For many years, the authors and other investigators have claimed that the use of multiple templates “naturally” increases the accuracy of homology modeling, presumably since it better captures the variability and divergence of natural structures. Although several individual such examples have been reported (Venclovas 2003), there have not really been any large scale studies that investigate if this is generally true, what extra information is really being extracted, and how it improves models—and not least, if there are cases when it causes problems. It has, for instance, been proposed that a good reason to use multiple templates is because it is nontrivial to identify the best out of two or more templates (Contreras-Moreira et al. 2003). However, this would mean that if it were possible to always select the better of two (or more) single-template models, the single-template performance would be superior or at least equal to the multiple-template model.
To gain insight into these questions we have examined to what extent multiple templates can improve quality, where the improvement comes from, and whether we can predict this potential for improvement before deciding whether to use multiple templates. We have used two standard programs (Nest and Modeller) that are designed to use multiple templates, and in addition (as a future test bed) developed a new multi-template builder, Pfrag, that can utilize multiple templates in two different ways, either by averaging high-scoring templates or by starting from the single highest-scoring template and then extending that model.
These four algorithms have been benchmarked using two different test sets, one set of difficult targets, where alignments were obtained from automatic servers during the CASP7 experiment, and an easier set, where alignments were obtained using standard sequence alignment algorithms as described by Wallner and Elofsson (2005). We show that for a significant number of cases Modeller actually manages to produce models that are better than any of the single-template-based models, but that the probability of producing a significantly worse model also increases. The other methods produce fewer improved models, but are also somewhat less likely to completely disrupt the structure. Therefore, we propose a method to select when to use multiple templates and when not to. We show that this method improves the performance of our Pcons algorithm used in CASP7 by 5% and also discuss other alternatives to predict the potential for model improvement.
Results
To analyze the performance of the four different methods tested, up to six of the highest-ranked alignments were fed to the model-building algorithms, and the resulting quality was evaluated from the change in TM score (Zhang and Skolnick 2004) averaged over all targets in each of the two data sets. Many other evaluation functions exist, such as LG score (Cristobal et al. 2001) and MaxSub (Siew et al. 2000). The TM score is useful since it exhibits quite high agreement with the results of human expert visual assessment (Zhang and Skolnick 2004). However, in addition to the TM score, model quality was also evaluated using GDT_TS (Zemla et al. 1997), which is the gold standard for evaluation in CASP. Both scoring functions provided virtually identical results, and the evaluations based on the GDT_TS score can be found in the Supplemental material.
Change in TM score with different number of alignments
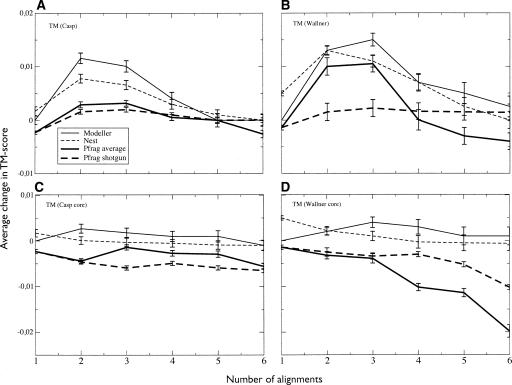
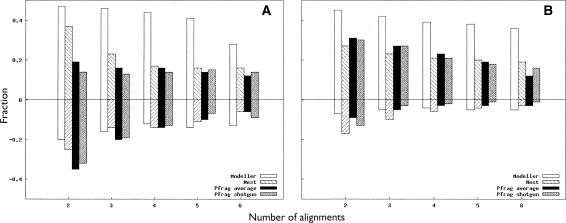
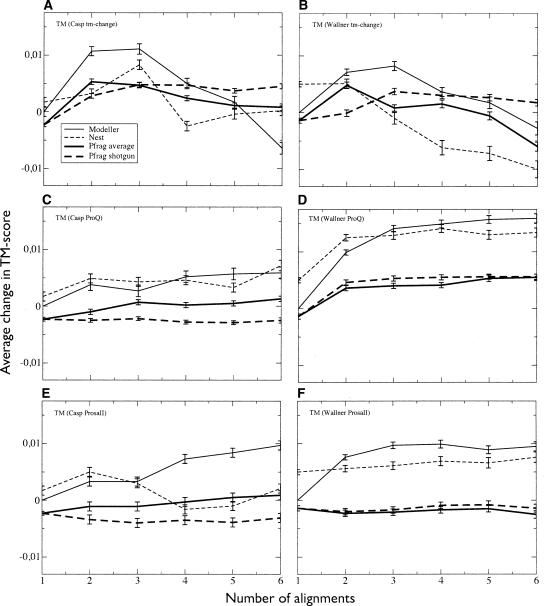
Figure 1 illustrates the change in the average TM score versus the number of alignments used for the four different methods. The left panel in Figure 1 shows the performance on the CASP7 set, and the right, the Wallner benchmark set that includes simpler targets (see Materials and Methods). The baseline for comparison is the highest-ranked single-template model built by Modeller; the reason for this choice is simply that the Pcons6 models were originally built with Modeller and thus served as a convenient point of reference.
Figure 1.
