Abstract
The complex of barnase (bn) and barstar (bs), which has been widely studied as a model for quantitative analysis of protein–protein interactions, is significantly destabilized by a single mutation, namely, bs Asp39 → Ala, which corresponds to a change of 7.7 kcal·mol−1 in the free energy of binding. However, there has been no structural information available to explain such a drastic destabilization. In the present study, we determined the structure of the mutant complex at 1.58 Å resolution by X-ray crystallography. The complex was similar to the wild-type complex in terms of overall and interface structures; however, the hydrogen bond network mediated by water molecules at the interface was significantly different. Several water molecules filled the cavity created by the mutation and consequently caused rearrangement of the hydrated water molecules at the interface. The water molecules were redistributed into a channel-like structure that penetrated into the complex. Furthermore, molecular dynamics simulations showed that the mutation increased the mobility of water molecules at the interface. Since such a drastic change in hydration was not observed in other mutant complexes of bn and bs, the significant destabilization of the interaction may be due to this channel-like structure of hydrated water molecules.
Keywords: protein–protein interaction, barnase, barstar, mutation, water-mediated interaction
Protein–protein interactions are a fundamental process in biological systems and are governed by several different factors, including hydrophobic and electrostatic interactions. Recent studies have shown that water-mediated indirect interactions are as important as direct interactions for maintaining the stability and specificity of interactions (Covell and Wallqvist 1997; Janin 1999; Kondo et al. 1999). A protein–protein interaction in water is affected not only by the hydrophobic effects of the surrounding water molecules but also by water molecules that bridge the interface between the proteins. The former are usually a major contributor to the stability of the complex by causing hydrophobic effects, whereas the latter stabilize or destabilize the interactions between proteins depending on the situation. Thus, it is generally more difficult to evaluate the effect of the bridging water molecules on the interactions between proteins.
The interaction between barnase (bn), an extracellular ribonuclease of Bacillus amyloliquefaciens, and its intracellular inhibitor barstar (bs), was shown to be an excellent system for investigating the interaction mediated by the penetrating water molecules because the complex of these two proteins has many water molecules at the interface (Buckle et al. 1994). Another important advantage of this system is the structural robustness of bn and bs. X-ray crystallography (Mauguen et al. 1982; Guillet et al. 1993b; Buckle et al. 1994; Ratnaparkhi et al. 1998) and NMR spectroscopy (Bycroft et al. 1991; Lubienski et al. 1993) showed that bn and bs barely changed their structures upon complex formation: The root-mean-square deviation (RMSD) between the structures before and after complex formation was <0.5 Å for all the Cα atoms of both the proteins. In addition, both the proteins exhibited very stable structures that resisted mutations; i.e., there was little change in the overall structures of bn and bs even after more than 130 and 30 mutations, respectively (Matouschek et al. 1990; Fersht and Serrano 1993; Nolting et al. 1995, 1997). Such structural robustness of bn and bs has provided a benchmark for theoretical studies on protein–protein interaction (Chen et al. 2003; Mintseris et al. 2005).
The kinetics and thermodynamics of binding between the two proteins was extensively investigated by using stopped-flow fluorescence measurements (Hartley 1993; Schreiber and Fersht 1993a, 1995; Schreiber et al. 1994, 1997; Vijayakumar et al. 1998; Frisch et al. 2001) and surface plasmon resonance measurements (Ikura et al. 2004). These results showed that the two proteins formed the complex extremely rapidly and that the complex was very stable: Its binding free energy was 19.0 kcal·mol−1, which corresponded to 1.3 × 10−14 M of the dissociation constant (Schreiber and Fersht 1993a).
Mutational studies indicated that several residues at the interface of the complex played important roles in the stabilization of the complex. With regard to bn-Lys27, bn-His102, bs-Asp35, and bs-Asp39, a mutation at any one these residues drastically increased the dissociation rate between the two proteins, while the association rates were hardly affected by the mutations (Schreiber and Fersht 1995). This suggested that electrostatic interactions between the charged groups of the proteins mainly contributed to the stability of the complex (Lee and Tidor 2001a,b). Among the abovementioned four residues, a mutation at bs-Asp39 most drastically destabilized the complex; i.e., the mutation bs-Asp39 → Ala (bs-D39A) increased the dissociation rate by 105, which corresponded to a decrease of 7.7 kcal·mol−1 in the binding free energy (Schreiber et al. 1994). X-ray crystallographic analysis of the complex, including a mutation on bn-Lys27, bn-His102, or bs-Asp35, indicated that a mutation at any of the three residues broke many intermolecular hydrogen bonds mediated by water molecules as well as the hydrogen bonds directly bridging both the proteins (Vaughan et al. 1999; Ikura et al. 2004). Although it is expected that the mutation bs-D39A will have similar effects on the hydrogen bonds, no structural evidence of the mutational effects has been available, as attempts to obtain good crystals of the particular complex have never been successful.
In the present study, we have successfully crystallized the mutant bs-D39A complex by introducing another mutation, bn-K98A, which changed the crystal packing, and determined the complex structure at 1.58 Å resolution by X-ray crystallography. Comparisons between the mutant and wild-type structures revealed that the structural changes of the protein molecules were localized around the mutational sites. On the other hand, water molecules at the interface were rearranged and redistributed into a channel-like structure, suggesting the complementarity of the deleted interactions with water molecules. Such high fluidity of water molecules at the interface was also confirmed by using molecular dynamics simulations. This hydrated structure of water molecules at the interface must be related to the destabilization of the interaction between the two proteins caused by the mutation.
