Abstract
Amylosucrase is a transglucosidase that catalyzes amylose-like polymer synthesis from sucrose substrate. About 60,000 amylosucrase variants from two libraries generated by the MutaGen random mutagenesis method were submitted to an in vivo selection procedure leading to the isolation of more than 7000 active variants. These clones were then screened for increased thermostability using an automated screening process. This experiment yielded three improved variants (two double mutants and one single mutant) showing 3.5- to 10-fold increased half-lives at 50°C compared to the wild-type enzyme. Structural analysis revealed that the main differences between wild-type amylosucrase and the most improved variant (R20C/A451T) might reside in the reorganization of salt bridges involving the surface residue R20 and the introduction of a hydrogen-bonding interaction between T451 of the B′ domain and D488 of flexible loop 8. This double mutant is the most thermostable amylosucrase known to date and the only one usable at 50°C. At this temperature, amylose synthesis by this variant using high sucrose concentration (600 mM) led to the production of amylose chains twice as long as those obtained by the wild-type enzyme at 30°C.
Keywords: amylosucrase, directed evolution, random mutagenesis, high-throughput screening, thermostability, amylose synthesis
Naturally available enzymes are usually not optimally suited for industrial applications. This incompatibility is often related to their lack of stability under process conditions. The use of high reaction temperatures may be advantageous regarding parameters like substrate and product solubility, viscosity, process speed, and microbial contamination. Moreover, high stability is often considered as an economic advantage to reduce the enzyme quantity used in industrial processes. For the past two decades, many methodologies describing protein thermostability improvement through the introduction of mutations have been reported, including comparison of naturally occurring homologous proteins, rational design, and directed evolution (Lehmann and Wyss 2001; van den Burg and Eijsink 2002; Eijsink et al. 2005). Sequence or structure comparisons of mesophilic enzymes with thermophilic relatives have indeed shown to be efficient approaches to select thermostabilizing mutations (Haney et al. 1999; Perl et al. 2000; Miyazaki et al. 2001; Hytonen et al. 2005; Ditursi et al. 2006). However, numerous protein sequences or reliable structural data are needed for this strategy to be effective. Other rational strategies for stability engineering based on tertiary-structure analysis usually aim at improving the packing of the hydrophobic core, extending networks of salt bridges and hydrogen bonds that will favorably enhance the enthalpic contribution or reduce the entropy of the unfolded state through salt or disulfide bridges, glycine substitutions, and introduction of prolines in turns (Vieille and Zeikus 2001; Eijsink et al. 2004). However, no general predictive rules enabling the design of proteins with increased stability can be proposed despite numerous successful experimental reports. Elucidating the structural basis of thermostability for a given protein and finding ways to improve it often appears as a specific task depending strongly on the structural context.
Directed evolution appears to be an efficient complementary approach to rational engineering. This methodology consists of using random mutagenesis and recombination methods to construct large libraries of protein variants, which are subsequently selected or screened for the desired trait. The identification of amino acids involved in specific functional features is thus allowed through the exploration of the entire protein sequence without a priori knowledge of its structure–activity relationships (Petrounia and Arnold 2000; Bloom et al. 2005). This strategy has shown to be a powerful engineering approach to tailor the properties of enzymes for a variety of industrial needs, such as increased thermostability, activity in organic solvents, and altered substrate specificity (Yuan et al. 2005). Noteworthy, is that the outcome of directed evolution experiments is critically dependent on how libraries are screened or selected. Therefore, use of this approach is mainly limited by the development of reliable high-throughput selection and screening systems enabling the isolation of the best mutant(s) among the available diversity.
Amylosucrase (AS) from Neisseria polysaccharea (EC 2.4.1.4) is a glucansucrase from the glycoside hydrolase (GH) family 13 that catalyzes the de novo synthesis of a water-insoluble amylose-like polymer from sucrose, a readily available and cheap agro-resource (Potocki de Montalk et al. 1999, 2000). Notably, the linear α-1,4 chains formed during the reaction precipitate into semicrystalline networks or aggregate when reaching critical concentration and length and then cannot be further elongated. At 30°C, the control of amylose chain precipitation is nicely monitored by the sucrose initial concentration, and this enables production of amylose with different morphologies and sizes (Potocki-Véronèse et al. 2005). Due to the availability and low cost of sucrose, AS is an attractive biocatalyst for amylose-like polymer synthesis. However, the development of industrial processes involving AS is limited by its low catalytic efficiency on sucrose alone (k cat = 1 s−1) and thermostability (t 1/2[50°C] = 3 min). Further application developments thus clearly require adapting AS to harsher reaction conditions. Rational improvement of these properties would benefit from comparisons with homologous proteins displaying different stability and activity characteristics. Nevertheless, such an approach is not possible in the case of AS since the only other described amylosucrase, isolated from mesophilic microorganism Deinococcus radiodurans, possesses similar stability and activity properties (Pizzut-Serin et al. 2005). Directed evolution techniques have already allowed the isolation of mutants displaying 1.5- to 4.2-fold increased activity compared to wild-type AS (van der Veen et al. 2004, 2006).