Average change in TM score for models built using different numbers of target-template pairs. Error bars indicate the standard error. The reported scores are for (A) full-length CASP7 models and (B) Wallner models. Panel C shows the length reduced CASP7 models and D shows the Wallner models. For both data sets, there is an increase in average TM score using two to three alignments. Modeller shows the most improvement of all programs for the CASP7 data (with a 0.0116 change in average score), while Nest and Modeller perform almost identically for the Wallner set. In contrast, Pfrag shotgun gives the best results using all six available alignments. Contrary to A and B, when only taking into account the residues already present in the first model (CASP7 in C and Wallner in D), the average TM score drops for both data sets and all programs except Modeller, indicating that Modeller actually can improve these core residues to a limited extent.
In general, Modeller appears to perform best when using two or three templates, which provides an average TM score improvement of just above 0.01 (for two templates) compared to a single template. However, when more alignments are used the performance gradually falls. Nest actually produces slightly better models than Modeller when using a single template. The behavior of Nest when using multiple templates is similar to Modeller, with a small increase when using two or three templates and then a gradual drop in average quality. For our Pfrag-average method, the largest improvement occurs when using three templates, while the Pfrag-shotgun model behaves differently, with the best results obtained when using all available templates.
The results for the larger, and easier, Wallner set are similar to those of the CASP7 set, but a few things are worth observing. First, the trend from the CASP7 data set that Nest builds slightly better single-template models persists in the larger data set. Also, Nest performs on par with Modeller using both two and three alignments, but both these programs then deteriorate more than for the CASP7 targets with an increasing number of alignments. The most likely explanation is that some of the lower-ranked alignments in this data set are of rather poor quality. While the improvement for Pfrag shotgun is never as high as for Modeller or Nest, it maintains the behavior of continuously improving with additional alignments.
However, as noted before, one of the major factors when using multiple templates is that regions not present in the highest-ranked target-template alignment can be added to the model, i.e., the length of the model increases. TM score and many other quality measures do not penalize incorrect regions, i.e., increasing the length of the model cannot decrease the average score. These improvements due to length can be considered rather trivial compared to the potential of improving local structure by better modeling variability and mutations in segments present already in the first model. Therefore, a more critical test of the ability of the different programs to actually improve upon the best single template in any given region is to take into account only those residues that are found in the best single-template model.
Thus, from this point (with the exception of Table 2) we only evaluate residues present in the highest-ranked single-template model, which we refer to as “core” residues. In Figure 1C it can be seen that Modeller is now the only program showing a slight improvement using two alignments. For Nest and Pfrag the models now get worse as more alignments are included. Unfortunately, this shows that the increase in TM score plotted in Figure 1A,B is largely an effect of the models becoming longer, not that we are able to discriminate between alternative local templates.
Table 2.
Changes in CASP7 performance for different methods
Chemical correctness
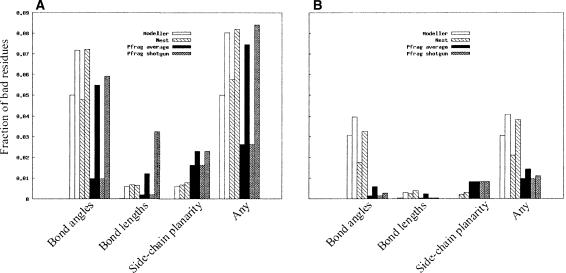
In our earlier study (Wallner and Elofsson 2005) we showed that all three programs (Modeller, SegMod, and Nest) produced models that were mostly chemically correct using single target-template alignments. Applying WHATCHECK (Hooft et al. 1996) and the same criteria as in the earlier study, the chemical correctness of single- and multiple-template models was investigated. Figure 2 shows that all four methods produce roughly the same amount of “bad” residues and that there are an increased number of such residues when multiple templates are used. However, in general it can be claimed that all methods are able to produce chemically correct models for a large majority of these test cases, and there are no obvious differences between the programs. In addition, all methods produce an equal (and low) fraction of knotted conformations (see Materials and Methods for details).
Figure 2.
Evaluation of chemical correctness of the models calculated using the WHATCHECK program for (A) CASP7 models and (B) Wallner models. The “Any” category is simply a union of the other three categories. For all methods and both data sets used, the chemical correctness is best with fewer alignments, but Pfrag (average and shotgun) seems to be most sensitive to the number of alignments using the CASP7 data set. For the Wallner data set, Pfrag (both versions) produces the most chemically correct models, possibly attributable to the energy minimization that follows initial model building. The left and right bars for each program correspond to models built with two and six template sequences, respectively.
Examples
To improve the understanding of how the different modeling methods perform, a large set of models was manually examined and a few selected successes and failures discussed below.