Results
Crystallographic analysis on the interactions in the mutant complex
Overall structure of the mutant complex
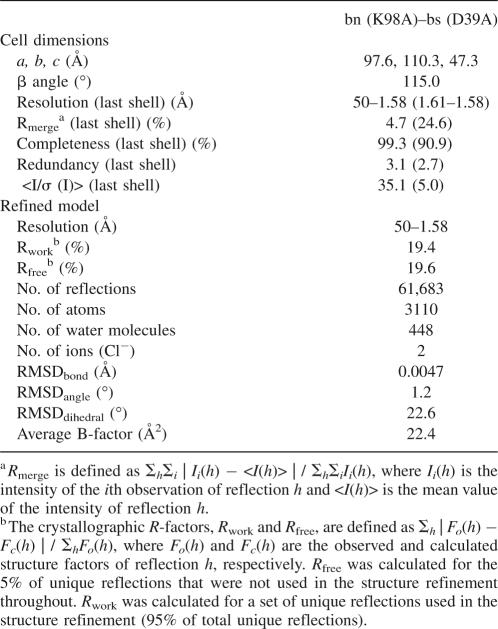
The crystal structure of the mutant complex bn (K98A)–bs (D39A) was determined at 1.58 Å resolution by X-ray crystallography (Table 1) to investigate the changes in intermolecular interactions caused by the mutation bs-D39A. This mutant complex crystallized in the monoclinic space group C2. There were two complexes in the asymmetric unit, and here we name them as the AB and CD complexes, where A and C were the bn chains and B and D were the bs chains (Fig. 1). Several residues of each complex were not detected: Electron density of Ala1–Gln2 in chain A, Glu64–Asn65 in chain B, Ala1 in chain C, and Glu57–Glu64 in chain D were completely or partially unresolved. We also observed N-terminal methionine residues in bs in chains B and D, as observed in other studies (Ratnaparkhi et al. 1998; Vaughan et al. 1999; Ikura et al. 2004). This methionine residue is assigned a residue number 0 to maintain the original numbering scheme. We detected two chloride ions bound to the CD complex: One chloride ion was located between the carboxamide Nδ2 of Asn58 and the indole nitrogen Nε1 of Trp71 in the bn chain C (Supplemental Fig. 1A), while the other was located between the guanidinium group Nη2 of Arg75 and the side-chain amino group Nζ of Lys78 in the bs chain D (Supplemental Fig. 1B). Although these ions were regarded as two water molecules at the initial stage of our refinement, they were finally judged as chloride ions based on their electron density, B-factors, and positively charged environments (Supplemental Fig. 1A,B). On the other hand, such strong density was not observed in the AB complex at the corresponding positions, although relatively weak electron density was observed at these positions. Thus, we suspected that these chloride ions had no biological significance (Mauguen et al. 1982; Ratnaparkhi et al. 1998).
Table 1.
Results of crystallographic data processing and structure refinement
Figure 1.
Crystal structure of the mutant complex between barnase (bn) and barstar (bs): bn (K98A)–bs (D39A). The two bn–bs complexes in the asymmetric unit viewed approximately down the twofold non-crystallographic axis between complexes AB and CD. Chains A and C represent bn (green), while chains B and D represent bs (blue). The mutational sites bn-K98A (white) and bs-D39A (red) were drawn with the space-fill model. (Two yellow spheres) Chloride ions.
The RMSDs between the two bn chains A and C and the two bs chains B and D were calculated at 0.28 Å, and 0.38 Å, respectively, by using the Cα atoms of bn 3–110 and bs 1–56 and 66–89 (Supplemental Table 1), showing that no significant structural differences were detected between two bn or two bs molecules. The orientations between the bn and bs chains, however, were subtly different between the two complexes AB and CD (Supplemental Fig. 1C): The RMSD between the two complexes AB and CD was 0.78 Å (Supplemental Table 1), corresponding to the 7° rotation of bs about an axis parallel to the interface plane.
Bn–bs interactions at the interface
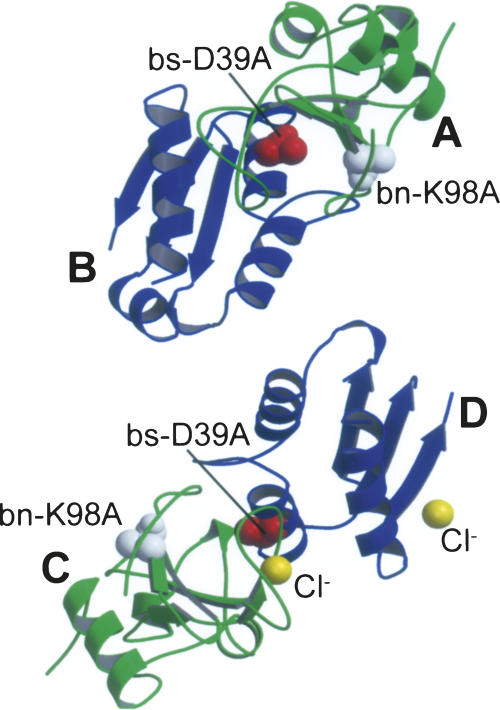
The residues involved in the bn–bs interface of the mutant complex were determined on the basis of the buried surface area. As a result, 20 residues of bn and 18 residues of bs were included in the interface: Lys27, Gln31, Trp35, Val36, Ala37, Ser38, Ile55, Phe56, Arg59, Glu60, Lys62, Glu73, Phe82, Arg83, Asn84, Ser85, Arg87, His102, Tyr103, and Gln104 of bn; and Tyr29, Gly31, Glu32, Asn33, Leu34, Asp35, Ala36, Trp38, Ala39, Thr42, Gly43, Trp44, Val45, Glu46, Tyr47, Val73, Glu76, and Glu80 of bs (Fig. 2A). The conformations in the interface of the two complexes in the asymmetric unit were almost identical to each other: The RMSD between the interfaces of the two complexes AB and CD was calculated at 0.52 Å by using all atoms constituting the interface (Supplemental Table 2).
Figure 2.
Potential surfaces of the interface of the bn–bs complex. This interface comprises 20 residues of bn (Lys27, Gln31, Trp35, Val36, Ala37, Ser38, Ile55, Phe56, Arg59, Glu60, Lys62, Glu73, Phe82, Arg83, Asn84, Ser85, Arg87, His102, Tyr103, and Gln104) and 16 residues of bs (Tyr29, Gly31, Asn33, Leu34, Asp35, Ala36, Trp38, Asp39 [Ala39], Thr42, Gly43, Trp44, Val45, Glu46, Tyr47, Val73, and Glu76). (A) Potential surfaces of the interface of the mutant complex without hydrated water molecules. The number written on the surface represents the number of amino acid residues of bn or bs. (B,C). Potential surfaces of the interface of the mutant (B) or wild-type (C) complex with hydrated water molecules. (Magenta spheres) “Edge” water molecules, (yellow spheres) “buried” water molecules, (cyan spheres) “bridging” water molecules. The number written on the spheres represents the number of the hydrated water molecule referred to in Supplemental Table 5. Surface color-coded according to electrostatic potential calculated by the Poisson-Boltzmann solver within GRASP2 and displayed within GRASP2 (Petrey and Honig 2003).
Two types of intermolecular interactions—hydrophobic and hydrophilic—were observed at the mutant interface. The hydrophobic interactions are characterized as nonspecific interactions between the hydrophobic side chains, while the hydrophilic interactions are characterized as specific interactions involving hydrogen bonds and salt bridges.