The present study describes the improvement of AS thermostability using directed evolution. We previously reported the optimization of a two-step in vivo selection and automated screening process enabling the search for AS variants with increased thermal stability (Emond et al. 2007). In the present study, we applied this procedure to screen two different AS mutant libraries generated by MutaGen, an original random mutagenesis technique based on the use of low-fidelity DNA polymerases (Bouayadi et al. 2002; Mondon et al. 2007; Emond et al. 2008). This work led to the isolation of three improved AS variants. Their properties are described and discussed further.
Results and Discussion
Isolation of thermostable amylosucrase variants
The wild-type AS whole encoding sequence was employed as a template to introduce genetic diversity using the MutaGen technique (Emond et al. 2008). This approach consists in generating random mutations by replicating the parental sequence with human error-prone DNA polymerases. Two AS variant libraries, named A and B, were constructed by utilizing two different polymerases, DNA polymerases β and η, respectively. Cloning and transformation of the mutagenized products to Escherichia coli TOP10 yielded ∼8 × 105 clones for library A and 1 × 105 for library B. Plasmid DNA isolated from these clones constituted the storage form of the libraries. The base mutation rates, determined through DNA sequencing of randomly chosen clones, were 1.7 and 10.9 mutations per kb for libraries A and B, respectively. These values were shown to be related to the amounts of active AS variants, which were 75% for library A and 20% for library B.
Transformation of library A to E. coli JM109 cells yielded 30,000 colonies, which were then subjected to selection with sucrose as the sole carbon source. Approximately 4500 active AS-expressing clones (47 microplates) were picked from these selective plates to inoculate small-volume cultures in a 96-well format that were stored at −20°C after growth. This library of individualized active AS variants was then screened for increased thermostability after a heat-treatment step at 50°C for 20 min. The same procedure was followed to isolate and screen 2700 active AS-expressing clones from library B.
Among the variants screened, two clones from library A and one from library B were found to be more thermostable compared to the wild-type AS and were thus retained for more-detailed characterization. The sequencing of the three selected variants revealed the mutations listed in Table 1. Both variants 1 and 2, isolated from library A, are double-mutants R20C/A451T and A170V/Q353L, respectively; each of them also contained an additional silent mutation. Mutant 3 isolated from library B is a single mutant, P351S, enduring three silent mutations.
Table 1.
Nucleotide substitutions and resulting amino acid replacements of the variants selected after screening for thermostability
Thermal resistance of wild-type and variant amylosucrases
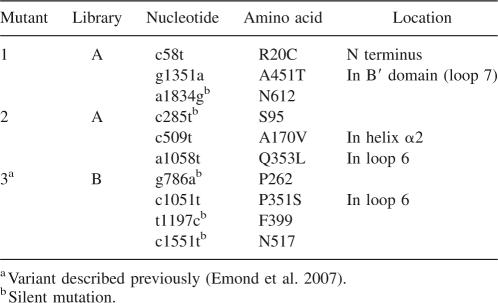
The selected AS variants were purified to electrophoretic homogeneity for further comparison with the wild-type enzyme. The stability of wild-type AS and selected variants were assessed by measuring their half-lives at 50°C (Fig. 1A). To determine their temperature dependency, initial specific activities were measured over a range of temperatures from 30°C to 55°C (Fig. 1B).
Figure 1.
Thermostability of AS and its variants. (A) Thermal stability of wild-type AS and variants R20C/A451T, A170V/Q353L, P351S, and A451T at 50°C. Wild-type and variant enzymes were incubated at 50°C, and at regular time intervals. Samples were removed and assayed for AS activity at 37°C. (B) Temperature dependency of wild-type AS and R20C/A451T, A170V/Q353L, P351S, and A451T variants. Specific activities were measured over a range of temperatures from 30°C to 55°C with 146 mM sucrose in 50 mM Tris-HCl pH 7.0.