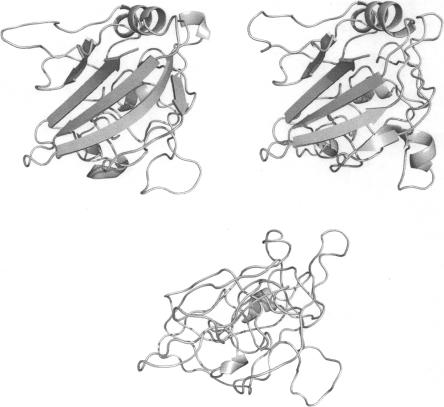
Figure 3 illustrates a Modeller model from the Wallner set using either of the two top single-template alignments (top left and right) or a multiple-template model using both alignments (bottom). This is a typical example of what happens when a program seems to fail to converge: There appear to be some constraints introduced from the multiple templates that make the program produce a suboptimal model. For this target, the Nest and Pfrag multiple-template models are similar to the corresponding single-template models, indicating that these programs manage to handle this particular case better (see figures in Supplemental material).
Figure 3.
An example of a program (in this case Modeller) failing to converge to a good model when more alignments are added. The TM score drops from 0.936 for the first single-template model (top left) and 0.930 for the second single-template model (top right), to 0.512 for the multi-template model (bottom), which also adopts a nonphysical conformation. This happens despite the fact that the two single-template models are quite similar (RMSD between the two single-template structures is 1.201). The same model built with Nest and Pfrag can be found in the Supplemental material. Molecular graphics were generated with PyMOL (DeLano Scientific).
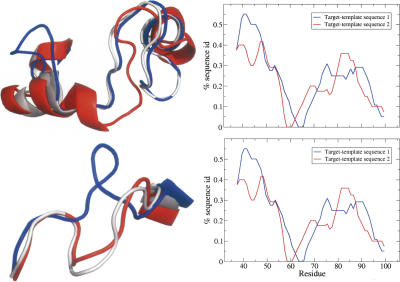
The second example (Fig. 4) also comes from the Wallner data set and shows a model built with Modeller that is improved using multiple templates. Here, the multi-template model is better than either of the two single-template models, because the program has chosen to follow different templates in different regions of the final model (Fig. 4, left panels). It can be seen from the bottom right panel in Figure 4 that the local sequence identity is important in this decision. Regions with a high local sequence identity to the template sequence have a lower RMSD. The region around residues 60–65 where the sequence identity is very low for both alignments also corresponds roughly to the region with the peak in RMSD between the multi-template model and the native structure (gray line in the top right panel in Fig. 4).
Figure 4.
Example of a model structure successfully alternating between templates, resulting in an overall better multi-template model. The per-residue RMSD (top right) shows how the multi-template model (gray) alternates between the two structures and in general stays closer to the one of the two (the first single-template model in blue and the second in red). A comparison can be made between regions of high RMSD in the top right panel and corresponding regions in the left panels. The bottom right panel shows that the local target-template sequence identity affects the modeling procedure (calculated using a 20-residue sliding window). A high local sequence identity corresponds to low RSMD-regions (top left). Overall RMSD between the multi-template model and single-template model 1 is 3.64, and 4.76 between the multi-template model and single-template model 2. In this case, the multi-template model is better (overall RMSD 3.3) than either of the two single-template models.
Discussion
From the results above it is quite clear that no significant average improvement is obtained for any of the tested methods when the increase in model length is ignored, which is somewhat striking. To improve a model, and not just increase its length, the modeling program needs to identify the best features of each of the target-template alignments and decide when to use one or another and how to combine them. An example of this type of algorithm has been published by Qian et al. (2004), who proposed using principal component vectors of variation between a set of template structures as degrees of freedom in refinement. If the modeling program is not capable of local discrimination or refinement, it is likely that the multiple-template model will rather resemble an “average” model, with a quality in between the corresponding single-template models. In that case it would be better to use the best single-template model if it could just be identified, and the only justification for the “average” model would be our shortcomings in selecting the best individual one (Contreras-Moreira et al. 2003). Finally, the multiple-target-template alignments might create conflicting constraints that make it harder for the modeling program to converge, and therefore the resulting model might be significantly worse than the corresponding single-template models.
However, when looking at the individual examples shown above, it is obvious that all four methods sometimes do improve models when multiple templates are used. Therefore, if it were possible to decide when to stop including multiple templates, it should be possible to build better models, at least on average. In addition, a better understanding of the factors that enable the modeling programs to create improved models might enable the development of even better modeling programs.
Which models are improved?
In Figure 5, models built from one or several target-template pairs are compared. Here, the multiple-template models are compared with all single-template models used. The fraction of multi-template models that are better or worse than all single-template models is reported. It can be seen that all methods sometimes produce both models that are better than the best of the single-template models and worse than all of the top-ranking ones. A similar ratio of models are improved using the Wallner set as in the CASP7 set, but fewer multiple-template models score lower than the best single-template models in the Wallner set. Obviously, as more alignments are included, a larger fraction of the multiple-alignment models fall into the intermediate region.
Figure 5.
Fraction of multiple-template models that are either better or worse than the top single-template models for different numbers of alignments. For both data sets ([A] CASP7 and [B] Wallner), the fraction of multiple-template models that is better than all single-template models for a given number of alignments decreases with increasing number of alignments. Also, the number of multiple-template models that are worse than all top-scoring single-template models decreases, as the single-template models built from alignments with a lower ranking are more likely to result in poor models.