Hydrophobic interactions
The following 13 hydrophobic residues were expected to contribute to the hydrophobic interactions at the bn–bs interface: Trp35, Val36, Ile55, Phe56, Phe82, and Tyr103 of bn; and Tyr29, Leu34, Trp38, Trp44, Val45, Tyr47, and Val73 of bs (Fig. 2A). However, in the case of the wild-type bn–bs complex, hydrated water molecules prevented the formation of a wide region of hydrophobic interactions because the poor complementarity between the interacting protein surfaces allowed many water molecules to penetrate (Buckle et al. 1994). The buried surface areas of the mutant complexes, which correlate with hydrophobic interactions (Janin and Rodier 1995), were calculated to be 1493 Å2 and 1444 Å2 for the complexes AB and CD, respectively (Supplemental Table 3), suggesting that the mutant complex had the “standard-size” interface of which the buried surface area was 1600 ± 400 Å2 (Lo Conte et al. 1999).
Hydrophilic interactions
The hydrophilic interactions facilitated by an extensive hydrogen bond network were observed at the interfaces of the mutant complexes (Supplemental Tables 4, 5). Eight intermolecular hydrogen bonds directly linking bn-K98A to bs-D39A were commonly observed for both complexes in the asymmetric unit, while an additional intermolecular hydrogen bond was observed only for the AB complex (Supplemental Table 4). The eight common hydrogen bonds constituted one hydrogen bond involving both charged donor and acceptor groups (bn-Arg59Nη1–bs-Glu76Oε2), six hydrogen bonds involving one charged partner, and one hydrogen bond between uncharged groups (bn-His102O–bs-Asn33Nδ2). A hydrogen bond bn-Gln31Nε2–bs-Thr42O was not observed for the CD complex, probably because of the heterogeneity at the edge of the bn–bs interface (Fig. 2A).
Many water molecules were observed at the bn–bs interface (Fig. 2B), which could be classified into three categories (Buckle et al. 1994): (1) Those bound to the edge of the interface (“edge”), (2) those buried in the surface cavity and bound to either bn or bs (“buried”), and (3) those mediating the interactions between both proteins through hydrogen bonds of bn–H2O–bs (“bridging”). In the present study, we observed 40 water molecules at the bn–bs interface that were commonly found at the corresponding positions of both complexes in the asymmetric unit: 17, 11, and 12 molecules were corresponding to the “edge,” “buried,” and “bridging” water molecules, respectively (Supplemental Table 5; Fig. 2B). The “edge” water molecules were sufficiently exposed to the solvent and have relatively large B-factors, suggesting they were uncorrelated with the stability of the complex (Buckle et al. 1994). The 11 “buried” water molecules were almost completely buried. Nine of them were bound only to bn, whereas the other two were bound only to bs. The 12 “bridging” water molecules, which were expected to mostly contribute to the stability of the complex formation, were buried relatively deep between the two proteins except for one (H2O266) whose ASA was similar to those of the “edge” water molecules. One molecule (H2O253) linked the main chains of the proteins, while two (H2O98 and H2O339) did the same for the side chains of the proteins. The remaining nine water molecules formed hydrogen bonds between the side chain of one protein and the main chain of the other protein. The B-factor values of the “buried” and “bridging” water molecules appeared to be uncorrelated with their accessible surface areas.
Discussion
In the present study, X-ray crystallographic analysis and molecular dynamics simulations fully elucidated the interactions between the mutant proteins bn (K98A) and bs (D39A). Although the mutation bs-D39A decreased the binding free energy by 7.7 kcal·mol−1, the mutant complex unexpectedly retained both of the hydrophobic and hydrophilic interactions similar to those found in the wild-type complex: The former was acquired by the extensive buried surface area (Supplemental Table 3), and the latter was formed with direct and water-mediated hydrogen bonds (Supplemental Tables 4–7).
Comparison of the mutant complex with the wild-type complex
To investigate the reason for the large decrease in the binding free energy, we compared the structure of the mutant complex with that of the wild type and analyzed the changes of the interactions caused by the mutation. Crystal structures of the wild-type complex have been independently determined by several groups (Guillet et al. 1993a; Buckle et al. 1994; Ikura et al. 2004), but all of them are virtually identical. Thus, we adopted the structure of the wild-type complex most recently reported (PDB ID: 1X1U) (Ikura et al. 2004), which contained three complexes in the asymmetric unit, i.e., the AD, BE, and CF complexes, where A, B, and C were the bn chains and D, E, and F were the bs chains. There were no significant structural differences detected among the three bn, three bs, or three complex molecules in the asymmetric unit: The average RMSDs for bn, bs, and complex were calculated at 0.30 Å, 0.63 Å, and 0.59 Å, respectively, by using the Cα atoms of bn 3-110 and bs 1-56 and 66-89. The interface of the wild-type complex consisted of 20 and 18 residues of bn and bs, respectively, which was the same as the mutant interface (Ikura et al. 2004).
Structural effects
Comparison of proteins of the mutant complex to the corresponding proteins of the wild-type complex (Supplemental Table 1) revealed that the mutation did not significantly change the structure of each component of the complex but caused a subtle change in the orientation of bs against bn; the average RMSDs for bn, bs, and complex were calculated at 0.51 Å, 0.75 Å, and 1.23 Å, respectively, whereas the mutant bs rotated by 8°–15° from the position of the wild-type bs about an axis perpendicular to the interface plane (Supplemental Fig. 1D). Similar variations in orientation between bn and bs were also observed between the two mutant complexes in the asymmetric unit even though their directions of rotation were different (Supplemental Fig. 1C). Given the fact that we could not find the significant difference in the structures between the mutant and wild-type interfaces (the average RMSD was only 1.28 Å for the Cα atoms of the interface residues), it is very unlikely that the small difference in orientation between bn and bs mentioned above is the main cause of the drastic decrease in the binding free energy.
Hydrophobic interactions
The mutant and wild-type complexes contained the same composition of the hydrophobic residues at the interface (Fig. 2A) because the mutation was not concerned with hydrophobic residues. However, the mutation slightly affected the side-chain conformations of the residues, resulting in the decrease in the buried surface area (Supplemental Table 3). The averaged buried surface area of the mutant complex was found to be 1469 Å2, while that of the wild-type complex was found to be 1558 Å2. The low binding free energy of the mutation complex might be partially explained by this decrease (92 Å2) in the buried surface area.