All variants isolated during screening exhibited higher half-lives at 50°C than their wild-type parent (Fig. 1A; Table 2). Whereas the wild-type enzyme denatured very rapidly (t 1/2 = 3 min; Table 2), R20C/A451T, A170V/Q353L, and P351S mutants displayed half-lives of 32, 10, and 24 min, respectively (Fig. 1A; Table 2). As shown in Figure 1B, the activity–temperature profile of the double-mutants R20C/A451T and A170V/Q353L was broadened in comparison to the wild-type enzyme. Stabilization of these variants was accompanied by an increase in the optimal temperature (40°C vs. 36°C for wild-type AS; Fig. 1B; Table 2), and their activity between 40°C and 50°C was 1.5–2.5 times higher than that of wild-type AS (Fig. 1B). Although no significant increase in optimal temperature was observed for the P351S mutant (Table 2), its specific activity between 45°C and 55°C was higher than that of the wild-type enzyme (Fig. 1B), thus confirming the gain of thermostability.
Table 2.
Stability properties of wild-type AS and its variants
Double-mutant R20C/A451T was the most stable enzyme at 50°C identified in this study (Table 2). Interestingly, a variant displaying the R20C substitution combined with the F598S mutation was previously isolated during a screen for more active AS variants (van der Veen et al. 2004, 2006). To analyze the contribution of the A451T substitution to the improvement of AS thermostability, we constructed the A451T single mutant; the resulting enzyme exhibited a half-life of 17 min at 50°C. In conclusion, the combination of the R20C and A451T substitutions clearly had a beneficial effect on the enzyme stability.
Structural basis of thermostabilization mechanisms
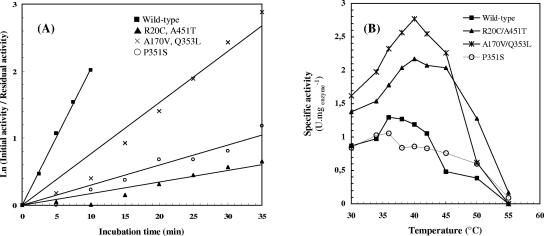
AS is a (β/α)8-barrel enzyme, which possesses the three common domains found in GH family 13: the characteristic barrel–catalytic A domain, a B domain between β-strand 3 and α-helix 3, and a C-terminal Greek key domain. Beside these common features, AS structure also contains two unique additional domains, namely, the N-terminal α-helical domain and a B′ domain between β-strand 7 and α-strand 7 of the catalytic barrel (Skov et al. 2001). The five mutations observed in the three thermostable variants isolated from screening are scattered all over the protein structure (Fig. 2; Table 1): three are located in the catalytic A domain (A170V, P351S, Q353L), one in the specific B′ domain (A451T), and another one in the N-terminal domain (R20C). Among these five point mutations, two of them (R20C and A170V) are found on the protein surface, while the three remaining ones (P351S, Q353L, and A451T) are located inside the enzyme structure but do not affect the catalytically essential residues of AS previously identified (Sarcabal et al. 2000). Most of the exchanges observed are located both in α-helices and in loops that connect β-strands with α-helices of the catalytic barrel, and none of them could have been predicted by rational analysis. To analyze the contribution of these newly introduced residues to AS thermostabilization, the three variants identified during screening were constructed in silico using the wild-type enzyme structure as a template. Generated 3D models of variants were subsequently subjected to an energy-minimization procedure. Although extensive molecular dynamics simulations would be needed to investigate thoroughly the protein conformation changes induced upon mutation, energy minimization could still provide some hints regarding the possible structural determinants likely to play a role in AS stabilization.
Figure 2.
Overview of mutation sites affecting amylosucrase thermostability. Secondary-structure elements are rendered in cartoon. Crystallographic maltoheptaose molecule is shown for reference, colored in yellow in AS to highlight the position of the catalytic pocket (PDB 1MW0).
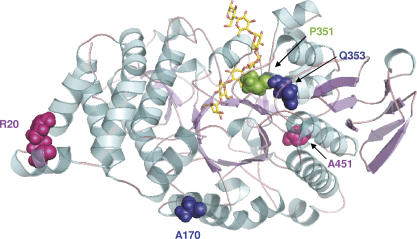
Double-mutant R20C/A451T was the most thermostable variant. Wild-type AS structural analysis revealed that R20 is a surface residue of the N domain, potentially involved in a salt-bridge interaction with both D13 and E24 (Fig. 3A). The electrostatic contribution to the free-energy change upon such salt-bridge formation has been shown to vary significantly, from being destabilizing to stabilizing to the protein structure, depending on their geometry location in the protein (Hendsch and Tidor 1994; Kumar and Nussinov 1999; Makhatadze et al. 2003). The geometric positioning of the interacting side chains in the salt bridges with respect to each other is a very important determinant of salt-bridge stability. Analysis of wild-type AS structure reveals that a network of salt bridges is likely to occur between R20–E24, R20–D13, and R10–D13 (Fig. 3A). On the other hand, average residue B factors (Reetz et al. 2006) also indicate that these surface residues belong to a poorly ordered region (B factors > 30), possibly indicating a high mobility. Examination of the variant-minimized structure indicates that the R20C substitution disrupts the D13–R20–E24 salt bridge observed in wild-type AS (Fig. 3B). This event is accompanied by a reorientation of D13 and E24 side chains, which can then form additional salt bridges with R10 and K25, respectively (Fig. 3B). The second residue mutated in the R20C/A451T variant is located at the end of the B′ domain. From visual inspection of the minimized R20C/A451T mutant structure, stabilization is proposed to be due to a favorable enthalpic hydrogen-bonding interaction of the introduced threonine side chain (A451T) with the Asp488 residue located in loop 8 of the catalytic core (Fig. 3C,D).