Modeller stands out from the other methods and clearly produces the largest number of improved models. However, when using more than three alignments in the CASP7 set, Modeller also produces slightly more deteriorated models. Taking this into account, Modeller seems to be the program that has the greatest potential to improve if it were possible to decide when to stop using multiple alignments.
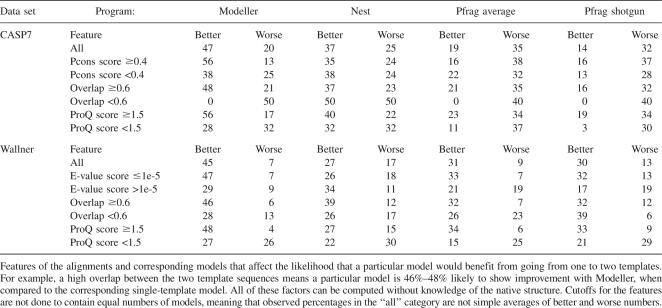
We have attempted to identify factors that determine when a model is improved and when it is not by comparing the first and second single-template model with the multiple-template model built from these two alignments (Table 1). It should be remembered that when we measure performance, only residues present in the first of the models are included, i.e., improvements due to a larger coverage are ignored. From Figure 6 it is evident that it is more likely to see an improvement with easy rather than hard models using Modeller. In particular, it is less likely that the model's quality will deteriorate. This could explain the difference between the CASP and Wallner data sets, as the latter has a higher fraction of easy targets. To a large extent, the lower fraction of models with decreased quality can be explained by the fact that the quality of the second model rarely is significantly worse than the first ranked model for good models (that would correspond to the empty area in the right corner of Fig. 6).
Table 1.
Factors affecting model quality (%), using “core” residues only
Figure 6.
Improvement with Modeller using two target-template pairs in (A) the CASP7 data set and (B), the Wallner data set. The figure shows the change in TM score relative to the best single-template model, and data are presented both for the second best single-template model as well as the second multi-template model. More multi-template models with a positive ▵-TM are found toward the right-hand side in both A and B, indicating that improvement with multiple templates is most prominent for easy models (high TM score). It is evident for the Wallner data set in particular that bad alignments in general produce bad models, as some of the second-ranking single-template models are quite bad, but also that some of the second-ranking models give scores higher than first-ranking models.
It was also observed that good models as judged by ProQ or significant Pcons/E-value scores are more likely to be improved. However, in particular for targets with low scores, fewer improve and more models deteriorate using multiple templates. Also, it seems that the chance of improving a multi-template model is greater if the template sequences are similar, as measured by their target sequence overlap. All these results indicate that multiple alignments are primarily useful when two good single-sequence alignments/models are combined. To a varying degree, the same trend can be seen for the other modeling programs.
In the modeling procedures of Modeller and Nest the sequence information is explicitly taken into account when using multiple templates. In Modeller the local sequence information is taken into account when the probability density functions (PDFs) are created from multiple templates, while in Nest the modeling is performed stepwise, and thereby the template with fewer local mutations might have more influence on the final structure. In the Pfrag-average method no such information is used, so both models are always given equal weight. Despite this, ∼15%–30% of the models are better than the best of the two individual models, which indicates that sometimes it is not only the ability to choose the right parts of a template that is important for using multiple templates.
Identification of improved models
The results above indicate that using multiple templates actually can improve the resulting models significantly, but also that it might cause the programs not to converge correctly. This would indicate that a strategy to identify convergence could provide a general tool to improve homology modeling using multiple target-template alignments. One of the reasons why a model deteriorates is if a particular program cannot resolve conflicting information from different alignments, and the ability to detect nonconverging models would be useful. Therefore, it would be interesting to examine automated procedures to decide when to stop including additional alignments. Although it might be possible to use the internal energy of the modeling programs, we tried to avoid this to make it generally applicable to other programs.
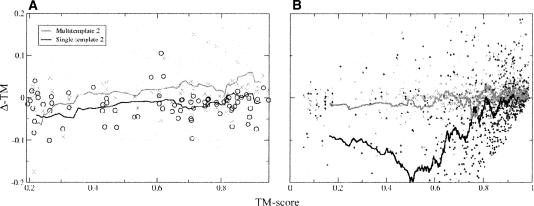
We have attempted two different approaches for this, as illustrated in Figure 7. In the first case we compared the structural similarity between the highest-ranked single-template model and the multiple-alignments-based model. Our results show that by comparing the difference in TM score between (1) the best single model and the first multiple-template model, (2) the first and second multiple-template models, and (3) the second and third multiple-template models, it is possible to improve the total cumulative TM score. We see an improvement when using a cutoff in TM score difference of 0.5. Modeller provided the best result, followed again by Nest and both Pfrag methods. Although the curves follow a similar trend without any cutoff rule, the overall performance is better. The improvement when using two or three alignments and Modeller or Nest is now larger and the gradual drop in performance with more alignments smaller. However, the performance of this method is still decreased when six alignments are used for most modeling programs.
Figure 7.