Hydrophilic interactions
The wild-type complex had 15 intermolecular hydrogen bonds at the interface (Supplemental Table 4). Eleven of them that directly link bn to bs were commonly observed for the three wild-type complexes in the asymmetric unit, while the remaining four hydrogen bonds were observed only for the two wild-type complexes. Comparison between the mutant and wild-type complexes revealed that the mutant complex lacked six of the hydrogen bonds. However, it was not because of the simple decrease of the hydrogen bonds. Four hydrogen bonds were newly formed in the mutant complex, while 10 hydrogen bonds observed in the wild-type complex were lost in the mutant complex. Four of the 10 lost hydrogen bonds were related to the mutational site bs-Asp39 (Supplemental Table 4): bn-Arg83Nη2–bs-Asp39Oδ1, bn-Arg87Nη2–bs-Asp39Oδ2, bn-His102Nε2–bs-Asp39Oδ2, and bn-Tyr103Oη–Asp39Oδ1. The other hydrogen bonds seemed to result from structural rearrangement of the side chains of the residues distant from the mutational site. Since the individual structures of bn and bs were barely altered by the mutation, such rearrangement of the side-chain conformations might be due to the alterations in the orientation between bn and bs caused by the mutation, as mentioned earlier. On the other hand, five hydrogen bonds were not affected by the mutation: bn-Arg59N–bs-Asp35Oδ1, bn-Arg59Nη1–bs-Glu76Oδ2, bn-Glu60N-bs–Asp35Oδ2, bn-Glu60Oε2–bs-Leu34N, and bn-His102O–bs-Asn33Nδ2. According to molecular dynamic simulations, only five and nine of the hydrogen bonds listed in Supplemental Table 4 were stable in the mutant and wild-type complexes, respectively (Supplemental Tables 4, 6), suggesting the stable hydrogen bonds were formed more in the wild-type complex than in the mutant complex. Therefore, the decrease in the direct hydrogen bonds must be also correlated to destabilization of the bn–bs complex.
There were 27 water molecules at the wild-type interface detected by the crystallographic analysis (Supplemental Table 7; Fig. 2C): 16 water molecules were commonly observed in the three complexes in the asymmetric unit, while 11 were observed only in the two complexes. These in the wild-type complex as well as the mutant complex were classified into the three categories (“edge,” “buried,” and “bridging”): Seven, eight, and 12 water molecules were corresponding to the “edge,” “buried,” and “bridging” water molecules, respectively. Comparison between hydrations of the mutant and wild-type interfaces showed that the mutation increased the number of the “edge” and “buried” water molecules, while the number of the “bridging” water molecules was not affected by the mutation. The increase of the “edge” water molecules was probably mostly due to the quality of diffraction data improved from 2.3 Å to 1.58 Å (Table 1), and unlikely to affect the stability of the complex significantly, as suggested in the previous study (Ikura et al. 2004). On the other hand, the “buried” and “bridging” water molecules were expected to correlate with the stability of the complex. The mutation increased the number of the “buried” water molecules from eight to 11: Two and one more water molecules were bound only to bn and bs, respectively, in the mutant complex.
The mutation, however, changed not only the number of the “buried” water molecules but also the distribution of water molecules. Seven water molecules of the wild-type complex, H2O7, H2O9, H2O39, H2O61, H2O66, H2O104, and H2O145, in the second category retained their positions after the mutation as H2O94, H2O76, H2O24, H2O36, H2O231, H2O56, and H2O47 of the mutant complex, respectively (Supplemental Table 5; Fig. 2B,C), while the last water molecule of the wild-type complex (H2O45) obviously changed the positions after the mutation and formed additional hydrogen bonds with bn-Arg59 and bs-Trp38 as a “bridging” water molecule (H2O340) in the mutant complex (Supplemental Fig. 3A). Of the four “buried” water molecules actually added into the mutant complex, a water molecule (H2O26) was located at the position corresponding to the side-chain oxygen atom Oδ2 of bs-Asp39 of the wild-type complex (Supplemental Fig. 3B) and bound only to bn, suggesting that this water molecule filled the cavity formed by the deletion of the side chain of bs-Asp39 and mimicked the two interactions formed by bs-Asp39, namely, bn-Arg87Nη2–bs-Asp39Oδ2 and bn-His102Nε2–bs-Asp39Oδ2 (Supplemental Table 4; Supplemental Fig. 3B). Two of the four “buried” water molecules of the mutant complex, H2O3 and H2O234, were corresponding to the “bridging” water molecules of the wild-type complex, H2O127 and H2O126, respectively (Supplemental Table 5; Supplemental Fig. 3C,D), implying that the mutation indirectly broke two water-mediated hydrogen bonds. The last “buried” water molecule (H2O55) of the mutant complex was located in a shallow cavity on the rim of the interface, though no water molecule was observed in the corresponding position of the wild-type complex.
Although the mutation did not change the total number of the “bridging” water molecules, they were rearranged on a large scale (Supplemental Table 5; Supplemental Fig. 3A,C–E). Not surprisingly, the largest difference was observed at the mutational site bs-D39A, where the side-chain oxygen atoms Oδ1 and Oδ2 of bs-Asp39 interacted with bn-Lys27, bn-Glu73, bn-His102, and bn-Tyr103 through the water-mediated interactions (H2O77 and H2O126) in the wild type (Supplemental Table 5; Supplemental Fig. 3D,E). In addition to the interactions related to bs-Asp39, the mutation bs-D39A also disturbed the interactions related to the preceding residue bs-Trp38. bs-Trp38Nε1 moved away from the water molecule (H2O24 of the wild-type complex) bound to bn-Ile55O and bn-Glu73Oε2 and built new linkages with bn-Ile55O and bn-Arg59Nη2 through other water-mediated hydrogen bonds (H2O340 of the mutant complex) (Supplemental Fig. 3A). In summary, the cavity created by the mutation bs-D39A allowed at least three water molecules to intrude into the interface (the number of the “buried” and “bridging” water molecules increased by three molecules in total) and then caused rearrangement of the other water molecules, including rearrangement of the existing water molecules.