Figure 3.
Comparison between wild-type AS (A,C) and double-mutant R20C/A451T (B,D).
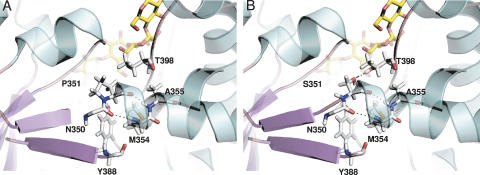
The second most thermostable variant, P351S, displays a half-life at 50°C that is increased by eightfold compared to wild-type AS, which is then of about same order as the R20C/A451T mutant. The drastic effect on thermostability of such a mutation at the loop connecting β-strand 6 with α-helix 6 of the catalytic barrel could increase the loop flexibility at this strategic region, which might then result in an entropic penalty. However, this negative contribution to the free-energy change might be compensated by a favorable enthalpic hydrogen-bonding interaction between the introduced serine residue and T398 of the catalytic barrel, which could also help to stabilize the protein core (Fig. 4A,B).
Figure 4.
Comparison between wild-type AS (A) and single-mutant P351S (B). For illustrative purposes, the maltoheptaose molecule from the 1MW0 structure is shown for reference (colored in yellow in the AS catalytic pocket).
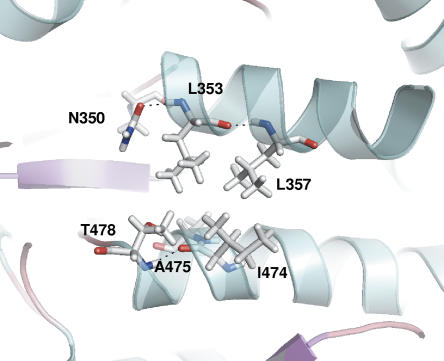
The double-mutant A170V/Q353L possesses a 3.5-fold increased half-life at 50°C compared to wild-type AS. Both mutations correspond to substitutions by more bulky hydrophobic side-chain residues. While A170 is a surface residue exposed to solvent of the catalytic barrel α-helix 2, Q353 belongs to loop 6 of the catalytic domain A and is located two positions away in the sequence from P351. Given that single mutants have not been constructed or characterized, it is difficult to assess the effect of individual mutations. Nonetheless, Figure 5 suggests that L353 is nicely embedded in a hydrophobic protein core, and such hydrophobic packing with nearby hydrophobic L357, A475, and I474 has been shown to be a major stabilizing factor for thermophiles (Vieille and Zeikus 2001).
Figure 5.
Representation of the hydrophobic region surrounding the Q353L mutation in double-mutant A170V/Q353L.
Amylose synthesis by thermostable amylosucrase variants
Amylose productions by the wild-type and the two most thermostable variants, namely the R20C/A451T and P351S mutants (Table 2), were carried out over a 24-h period using either 100 or 600 mM sucrose at two different reaction temperatures, 30°C or 50°C.
When using the P351S mutant at 30°C and 50°C, the amount of produced amylose was insufficient to visualize any precipitate in the final reaction media and to complex iodine. Therefore, this mutant, which appeared to be prone to aggregation in long-term reactions, cannot be considered as exploitable for amylose synthesis.
HPAEC-PAD analysis of the reaction products showed that the wild-type AS and the R20C/A451T double mutant produced the same compounds from 100 and 600 mM initial sucrose, namely, glucose, fructose, turanose, trehalulose, malto-oligosaccharides of increasing size, and amylose. This indicates that mutations did not alter the enzyme specificity.
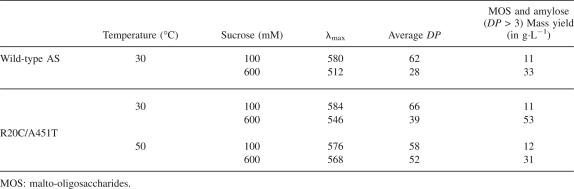
At 30°C, the average degree of polymerization (DP) of the amylose formed was dependent on the sucrose concentration for both the wild-type and the mutant enzyme, the average chain length decreasing with the increase of sucrose concentration, as previously shown (Potocki-Véronèse et al. 2005). However, from 600 mM sucrose, the amylose production yield of the mutant (53 g/L) increased significantly compared to that of the wild type (33 g/L). Due to its best thermostability and specific activity at 30°C, the mutant was able to consume more sucrose than was the wild type after 24 h reaction (Table 3).