Changes in quality for the CASP7 (left panels) and the Wallner sets (right panels) using different methods of selecting the best model out of a given number of multi-template models with only core residue included. For panels A and B, no more alignments were added when the change in TM score between two consecutive models was above 0.5. Using ProQ as an energy function to select the best possible model for each target (CASP7 is C and Wallner D) gives an almost monotonous increase in model quality, since there are increasingly more models to choose from. The same trend (except for Nest in E) can be observed using ProsaII for both data sets (CASP7 in E and Wallner in F).
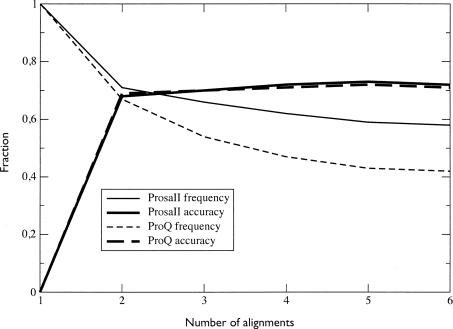
An alternative idea is to use a model quality assessment program (MQAP) (Wallner and Elofsson 2007) to select the best model out of a set of predictions. We tested MQAPs, ProsaII (Sippl 1993), and ProQ (Wallner and Elofsson 2003) to pick one of the models built from one to six multiple target-template alignments pairs. Using this, most methods show an almost monotonous increase with the number of included alignments (Fig. 7). The only exception is when using Nest and ProsaII on the CASP7 benchmark set. This indicates that although Pcons is a better method than any of these MQAPs to detect the best models (Wallner and Elofsson 2007), the MQAPs are somewhat capable of automatically deselecting models that did not converge. It is also clear that Modeller in general is the method that gains most from including multiple sequence alignments. However, because Nest produces slightly better single-template models, the final performance difference between the two programs is quite small. By using a MQAP to select models, an automated pipeline relies heavily on the ability of the program to select good models. The accuracy of ProQ and ProsaII in this work is in approximately 70%, meaning that when either of the programs identifies another model than the first as being the best, they are successful in 70% of the cases (Fig. 8). In this light, other MQAPs, such as the most recent work of Qiu et al. (2007), are interesting. It should also be noted that inclusion of the single-template models for alignments two to six to the procedure did not improve the performance (data not shown).
Figure 8.
Fraction of models (%) selected by either ProsaII or ProQ that are not the first-ranked single-template model in the Wallner data set and the accuracy in making that selection, i.e., how many of the selected models were actually better than the first-ranked single template. Both MQAPs show a similar accuracy level, but ProQ selects on average slightly more models that are based on multiple templates.
Improvement in CASP7 results
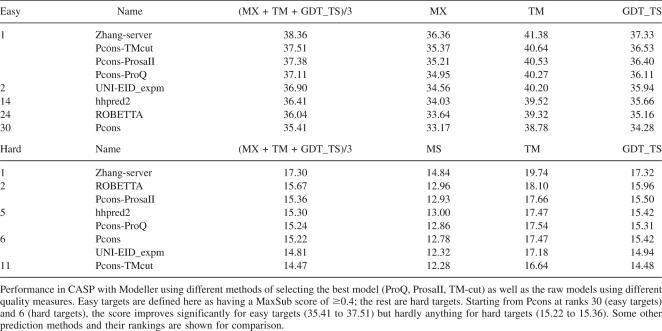
Finally, we examined what effect the inclusion of multiple-template alignments would have had on the Pcons performance in CASP7. Using Modeller and the three different strategies for inclusion of multiple sequence alignments, the performance of Pcons was tested on the CASP7 targets and compared with all other automatic predictions (Table 2). Without multiple sequence information or ROBETTA models the performance of Pcons would have been intermediate (rank 6) for the hard targets and quite bad for the easy targets (rank 30). However, the performance difference compared to all other methods, except the top-ranked Zhang server, is quite small. For the easy targets the three multiple target-template alignment methods would have performed better than all other methods except Zhang server, i.e., rank 2. The improvement is on the order of 5%, partly due to the fact that the models increase in size (∼35% of the improvement). For the hard targets the improvement is marginal for the ProsaII and ProQ methods, while the TM-cutoff method actually performs worse than the single-template approach, again indicating that currently the most useful application of multiple template alignments is for easier targets. To put numbers in perspective, the top 20 groups in the latest round of CASP, leaving out the Zhang server, all score within 6% of each other, clearly illustrating the significance of a seemingly modest increase of 5%.
Conclusions
Automatic inclusion of multiple templates for large-scale modeling definitely has potential to improve the average quality of models. The main effect is simply that the sequence coverage is improved, which increases the size of the produced model and most standard scores. However, when this size increase is ignored the picture is less rosy; none of the methods studied here manages to significantly improve average quality of the “core residues” present already in the single-template model. It is also noteworthy that multiple templates are not efficient for “averaging” information—in fact, the resulting quality typically drops when more than a handful of templates are included.