On the other hand, molecular dynamic simulations indicated that the total number of stable water molecules detected at the interface was not affected by the mutation (Supplemental Table 7). The attributes of water molecules, however, were completely different between the mutant and wild-type complexes (Supplemental Table 7; Supplemental Fig. 2): Two, five, and seven of the water molecules were corresponding to the “edge,” “buried,” and “bridging” water molecules, respectively, in the wild-type complex, whereas three, nine, and two were corresponding to the “edge,” “buried,” and “bridging” water molecules, respectively, in the mutant complex. In particular, the number of the “buried” and “bridging” water molecules were significantly different between the mutant and wild-type complexes: Five more “bridging” water molecules were detected in the wild-type complex than in the mutant complex, while four more “buried” water molecules were detected in the mutant complex than in the wild-type complex (Supplemental Table 7). Such a difference in the number of water molecules suggests that the wild-type complex was more stabilized by the water-mediated hydrogen bonds than the mutant complex.
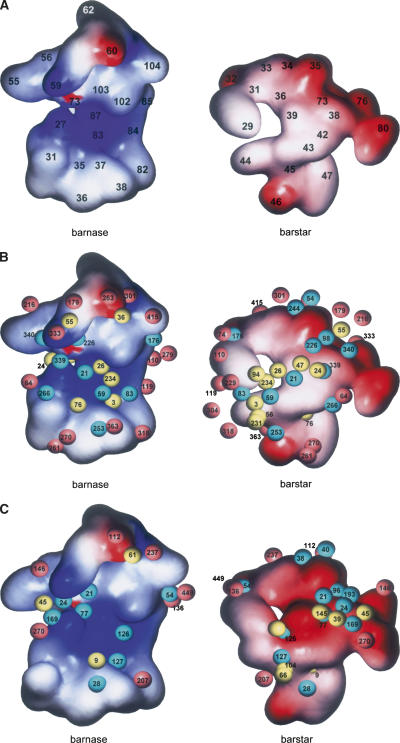
Regarding the lifetime of water-mediated hydrogen bonds, the water-mediated hydrogen bonds as well as the direct hydrogen bonds in general have rather long lifetimes if they were observed by the crystallographic analysis (Higo and Nakasako 2002; Ikura et al. 2004). In the present study, however, the water-mediated hydrogen bonds did not always succeed for a long time (500 ps) (Supplemental Tables 5, 7): Of the 12 “bridging” water molecules detected by the crystallographic analysis (Supplemental Table 5), only two water-mediated interactions were retained during >500 ps in the mutant complex, whereas seven interactions succeeded in the wild-type complex. This suggests that the hydrated water molecules have higher mobility inside the mutant complex than in the wild-type complex. Snapshots of the molecular dynamics simulations clearly showed that more water molecules penetrated into the mutant interface than the wild-type interface, though the total number of hydrated water molecules was similar between the mutant and wild-type complexes (Supplemental Fig. 2). They also showed that the penetrating water molecules formed a channel-like structure in the mutant complex, which suggested the flow of hydrated water molecules inside the interfaces (Fig. 3). Contrastingly, in the wild-type complex, bs-Asp39 stopped the flow of hydrated water molecules by means of the direct hydrogen-bonding network (Supplemental Table 4; Supplemental Fig. 2B). According to the analogy of the water channel, the water gate was closed in the wild-type complex, while it was opened in the mutant complex.
Figure 3.
The channel-like structure of hydrated water molecules observed in a 600-ps snapshot of the molecular dynamics simulation of the mutant complex. (Green) Molecular surface of bn, (pink) molecular surface of bs, (blue spheres) molecular surfaces of a part of hydrated water molecules (see Supplemental Fig. 2A).
In conclusion, the mutation bs-D39A drastically changed the hydrogen-bonding network at the interface, and then it significantly destabilized the hydrophilic interactions as well as the hydrophobic interactions. The change of the hydrogen-bonding network was mainly due to the rearrangement of hydrated water molecules, resulting in the channel-like structure. Such a channel-like structure of the hydrated water molecules must be closely connected with the destabilization of the mutant complex, although it might be the result and not the cause of the destabilization.
Comparison with other mutant complexes
In the previous study, we observed changes in the hydrogen-bond networks caused by the mutations bs-Asp35 → Ala (bs-D35A), bs-Glu76 → Ala (bs-E76A), and bs-Glu80 → Ala (bs-E80A) (Ikura et al. 2004). Glu76 and Glu80 of bs were located at the edge of the interface of the complex, and the mutations bs-E76A and bs-E80A hardly affected the hydrogen-bond network at the interfaces. Contrastingly, the Asp35 of bs was located at the interface and facilitated the hydrogen-bond network. The mutation bs-D35A accelerated the dissociation rate by 1000, corresponding to a decrease of 4.5 kcal·mol−1 in the binding free energy (Schreiber and Fersht 1995). According to the crystallographic analysis of the mutant complex bn (Q2A)–bs (D35A), an additional single water molecule was buried at the interface and mimicked the interaction formed by bs-Asp35, namely, bn-Arg59N–bs-Asp35Oδ1. No rearrangement of hydrated water molecules, however, was observed in this mutant complex. Although additional water molecules were also trapped at the interface in the other mutant complexes, namely, bn (H102A)–bs (Y29F), bn (K27A)–bs (D35A), and bn (K27A)–bs (T42A), these mutant complexes retained hydrogen-bond networks that were identical to that of the wild-type complex (Vaughan et al. 1999). To our knowledge, only the mutation bs-D39A drastically changed the hydrogen-bond network at the interface, suggesting that the side chain of this residue played the most important role for the hydrogen-bonding network.
Comparison with theoretical analysis of bn–bs interaction
There were several attempts to theoretically analyze the bn–bs interaction so far (Chong et al. 1998; Vijayakumar et al. 1998; Lee and Tidor 2001a,b; Dong et al. 2003; Wang et al. 2004; Spaar et al. 2006; Ababou et al. 2007). These theoretical approaches indicated that the interaction between bn and bs was mainly driven by electrostatic effects. In fact, the mutations on the charged residues, bn-Lys27, bn-His102, bs-Asp35, and bs-Asp39, significantly decreased the stability of the bn–bs complexes (Schreiber and Fersht 1995). Although earlier theoretical works were limited to only qualitative descriptions of the tight binding between bn and bs (Chong et al. 1998; Lee and Tidor 2001a,b), some of the recent works more quantitatively estimated the binding free energy and the change in the energy by mutations on the charged residues (Vijayakumar et al. 1998; Dong et al. 2003; Wang et al. 2004; Spaar et al. 2006; Ababou et al. 2007). At least three of these studies directly analyzed the present mutation, bs-D39A (Dong et al. 2003; Wang et al. 2004; Ababou et al. 2007). Dong et al. (2003) showed that the experimental binding stability of the bn–bs complexes was well described by the Poisson-Bolzmann model with the dielectric boundary specified as the van der Waals (vdW) surface of the protein. In this model, the effects of structural fluctuations and transient exposure to solvent were realized by using the dielectric map generated from the vdW surface. They estimated the changes in the energy by 17 mutations on the bn–bs complex, and obtained the successful results about the 11 mutations. The calculations for six single mutations including our mutation bs-D39A, however, revealed substantial discrepancies between the theoretical and experimental results. On the other hand, Wang et al. (2004) applied both the Poisson-Bolzmann electrostatic calculations and a semi-empirical method, COMBINE, to evaluate the binding energy of the mutant and wild-type bn–bs complexes. In the COMBINE analysis, the weighting parameters, which determine the contribution from each interaction energy term, were obtained by partial least-squares analysis of a training set consisting of the experimental data (Wang et al. 2004). When the wild-type complex and the 64 mutant complexes were analyzed by COMBINE, 54 of the complexes were precisely predicted. For the rest of the mutant complexes, including bs-D39A, bn-R59A/bs-D39A, and bn-K27A/bs-D39A, however, they were faced with large discrepancies between the experimental and predicted binding energies. Therefore, both results of the two studies suggest that the mutational effects of bs-D39A are difficult to estimate. If the rearrangement of the hydrogen-bonding network caused by the mutation bs-D39A is taken into consideration, their methods might be improved.