Table 3.
Analysis of products synthesized by wild-type AS and R20C/A451T variant
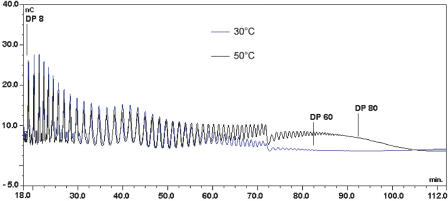
At 50°C, the wild-type enzyme was not able to consume any sucrose because of its poor thermostability. Indeed, the R20C/A451T mutant is the only amylosucrase that remains functional at this temperature. The effect of sucrose concentration on amylose chain length was less pronounced than that observed at 30°C. Amylose (DP > 3) with an average DP of 58 and 52 was synthesized from 100 and 600 mM sucrose, respectively (Table 3). Notably, from 600 mM sucrose, we observed that the average chain length of the products formed by the mutant increased with temperature. As shown in Figure 6, the longest α-1,4 chain synthesized at 30°C and visualized with HPAEC profiles corresponded to a DP of 61, while synthesis performed at 50°C yielded longer and more concentrated chains.
Figure 6.
HPAEC profile of the amylose chains synthesized by the R20C/A451T amylosucrase variant after a 24-h reaction in 600 mM initial sucrose at 30°C (blue) or 50°C (black).
We have previously shown that the amylose produced from 100 mM sucrose at 30°C by the wild-type AS is organized in semicrystalline networks (Potocki-Véronèse et al. 2005). Conversely, amylose synthesized from 600 mM sucrose forms 94% type-B crystalline aggregates (Potocki-Véronèse et al. 2005). The reaction temperature increase from 30°C to 50°C did not significantly influence the weak self-assembling properties of the amylose chains produced from 100 mM sucrose but probably reduced more severely the self-assembly of the chains synthesized from 600 mM sucrose, which were more accessible to enzyme elongation. The result is that a higher yield (31 g/L) of long amylose chains can be synthesized with the thermostable R20C/A451T mutant.
Conclusions
We have described the identification of thermostable variants of N. polysaccharea amylosucrase using a combinatorial engineering approach based on the construction of random mutant libraries, subsequently selected and screened for the desired trait. This strategy led to the identification of three AS variants displaying 3.5- to 10-fold increased half-lives at 50°C compared to the parent wild-type enzyme. Among them, the double-mutant R20C/A4551T is the most thermostable amylosucrase characterized to date. This new variant was able to catalyze at 50°C the high-yield synthesis of amylose chains, probably displaying different self-assembling properties than those produced at 30°C. Resulting physicochemical properties will be further investigated. In addition, a preliminary in silico comparative analysis between the wild-type AS structure and the modeled variants allowed the highlighting of molecular features possibly implicated in thermostability enhancement. To further optimize amylosucrase, the combination of the mutations identified here with substitutions involved in enzyme efficiency and/or specificity obtained through rational and combinatorial engineering will be investigated in a future study.
Materials and Methods
Bacterial strains and plasmids
One Shot TOP10 E. coli (Invitrogen) was used for transformation of ligation mixtures. E. coli JM109 (Promega) was used to screen AS variants and for large-scale production of the selected mutants. Plasmid pGST-AS, derived from pGEX-6P-3 (GE Healthcare Biosciences), was used for the construction, selection, and screening of AS random mutagenesis libraries and for the production of glutathione S-transferase (GST)–amylosucrase fusion proteins (Potocki de Montalk et al. 1999). E. coli cells were grown on LB (agar) with 100 μg/mL ampicillin. For expression of amylosucrase in E. coli JM109 media were supplemented with 1 mM isopropyl-β-d-thiogalactose (IPTG).
DNA manipulations
Restriction endonucleases and DNA-modifying enzymes were purchased from New England Biolabs and used according to the manufacturer's instructions. DNA purification was performed using QIAQuick (PCR purification and gel extraction) and QIASpin (miniprep and maxiprep) (Qiagen). DNA sequencing was carried out using the dideoxy chain-termination procedure (v3.1 Cycle Sequencing Kit; Applied Biosystems) by MilleGen sequencing service (France).