A more detailed analysis shows that all methods actually do produce individual models that are better, but also those worse than the top-ranked single-template models. Therefore, if it was possible to assess when models improve and when they do not, it should be possible to improve the average model quality. Indeed we show that using either measure to identify convergence problems or by using a MQAP (ProQ or ProsaII), it is quite straightforward to obtain models whose average quality improves by 5%. If this method had been applied to the Pcons algorithm in CASP7, the rank for the easier targets would have improved from 30th to 2nd. By analyzing a number of factors that could influence the performance of the multi-template algorithms, it is clear that the single most important property is that both the first and second individual alignment used for the multi-template model are of high quality. To conclude, it is not obvious that multiple templates a priori improve models in all cases, but it is clear that at least Modeller sometimes is able to select the best regions out of the two alignments and combine them into an improved structure. Also, from the Pfrag results, it is clear that homology modeling with multiple templates is a tricky business.
Materials and Methods
Data sets
Two test sets were employed for this study. Both these were created to represent realistic problems in large-scale homology modeling. For each data set, the alignments were ranked and most comparisons are made with the first-ranked model. For each target, from one up to six of the highest-ranked target-template alignments were used as an input to the different homology modeling programs.
The smaller CASP7 set consists of the targets from the recent CASP7 event (Moult et al. 2007). For this data set, alternative target alignments were obtained from different servers around the world by the Pcons.net Web server (Wallner et al. 2007). These target-template alignments were ranked based on the Pcons consensus score (Lundström et al. 2001; Wallner and Elofsson 2005) for the corresponding single-template model, i.e., not using any information about the alignments themselves. Pcons will report two pieces of information, a global quality score, reflecting the overall protein structure quality, and a local per-residue score, reflecting the quality of each residue in the model. Here, the alignment with the highest global Pcons score is selected as alignment number one, the second highest as number two, etc. This means that several alignments could be based on the same template, and in those cases differ only in the positions of, e.g., alignment gaps.
The larger data set is based on a previously used benchmark consisting of 1037 models (Wallner and Elofsson 2006). This set consists of alignments between protein sequences with known three-dimensional structure belonging to the same family according to SCOP (Murzin et al. 1995). The structures should have a resolution better than 3 Å and an R-factor less than 0.25. The alignments were constructed using rpsblast to search against the profile library of the Pcons.net server. All reported E-values are those given by rpsblast (Altschul et al. 1997). Also, all target-template pairs with a sequence identity >80% were removed, since it was judged that in such cases the best single-template model would be difficult to improve upon. Further, any possible improvement would be small and hence more difficult to detect. The alignments for each target were then ranked based on the reported E-value. The majority of alignments are quite similar, and 90% of the second-ranking templates have an E-value <1e-5. For ∼70% of the targets six or more template sequences with an E-value <1e-5 were found. For the rest, the six alignments with the lowest E-value were used anyway, to get a complete set of alignments (and because the E-value is not always a perfect measure of alignment quality). This set is referred to as the “Wallner” data set. It should also be noted that the aim of this work is to study multiple templates in the context of automated modeling. While different templates might introduce constraints that can be potentially difficult for a particular program to resolve, the possibilities for manual inspection of such constraints are very limited here.
Modeling methods
In our previous benchmark of homology modeling methods it was shown that three programs (Modeller, Nest, and SegMod) provided very similar performance and were better than alternatives. Of these, Nest and Modeller can utilize multiple target-template alignments.
Modeller
Modeller (Sali and Blundell 1993) is perhaps the most frequently used homology-modeling program. It was one of the first fully automated programs and it is also relatively fast, making it suitable, e.g., for whole-genome modeling (Marti-Renom et al. 2000; Pieper et al. 2004). Models are derived by optimizing spatial restraints derived from the alignment and expressed as PDFs for the different types of restraints. The PDFs restrain CA–CA and backbone N–O distances, as well as backbone and side-chain dihedral angles for different residue types. Minimizing violations of these restraints generates the model. When multiple templates are used, Modeller will automatically combine the target-template alignments using information about the local sequence identity and the structural differences to guide the modeling. This study used Modeller version 9.1, which is available from http://salilab.org/modeller/.
Nest
Nest (Petrey et al. 2003) is the core program within the Jackal Modeling Package and uses an artificial evolution method. To build models, changes from the template structure such as residue mutations, insertions, and deletions are made one at a time. After each change, a torsion energy minimizer is applied and energy calculated based on a simplified potential function. The alteration that produces the most favorable change in energy is accepted and the process repeated until the target sequence is completely modeled. When multiple alignments are used, the changes that provide the smallest evolutionary change to the target sequence are applied first. The Jackal Package can be downloaded from http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:Jackal.
Pfrag—an extended version of the SegMod algorithm
SegMod/ENCAD is a combination of a segment-matching algorithm (SegMod) (Levitt 1992) and energy minimization routines (ENCAD) (Levitt 1983). SegMod is based on a database of fragments from known protein structures. First, the aligned coordinates are copied to the target structure and then it tries to bridge the gaps by breaking down the target structure into a set of short segments and searches the database for segments that match the framework of the target structure. The matching is based on three criteria: sequence similarity, conformational similarity, and compatibility with the target structure using van der Waals interactions. The final model is then energy minimized using ENCAD. SegMod/ENCAD is available upon request from michael.levitt@stanford.edu.