Last year, Ababou et al. (2007) applied a semi-empirical divide-and-conquer methodology coupled with a dielectric continuum model to determine the contribution of electrostatic, polarization (POL), and charge transfer (CT) to the bn–bs interactions. Molecular dynamic simulation was also performed to account for the dynamic behavior of the complex. They then determined the change in the binding energy by 13 mutations, including bs-D39A. According to the investigators, their theoretical results were in good agreement with the experimental results, except for several specific mutations. However, since both the POL and CT effects could not be directly compared with the experimental binding energies, these were scaled by the appropriate factors, without a scientific basis. Therefore, whether or not the theoretical results show good agreement with the experimental data depends absolutely on the scaling factors.
Furthermore, Ababou et al. (2007) concluded that water molecules at the interface contributed little to the binding energy of the bn–bs complex by analyzing the POL and CT effects with and without six buried water molecules. As shown in the present study, however, the contribution of the water-mediated hydrogen bonds to the stability of the bn–bs complex is dependent on both the position of the hydrogen bond and the kind of mutation (Supplemental Tables 5, 7). It must be too early to neglect the effects of water-mediated interaction between bn and bs at this stage of theoretical approaches.
Conclusions
Both the crystallographic analysis and molecular dynamics simulations indicate that the mutation bs-D39A significantly decreased the hydrophobic and hydrophilic interactions between bn and bs. Drastic changes of the hydrophilic interactions by the mutation were observed in the water-mediated hydrogen bonds as well as the direct hydrogen bonds. Several water molecules penetrated the cavity created by the mutation, caused the rearrangement of hydrated water molecules at the interface, and then redistributed the water molecules into a channel-like structure that penetrated through the interface (Fig. 3). The significant destabilization of the interaction between bn and bs may be due to this channel-like structure of hydrated water molecules.
Materials and Methods
Mutation, protein expression, and purification
The expression plasmids of wild-type bn and the double-mutant bs C40/82A were gifts from Professor Sir A.R. Fersht (University of Cambridge, United Kingdom). Since the mutant bs C40/82A is regarded as the reduced form of wild-type bs, the complex between wild-type bn and bs C40/82A has been analyzed as the wild-type complex in a number of studies (Schreiber and Fersht 1993a,b, 1995; Buckle et al. 1994; Schreiber et al. 1994, 1997; Vaughan et al. 1999; Frisch et al. 2001; Ikura et al. 2004). Thus, we followed such convention in the present study.
Initially, we attempted to crystallize the complex formed by wild-type bn and mutant bs D39A/C40A/C82A by widely screening the crystallizing conditions. Crystals of this complex, however, were never obtained, probably because the low binding constant of the complex dissociated the mutant bs from bn under any crystallizing conditions, including the conditions used for crystallizing the other mutant complexes. Therefore, we attempted to find more moderate crystallizing conditions by changing the crystal packing observed in the crystals of the other mutant complexes. Based on the known crystal structure of bn–bs complexes, we designed a series of single mutants that would disturb the crystal packing but not affect the complex formation, in the hope that the mutant complex crystallized in a different crystal form. Among those mutants, the mutant bn-K98A produced well-diffraction crystals without changing its function in terms of the binding to bs: Surface plasmon resonance measurements showed that the equilibrium dissociation constant (K d) of this mutant bn with bs C40A/C82A was 1.4 × 10−10 M, similar to the K d of the wild-type complex, 1.3 × 10−10 M (Ikura et al. 2004). The crystals made it possible for the first time to analyze the bs-D39A mutant complex in detail. Site-directed mutagenesis was performed for bn and the double-mutant bs by using QuikChange (Stratagene). The expression and purification of the proteins have been described elsewhere (Serrano et al. 1990; Schreiber and Fersht 1993b). Further, the mutation D39A/C40A/C82A on this double-mutant bs was not described as a triple mutation, but as a single mutation D39A in order to avoid verbiage.
Crystallization and structure determination
The crystals used for the X-ray diffraction studies were grown at 20°C by vapor diffusion in the hanging-drop configuration. Protein concentrations were 1.0 mM for the complexes formed by a 1:1 mixture of bn and bs, and 1 or 2 μL of the protein solution was mixed with an equal volume of an equilibrating buffer in a well in a sealed chamber containing 500 μL of the equilibrating buffer. The equilibrating buffer was 0.1 M 2-[4-(2-Hydroxyethyl)-1-piperadinyl] ethansulfonic acid (HEPES) (pH 7.0) containing 20% polyethylene glycol (PEG) 6000, and 1.0 M lithium chloride. The crystals were flash-frozen using mother liquor supplemented with 20% glycerol. Diffraction data for the mutant complex were collected at beamline BL-6A at the Photon Factory and were integrated and scaled with HKL2000 (HKL Research, Inc.). The space group is C2 with unit cell dimensions a = 97.6 Å, b = 110.3 Å, c = 47.3 Å, and β = 115.0°, with two complexes per asymmetrical unit. The structure was solved by molecular replacement by using the crystal structure of the wild-type complex reported by Buckle et al. (1994) (PDB code: 1BRS) as the search model in CNS (Brünger et al. 1998). Refinement was performed with CNS and XtalView (McRee 1999).