Construction, selection, and screening of AS random mutagenesis libraries
Two libraries of AS variants, named A and B, were constructed by replicating the wild-type AS encoding sequence with human Pol β and Pol η, respectively, following the MutaGen technique patented by MilleGen (Bouayadi et al. 2002; Mondon et al. 2007). A detailed description of the experimental procedure is reported elsewhere (Emond et al. 2008). AS libraries were cloned into pGEX-6P-3. Ligation products were first transformed in E. coli TOP10 to create the libraries. After plasmid extraction from the resulting colonies, the library was then expressed in E. coli JM109 for selection and screening.
A selection step was performed in order to isolate colonies expressing an active AS variant. After transformation, E. coli JM109 cells were plated on solid LB containing 100 μg/mL ampicillin. After growth, the colonies were scraped and diluted in physiological water to an OD600 of 10−4. The clones were then subjected to selection pressure by plating them onto solid M9 medium (42 mM Na2HPO4, 24 mM KH2PO4, 9 mM NaCl, 19 mM NH4Cl, 1 mM MgSO4, 0.5 μg/mL thiamine) containing 50 g/L sucrose as the sole carbon source and 100 μg/mL ampicillin. The plates were incubated for 3–5 d at 30°C, to obtain sufficient cell growth linked to active amylosucrase production.
Colonies expressing active AS variants were picked using a colony picker (Genetix) and transferred to sterile 96-well microplates (Nunc Brand) filled with 150 μL of LB medium. After 24 h of growth at 30°C with horizontal shaking (250 rpm) in an incubator shaker (Kühner), microplate wells were filled with 100 μL of sterile glycerol 30% (v/v) and stored at −20°C. These microplates are referred to as storage microplates.
The screening procedure was performed as previously described (Emond et al. 2007). Briefly, the storage microplates were thawed and replicated to inoculate cultures in 96-well sterile microplates referred to as starter microplates (150 μL of LB per well) grown for 24 h at 30°C under horizontal shaking. These starter cultures were then replicated as start cultures for recombinant AS production in sterile 96-well microplates (per well: 200 μL LB, 1 mM IPTG), which were incubated at 30°C for 24 h under horizontal shaking. After growth, lysozyme was added to a final concentration of 0.5 g/L and microplate cultures were frozen at −80°C for 8–12 h. After thawing for 1 h at room temperature, AS variants were assayed for thermostability on an integrated robotic TECAN Genesis RSP-200 platform. Eighty μL of crude cell extracts were transferred in a nonsterile 96-well microplate (Barloworld Scientific) and subsequently submitted to a heat-treatment step (20 min, 50°C). Enzymatic reaction was then performed by adding 80 μL of sucrose to a final concentration of 146 mM followed by incubation at 37°C for 4 h. Reducing sugar production was measured following a miniaturized dinitrosalicylic acid (DNS) assay described previously (Emond et al. 2007).
Site-directed mutagenesis
Site-directed mutagenesis was carried out with the QuikChange Site-Directed Mutagenesis kit (Stratagene) as previously described (Sarcabal et al. 2000) to introduce the A451T substitution into the wild-type AS encoding sequence. A KpnI restriction site was also introduced silently to screen the correct clones. Plasmid pGST-AS (Potocki de Montalk et al. 1999) was used as a template. The pair of primers used for mutagenesis inverted PCR was (KpnI restriction site is underlined, and mutated bases are indicated in bold): AS-A451T-for, 5′-TGTCAGTGGTACC ACCGCGGCATTGGTCGGCT-3′; AS-A451T-rev, 5′-AGCCGACCAATGCCGCGGTG GTACCACTGACA-3′. The resulting clones were confirmed by DNA sequencing.
Production, purification, and activity assay of wild-type and variant AS
E. coli carrying the recombinant plasmid containing wild-type or mutated AS encoding sequence was grown on LB containing 100 μg/mL ampicillin and 2 mM IPTG for 15 h. The cells were harvested by centrifugation, resuspended, and concentrated to an OD600 of 80 in PBS buffer (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3). The intracellular enzyme was extracted by sonication, and the lysate supernatant was used as the source for enzyme purification. Overexpression was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of wild-type and mutated enzymes was performed as previously described (Potocki de Montalk et al. 1999) by affinity chromatography of the GST/AS fusion protein on glutathione–Sepharose 4B (GE Healthcare Biosciences). The fusion protein solution was subjected to proteolysis to remove the GST-tag, using the PreScission protease (GE Healthcare Biosciences). The purified AS was finally eluted in PreScission buffer (50 mM Tris-HCl pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol). The enzyme was concentrated using Centricon-10 filters (Amicon, Millipore).