We are extending the original SegMod algorithm into a GPL-licensed modeling program called Pfrag (http://pfrag.cbr.su.se/), which will be described in detail in future work. Multiple target-template alignments can be used in two different ways in Pfrag, both rather simplistic. For each target-template pair in the input alignment, we first build single-template models. Then, in the first method (Pfrag average) a model is constructed using the SegMod algorithm with the target coordinates of each residue not found in the first model being the average coordinates after an optimal superposition of the single-template models. This is in the same spirit as the original SegMod algorithm, which by default builds 10 independent models of each target and then averages them. More elaborate schemes are certainly possible, but this method serves as a basis for future improvements. To verify that this way of averaging coordinates does not introduce knotted or otherwise unphysical conformations, the output from Pfrag was screened for knots using the http://knots.mit.edu server (Kolesov et al. 2007). The fraction of knots for Pfrag models is between 0% and 2%, well comparable with Modeller and Nest (between 0% and 3%), at least for this data set.
In our second approach (Pfrag shotgun), the building starts from the highest-ranking single-template model and in the next step it is extended with residues that are missing in the first model but exist in models built from other target-template pairs. The idea behind this is to test if averaging extensions or selection between models primarily improves current multi-template modeling. To this end, Pfrag iterates over a given number of single-template models, and (yet again after a structural superposition of the models) uses any coordinates from the other models in order of their ranking for parts of the model that are not found in the first. In addition, it looks at the local residue quality scores in the first model and replaces any residues with a low local quality score with the corresponding residue in another model with a higher local quality score. This way, a model is produced in a similar way as in the 3D-shotgun method (Fischer 2003), with coordinates and fragments taken from different sources. For both approaches the models are energy minimized using the ENCAD force field (Levitt 1983).
Evaluation
TM score measures the structural similarity between two structures. In many instances it is preferred over other measures such as RMSD, since it does not overly penalize a model that is bad in only a small part of the overall structure. The TM score runs between 0 and 1, where a score from 0 to roughly 0.2 is considered a random hit, and a score above 0.4 is meaningful (Zhang and Skolnick 2004). To get rid of the length dependence in that longer models will get higher TM scores, we made a reduction of models to only include residues present in the highest-ranked single-template model. We refer to these residues as “core” residues. This way, it is possible to assess how much of the improvement in model quality is due to the increased length of models, and how much the programs improve already existing parts of a structure using several templates. For reference, alternative evaluation methods such as LGscore (Cristobal et al. 2001), MaxSub (Siew et al. 2000), or GDT_TS (Zemla et al. 1997) provided virtually identical results.
Comparison with CASP7 results
The baseline for a comparison of automatic modeling methods in CASP7 was the Pcons method. However, during CASP7, models from the ROBETTA server were also included in the consensus predictions of Pcons, but because the ROBETTA models do not come with alignments, they could not be used as input to the different modeling programs. Therefore, a version of Pcons ignoring the ROBETTA models was used as a baseline of CASP7 performance. The performance of Pcons without ROBETTA models is just slightly worse than the performance including ROBETTA, in particular for the harder targets (data not shown), and the expected improvements in CASP7 based on these slightly lower numbers (see Table 2).
Acknowledgments
Michael Levitt is kindly acknowledged for providing the SegMod code as a template for the work in this study, as well as stimulating discussion, and Anna Johansson for critically reading the manuscript. This work was supported by the Carl Trygger Foundation and the Swedish Foundation for Strategic Research to E.L., and the Swedish Research Council to A.E. and E.L. The EU 6th Framework Program is gratefully acknowledged for support to the EMBRACE project, contract LSHG-CT-2004-512092.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Arne Elofsson, Center for Biomembrane Research, Department of Biochemistry and Biophysics, Stockholm University, SE-106 91 Stockholm, Sweden; e-mail: arne@sbc.su.se; fax: 46-8-153679.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073344908.