Definition of hydrogen bond
The traditional definition of the hydrogen bond (Pauling et al. 1951) may be revised by recent database analysis on hydration for biomolecules (HHDB) (Niimura et al. 2004). According to the database, we applied the following definition of the hydrogen bond in the present study: The hydrogen bond between the functional groups of proteins was defined with distance and angle between donor and acceptor,
2.3 Å < distance between donor and acceptor < 3.6 Å, and
120° < angle N–O=C, etc., < 180°,
while the hydrogen bond between protein and water was defined only with the distance. Since no hydrogen bonds searched by the above criteria had less than the distance of 2.5 Å, the lower limit of 2.5 Å gives the same results.
Categories of hydrating water molecules
Many hydrating water molecules were found at the bn–bs interface of the present mutant complex as well as the wild-type and other mutant complexes. These water molecules were suggested to contribute to the bn–bs interactions in the manner as described in the wild-type complex (Buckle et al. 1994). Therefore, according to Buckle et al. (1994), water molecules hydrating to the bn–bs interface were classified into three categories: (1) water molecules bound to the edge of the interface, (2) water molecules buried in the surface cavity and bound to either bn or bs, and (3) water molecules mediating the interactions between both proteins through hydrogen bonds of bn–H2O–bs. In this definition, water molecules in the first category were hydrogen bonding to either bn or bs, and their solvent-accessible surface area (ASA) was >10 Å2, whereas water molecules in the second category were also hydrogen bonding to either bn or bs, but their ASA was <10 Å2. On the other hand, water molecules in the third category were hydrogen bonding to both bn and bs irrespective of their ASA. In this study, we described the water molecules in the first, second, and third categories as the “edge,” “buried,” and “bridging” water molecules, respectively, in order to avoid verbiage.
Data deposition
The coordinates have been deposited in the Brookhaven Protein Data Bank with accession number 2ZA4.
Acknowledgments
We thank Professor Sir A.R. Fersht for providing the expression plasmids of bn and bs. We thank Professors K. Kuwajima, M. Nakasako, and T. Azuma for permitting the use of experimental facilities. We thank Professor A. Kitao and Dr. Y. Joti for permitting the use of the high-performance parallel computer.
Footnotes
Supplemental material: see www.proteinscience.org
Reprint requests to: Nobutoshi Ito, Laboratory of Structural Biology, School of Biomedical Science, Tokyo Medical and Dental University, 1-5-45 Yushima, Bunkyo-ku, Tokyo 113-8510, Japan; e-mail: ito.str@tmd.ac.jp; fax: 81-3-5803-4594.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.073322508.
References
- Ababou, A., van der Vaart, A., Gogonea, V., Merz K.M., Jr Interaction energy decomposition in protein–protein association: A quantum mechanical study of barnase–barstar complex. Biophys. Chem. 2007;125:221–236. doi: 10.1016/j.bpc.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Buckle, A.M., Schreiber, G., Fersht, A.R. Protein–protein recognition: Crystal structural analysis of a barnase–barstar complex at 2.0 Å resolution. Biochemistry. 1994;33:8878–8889. doi: 10.1021/bi00196a004. [DOI] [PubMed] [Google Scholar]
- Bycroft, M., Ludvigsen, S., Fersht, A.R., Poulsen, F.M. Determination of the three-dimensional solution structure of barnase using nuclear magnetic resonance spectroscopy. Biochemistry. 1991;30:8697–8701. doi: 10.1021/bi00099a030. [DOI] [PubMed] [Google Scholar]
- Chen, R., Mintseris, J., Janin, J., Weng, Z. A protein–protein docking benchmark. Proteins. 2003;52:88–91. doi: 10.1002/prot.10390. [DOI] [PubMed] [Google Scholar]
- Chong, L.T., Dempster, S.E., Hendsch, Z.S., Lee, L.P., Tidor, B. Computation of electrostatic complements to proteins: A case of charge stabilized binding. Protein Sci. 1998;7:206–210. doi: 10.1002/pro.5560070122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covell, D.G., Wallqvist, A. Analysis of protein–protein interactions and the effects of amino acid mutations on their energetics. The importance of water molecules in the binding epitope. J. Mol. Biol. 1997;269:281–297. doi: 10.1006/jmbi.1997.1028. [DOI] [PubMed] [Google Scholar]
- Dong, F., Vijayakumar, M., Zhou, H.X. Comparison of calculation and experiment implicates significant electrostatic contributions to the binding stability of barnase and barstar. Biophys. J. 2003;85:49–60. doi: 10.1016/S0006-3495(03)74453-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht, A.R., Serrano, L. Principles of protein stability derived from protein engineering experiments. Curr. Opin. Struct. Biol. 1993;3:75–83. [Google Scholar]
- Frisch, C., Fersht, A.R., Schreiber, G. Experimental assignment of the structure of the transition state for the association of barnase and barstar. J. Mol. Biol. 2001;308:69–77. doi: 10.1006/jmbi.2001.4577. [DOI] [PubMed] [Google Scholar]
- Guillet, V., Lapthorn, A., Fourniat, J., Benoit, J.P., Hartley, R.W., Mauguen, Y. Crystallization and preliminary X-ray investigation of barstar, the intracellular inhibitor of barnase. Proteins. 1993a;17:325–328. doi: 10.1002/prot.340170309. [DOI] [PubMed] [Google Scholar]
- Guillet, V., Lapthorn, A., Hartley, R.W., Mauguen, Y. Recognition between a bacterial ribonuclease, barnase, and its natural inhibitor, barstar. Structure. 1993b;1:165–176. doi: 10.1016/0969-2126(93)90018-c. [DOI] [PubMed] [Google Scholar]
- Hartley, R.W. Directed mutagenesis and barnase–barstar recognition. Biochemistry. 1993;32:5978–5984. doi: 10.1021/bi00074a008. [DOI] [PubMed] [Google Scholar]
- Higo, J., Nakasako, M. Hydration structure of human lysozyme investigated by molecular dynamics simulation and cryogenic X-ray crystal structure analyses: On the correlation between crystal water sites, solvent density, and solvent dipole. J. Comput. Chem. 2002;23:1323–1336. doi: 10.1002/jcc.10100. [DOI] [PubMed] [Google Scholar]