The protein content was determined by the micro-Bradford method, using the Bio-Rad reagent (Bio-Rad Laboratories) and bovine serum albumin as a standard (Bradford 1976). Specific amylosucrase activity was measured at 30°C with 146 mM sucrose by HPLC analysis using the procedure described below (carbohydrate analysis).
Thermostability of wild-type and mutant amylosucrase
Half-lives of thermal inactivation were determined for purified wild-type and variant GST-AS by incubating the enzymes (∼500 mg/L) at 50°C for various time intervals. Initial and residual activities were measured at 37°C with 146 mM sucrose in PBS buffer at pH 7.3 using the DNS method to quantify the released reducing sugar. The first-order rate constant, k d, of irreversible thermal denaturation was obtained from the slope of the plots of ln (initial activity/residual activity) versus time, and the half-lives (t 1/2) were calculated as ln 2/k d.
Activity–temperature profile of wild-type and mutant amylosucrases
Initial specific activities of the purified wild-type and mutant amylosucrases were measured at temperatures ranging from 30°C to 55°C, in the presence of 146 mM sucrose and 50 mM Tris, pH 7.0. Fructose and glucose concentrations were determined by HPLC using the procedure described below. It was checked for all tested enzymes that the ratio between the glucose (released by sucrose hydrolysis) and the fructose production rates (amount of free glucose or fructose released versus time) was independent of the reaction temperature.
In vitro synthesis of amylose and oligosaccharides
Reactions were performed using the wild-type AS and the double-mutant R20C/A451T at 1 U/mL (activity measured at 30°C on 146 mM sucrose with 0.1 g/L glycogen in 50 mM Tris-HCl, pH 7.0). Reaction using the P351S variant was carried out at 0.5 U/mL since this enzyme was prone to aggregate at higher concentrations. Synthesis reactions were performed for 24 h at 30°C and 50°C in 50 mM Tris-HCl, pH 7.0, containing 100 or 600 mM sucrose. For carbohydrate analysis, samples collected at the beginning and at the end of the synthesis were heated at 90°C during 5 min to stop the reaction.
Carbohydrate analysis
To determine the temperature dependency of wild-type and variant amylosucrases, glucose and fructose concentrations were measured using a refractometer (Dionex), after separation through a Ca2+ column (Bio-Rad) and elution at 80°C with water at 0.6 mL/min flow rate.
After 24 h reaction using sucrose as a substrate, the final media composition contained a soluble fraction (monosaccharides and oligosaccharides) and a precipitate (longer α-1,4 linked linear chains). Quantification of the products contained in the final reaction media was performed by high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD). For the determination of the reaction yields by quantification of monosaccharides and oligosaccharides, the reaction media supernatants were diluted in water to 17 mg/kg of total sugars. For analysis of the longer amylose chains, the final reaction media (containing both soluble and insoluble products) was solubilized in 1 M KOH to a final polymer content of 10 g/L. Separation was performed on a 4 × 250 mm Dionex CarboPac PA100. The flow rate of 150 mM NaOH was 1.0 mL/min, and an acetate gradient was applied as follows: 0–20 min, 6–196 mM; 20–70 min, 196–303.5 mM; 70–134 min, 303.5–450 mM; 135–140 min, 6 mM. Detection was performed by use of a Dionex ED40 module with a gold working electrode and a Ag/AgCl pH reference. Amylose chain (DP > 3) yields were determined as the difference between the amount of sucrose consumed and the amount of glucosyl residues incorporated into the soluble products (glucose, sucrose isomers, maltose, and maltotriose).
Iodine binding properties
The iodine reactivity of the amyloses produced by the wild-type AS and the double-mutant R20C/A451T were measured. The final reaction media (20 μL) were solubilized with 20 μL of 2 M KOH, neutralized with 200 μL of 0.1 N HCl, and mixed with 32 μL of aqueous iodine solution [2% (w/v) KI and 0.2% (w/v) I2] and 8 μL of 0.1 N HCl in microtiter plates. The maximum absorption wavelength (λmax) of the iodine/amylose complexes was determined by recording spectra between 500 and 690 nm at 2-nm steps, on a Sunrise spectrophotometer (Tecan). The corresponding average DP was calculated from the following equation previously established for DP 28–70 amylose chains (John et al. 1983):
Computational modeling of AS variants
In silico construction of AS mutants was performed using the coordinates of the AS crystallographic structure (Protein Data Bank entry: 1MW0). Hydrogens were added to each atom of the AS protein using the InsightII molecular modeling package (Accelrys) running on a Silicon Graphics O2 workstation. The crystallographic water molecules and bound substrate were removed from the structure. AMBER9 software (http://amber.scripps.edu/) was subsequently used to optimize the energy of the protein structure in the gas phase using a combination of steepest-descent and conjugated gradient-minimization steps.