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos, C., Yeates, T.O. Verification of protein structures: Patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Moreira, B., Fitzjohn, P.W., Bates, P.A. In silico protein recombination: Enhancing template and sequence alignment selection for comparative protein modelling. J. Mol. Biol. 2003;328:593–608. doi: 10.1016/s0022-2836(03)00309-7. [DOI] [PubMed] [Google Scholar]
- Cristobal, S., Zemla, A., Fischer, D., Rychlewski, L., Elofsson, A. A study of quality measures for protein threading models. BMC Bioinformatics. 2001;2:5. doi: 10.1186/1471-2105-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton, J.A., Jackson, R.M. An evaluation of automated homology modelling methods at low target template sequence similarity. Bioinformatics. 2007;23:1901–1908. doi: 10.1093/bioinformatics/btm262. [DOI] [PubMed] [Google Scholar]
- Eisenberg, D., Luethy, R., Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Fischer, D. 3D-SHOTGUN: A novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51:434–441. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- Honig, B. Protein folding: From the levinthal paradox to structure prediction. J. Mol. Biol. 1999;293:283–293. doi: 10.1006/jmbi.1999.3006. [DOI] [PubMed] [Google Scholar]
- Hooft, R.W., Vriend, G., Sander, C., Abola, E.E. Errors in protein structures. Nature. 1996;381:272. doi: 10.1038/381272a0. [DOI] [PubMed] [Google Scholar]
- Kolesov, G., Virnau, P., Kardar, M., Miny, L.A. Protein knot server: Detection of knots in protein structures. Nucleic Acids Res. 2007;35:W425–W428. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt, M. Molecular dynamics of native protein. I. Computer simulation of trajectories. J. Mol. Biol. 1983;168:595–617. doi: 10.1016/s0022-2836(83)80304-0. [DOI] [PubMed] [Google Scholar]
- Levitt, M. Accurate modeling of protein conformation by automatic segment matching. J. Mol. Biol. 1992;226:507–533. doi: 10.1016/0022-2836(92)90964-l. [DOI] [PubMed] [Google Scholar]
- Lundström, J., Rychlewski, L., Bujnicki, J., Elofsson, A. Pcons: A neural network based consensus predictor that improves fold recognition. Protein Sci. 2001;10:2354–2365. doi: 10.1110/ps.08501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti-Renom, M.A., Stuart, A.C., Fiser, A., Sanchez, R., Melo, F., Sali, A. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- Moult, J. A decade of CASP: Progress, bottlenecks and prognosis in protein structure prediction. Curr. Opin. Struct. Biol. 2005;15:285–289. doi: 10.1016/j.sbi.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Moult, J., Fidelis, K., Kryshtafovych, A., Rost, B., Hubbard, T., Tramontano, A. Critical assessment of methods of protein structure prediction-round VII. Proteins. 2007;69(Suppl):3–9. doi: 10.1002/prot.21767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzin, A.G., Grenner, S.E., Hubbard, T., Chothia, C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- Ohlson, T., Elofsson, A. ProfNet, a method to derive profile–profile alignment scoring functions that improves the alignments of distantly related proteins. BMC Bioinformatics. 2005;6:253. doi: 10.1186/1471-2105-6-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlson, T., Wallner, B., Elofsson, A. Profile–profile methods provide improved fold-recognition. A study of different profile–profile alignment methods. Proteins. 2004;57:188–197. doi: 10.1002/prot.20184. [DOI] [PubMed] [Google Scholar]
- Petrey, D., Xiang, Z., Tang, C.L., Xie, L., Gimpelev, M., Mitros, T., Soto, C.S., Goldsmith-Fischman, S., Kernytsky, A., Schlessinger, A., et al. Using multiple structure alignments, fast model building, and energetic analysis in fold recognition and homology modeling. Proteins. 2003;53(Suppl):430–435. doi: 10.1002/prot.10550. [DOI] [PubMed] [Google Scholar]
- Pieper, U., Eswar, N., Braberg, H., Madhusudhan, M.S., Davis, F.P., Stuart, A.C., Mirkovic, N., Rossi, A., Marti-Renom, M.A., Fiser, A., et al. MODBASE: A database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32:D217–D222. doi: 10.1093/nar/gkj059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian, B., Ortiz, A.R., Baker, D. Improvement of comparative model accuracy by free-energy optimization along principal components of natural structural variation. Proc. Natl. Acad. Sci. 2004;101:15346–15351. doi: 10.1073/pnas.0404703101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qui, J., Sheffler, W., Baker, D., Noble, W.S. Ranking predicted protein structures with support vector regression. Proteins. 2007;71:1175–1182. doi: 10.1002/prot.21809. [DOI] [PubMed] [Google Scholar]
- Rychlewski, L., Jaroszewski, L., Li, W., Godzik, A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali, A., Blundell, T.L. Comparative modelling by statisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Siew, N., Elofsson, A., Rychlewski, L., Fischer, D. MaxSub: An automated measure to assess the quality of protein structure predictions. Bioinformatics. 2000;16:776–785. doi: 10.1093/bioinformatics/16.9.776. [DOI] [PubMed] [Google Scholar]
- Sippl, M.J. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Venclovas, C. Comparative modeling in CASP5: Progress is evident, but alignment errors remain a significant hindrance. Proteins. 2003;53(Suppl):S380–S388. doi: 10.1002/prot.10591. [DOI] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. Can correct protein models be identified? Protein Sci. 2003;12:1073–1086. doi: 10.1110/ps.0236803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. All are not equal: A benchmark of different homology modeling programs. Protein Sci. 2005;14:1315–1327. doi: 10.1110/ps.041253405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Sci. 2006;15:900–913. doi: 10.1110/ps.051799606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallner, B., Elofsson, A. Prediction of global and local model quality in CASP7 using Pcons and ProQ. Proteins. 2007;69(Suppl):184–193. doi: 10.1002/prot.21774. [DOI] [PubMed] [Google Scholar]
- Wallner, B., Larsson, P., Elofsson, A. Pcons.net: Protein structure prediction meta server. Nucleic Acids Res. 2007;35:W369–W374. doi: 10.1093/nar/gkm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, G., Dunbrack, R.L. Scoring profile-to-profile sequence alignments. Protein Sci. 2004;13:1612–1626. doi: 10.1110/ps.03601504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemla, A., Venclovas, C., Reinhardt, A., Fidelis, K., Hubbard, T.J. Numerical criteria for the evaluation of ab initio predictions of protein structure. Proteins. 1997;1(Suppl):140–150. doi: 10.1002/(sici)1097-0134(1997)1+<140::aid-prot19>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Skolnick, J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]