- Ikura, T., Urakubo, Y., Ito, N. Water-mediated interaction at a protein–protein interface. Chem. Phys. 2004;307:111–119. [Google Scholar]
- Janin, J. Wet and dry interfaces: The role of solvent in protein–protein and protein–DNA recognition. Structure. 1999;7:R277–R279. doi: 10.1016/S0969-2126(00)88333-1. [DOI] [PubMed] [Google Scholar]
- Janin, J., Rodier, F. Protein–protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- Kondo, H., Shiroishi, M., Matsushima, M., Tsumoto, K., Kumagai, I. Crystal structure of anti-Hen egg white lysozyme antibody (HyHEL-10) Fv-antigen complex. Local structural changes in the protein antigen and water-mediated interactions of Fv-antigen and light chain–heavy chain interfaces. J. Biol. Chem. 1999;274:27623–27631. doi: 10.1074/jbc.274.39.27623. [DOI] [PubMed] [Google Scholar]
- Lee, L.P., Tidor, B. Barstar is electrostatically optimized for tight binding to barnase. Nat. Struct. Biol. 2001a;8:73–76. doi: 10.1038/83082. [DOI] [PubMed] [Google Scholar]
- Lee, L.P., Tidor, B. Optimization of binding electrostatics: Charge complementarity in the barnase–barstar protein complex. Protein Sci. 2001b;10:362–377. doi: 10.1110/ps.40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte, L., Chothia, C., Janin, J. The atomic structure of protein–protein recognition sites. J. Mol. Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Lubienski, M.J., Bycroft, M., Jones, D.N., Fersht, A.R. Assignment of the backbone 1H and 15N NMR resonances and secondary structure characterization of barstar. FEBS Lett. 1993;332:81–87. doi: 10.1016/0014-5793(93)80489-h. [DOI] [PubMed] [Google Scholar]
- Matouschek, A., Kellis J.T., Jr, Serrano, L., Bycroft, M., Fersht, A.R. Transient folding intermediates characterized by protein engineering. Nature. 1990;346:440–445. doi: 10.1038/346440a0. [DOI] [PubMed] [Google Scholar]
- Mauguen, Y., Hartley, R.W., Dodson, E.J., Dodson, G.G., Bricogne, G., Chothia, C., Jack, A. Molecular structure of a new family of ribonucleases. Nature. 1982;297:162–164. doi: 10.1038/297162a0. [DOI] [PubMed] [Google Scholar]
- McRee, D.E. XtalView/Xfit—a versatile program for manipulating atomic coordinates and electron density. J. Struct. Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- Mintseris, J., Wiehe, K., Pierce, B., Anderson, R., Chen, R., Janin, J., Weng, Z. Protein–protein docking benchmark 2.0: An update. Proteins. 2005;60:214–216. doi: 10.1002/prot.20560. [DOI] [PubMed] [Google Scholar]
- Niimura, N., Chatake, T., Kurihara, K., Maeda, M. Hydrogen and hydration in proteins. Cell Biochem. Biophys. 2004;40:351–369. doi: 10.1385/CBB:40:3:351. [DOI] [PubMed] [Google Scholar]
- Nolting, B., Golbik, R., Fersht, A.R. Submillisecond events in protein folding. Proc. Natl. Acad. Sci. 1995;92:10668–10672. doi: 10.1073/pnas.92.23.10668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolting, B., Golbik, R., Neira, J.L., Soler-Gonzalez, A.S., Schreiber, G., Fersht, A.R. The folding pathway of a protein at high resolution from microseconds to seconds. Proc. Natl. Acad. Sci. 1997;94:826–830. doi: 10.1073/pnas.94.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauling, L., Corey, R.B., Branson, H.R. The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. 1951;37:205–211. doi: 10.1073/pnas.37.4.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrey, D., Honig, B. GRASP2: Visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- Ratnaparkhi, G.S., Ramachandran, S., Udgaonkar, J.B., Varadarajan, R. Discrepancies between the NMR and X-ray structures of uncomplexed barstar: Analysis suggests that packing densities of protein structures determined by NMR are unreliable. Biochemistry. 1998;37:6958–6966. doi: 10.1021/bi972857n. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Fersht, A.R. Interaction of barnase with its polypeptide inhibitor barstar studied by protein engineering. Biochemistry. 1993a;32:5145–5150. doi: 10.1021/bi00070a025. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Fersht, A.R. The refolding of cis- and trans-peptidylprolyl isomers of barstar. Biochemistry. 1993b;32:11195–11203. doi: 10.1021/bi00092a032. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Fersht, A.R. Energetics of protein–protein interactions: Analysis of the barnase–barstar interface by single mutations and double-mutant cycles. J. Mol. Biol. 1995;248:478–486. doi: 10.1016/s0022-2836(95)80064-6. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Buckle, A.M., Fersht, A.R. Stability and function: Two constraints in the evolution of barstar and other proteins. Structure. 1994;2:945–951. doi: 10.1016/s0969-2126(94)00096-4. [DOI] [PubMed] [Google Scholar]
- Schreiber, G., Frisch, C., Fersht, A.R. The role of Glu73 of barnase in catalysis and the binding of barstar. J. Mol. Biol. 1997;270:111–122. doi: 10.1006/jmbi.1997.1080. [DOI] [PubMed] [Google Scholar]
- Serrano, L., Horovitz, A., Avron, B., Bycroft, M., Fersht, A.R. Estimating the contribution of engineered surface electrostatic interactions to protein stability by using double-mutant cycles. Biochemistry. 1990;29:9343–9352. doi: 10.1021/bi00492a006. [DOI] [PubMed] [Google Scholar]
- Spaar, A., Dammer, C., Gabdoulline, R.R., Wade, R.C., Helms, V. Diffusional encounter of barnase and barstar. Biophys. J. 2006;90:1913–1924. doi: 10.1529/biophysj.105.075507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan, C.K., Buckle, A.M., Fersht, A.R. Structural response to mutation at a protein–protein interface. J. Mol. Biol. 1999;286:1487–1506. doi: 10.1006/jmbi.1998.2559. [DOI] [PubMed] [Google Scholar]
- Vijayakumar, M., Wong, K.Y., Schreiber, G., Fersht, A.R., Szabo, A., Zhou, H.X. Electrostatic enhancement of diffusion-controlled protein–protein association: Comparison of theory and experiment on barnase and barstar. J. Mol. Biol. 1998;278:1015–1024. doi: 10.1006/jmbi.1998.1747. [DOI] [PubMed] [Google Scholar]
- Wang, T., Tomic, S., Gabdoulline, R.R., Wade, R.C. How optimal are the binding energetics of barnase and barstar? Biophys. J. 2004;87:1618–1630. doi: 10.1529/biophysj.104.040964. [DOI] [PMC free article] [PubMed] [Google Scholar]