To assess the contribution of the individual amino acids to the thermostability of AS, we constructed the AS mutants, in which the amino acids were individually replaced in silico with the corresponding amino acids observed in isolated improved mutants. Starting from the optimized AS structure, site mutations were modeled using the Biopolymer module of the InsightII software package (Accelrys). The conformation of the mutated residue side chain was optimized by manually selecting a low-energy conformation from a side-chain rotamer library. Steric clashes (van der Waals overlap) and nonbonded interaction energies (Coulombic and Lennard-Jones) were evaluated for the different side-chain conformations. All mutants were subsequently minimized following the same energy-minimization procedure described for AS and using the last available version of AMBER9 molecular mechanics force field (Pearlman et al. 1995). All drawings were performed using PyMOL software (DeLano Scientific).
Acknowledgments
The authors are grateful to Pascale Klopp and Nathalie Souyris for technical support in the construction of AS libraries and Dr. Cécile Albenne for helpful assistance during the screening experiments. S.E. was financially supported by MilleGen SA and l'Association Nationale de la Recherche Technologique (France).
Footnotes
Reprint requests to: Magali Remaud-Simeon, Laboratoire d'Ingénierie des Systèmes Biologiques et des Procédés, Institut National des Sciences Appliquées de Toulouse, 135 avenue de Rangueil, F-31400 Toulouse, France; e-mail: magali.remaud@insa-toulouse.fr; fax: +33-561-559-400.
Abbreviations: t 1/2, half-life of thermal inactivation; DP, mean degree of polymerization; λmax, maximum absorption wavelength; HPAEC-PAD, high-performance anion-exchange chromatography with pulsed amperometric detection.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.083492608.
References
- Bloom, J.D., Meyer, M.M., Meinhold, P., Otey, C.R., MacMillan, D., Arnold, F.H. Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 2005;15:447–452. doi: 10.1016/j.sbi.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Bouayadi, K., Kharrat, H., Louat, T., Servant, L., Cazaux, C., Hoffmann, J.S. Use of mutagenic polymerase for producing random mutations. [May 16, 2002];2002 Patent WO/2002/038756.
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Ditursi, M.K., Kwon, S.J., Reeder, P.J., Dordick, J.S. Bioinformatics-driven, rational engineering of protein thermostability. Protein Eng. Des. Sel. 2006;19:517–524. doi: 10.1093/protein/gzl039. [DOI] [PubMed] [Google Scholar]
- Eijsink, V.G., Bjork, A., Gaseidnes, S., Sirevag, R., Synstad, B., van den Burg, B., Vriend, G. Rational engineering of enzyme stability. J. Biotechnol. 2004;113:105–120. doi: 10.1016/j.jbiotec.2004.03.026. [DOI] [PubMed] [Google Scholar]
- Eijsink, V.G., Gaseidnes, S., Borchert, T.V., van den Burg, B. Directed evolution of enzyme stability. Biomol. Eng. 2005;22:21–30. doi: 10.1016/j.bioeng.2004.12.003. [DOI] [PubMed] [Google Scholar]
- Emond, S., Mondon, P., Pizzut-Serin, S., Douchy, L., Crozet, F., Bouayadi, K., Kharrat, H., Potocki-Véronèse, G., Monsan, P., Remaud-Simeon, M. A novel random mutagenesis approach using human mutagenic DNA polymerases to generate enzyme variant libraries. Protein Eng. Des. Sel. 2008;21:267–274. doi: 10.1093/protein/gzn004. [DOI] [PubMed] [Google Scholar]
- Emond, S., Potocki-Véronèse, G., Mondon, P., Bouayadi, K., Kharrat, H., Monsan, P., Remaud-Simeon, M. Optimized and automated protocols for high-throughput screening of amylosucrase libraries. J. Biomol. Screen. 2007;12:715–723. doi: 10.1177/1087057107301978. [DOI] [PubMed] [Google Scholar]
- Haney, P.J., Badger, J.H., Buldak, G.L., Reich, C.I., Woese, C.R., Olsen, G.J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. 1999;96:3578–3583. doi: 10.1073/pnas.96.7.3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendsch, Z.S., Tidor, B. Do salt bridges stabilize proteins? A continuum electrostatic analysis. Protein Sci. 1994;3:211–226. doi: 10.1002/pro.5560030206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytonen, V.P., Maatta, J.A., Nyholm, T.K., Livnah, O., Eisenberg-Domovich, Y., Hyre, D., Nordlund, H.R., Horha, J., Niskanen, E.A., Paldanius, T., et al. Design and construction of highly stable, protease-resistant chimeric avidins. J. Biol. Chem. 2005;280:10228–10233. doi: 10.1074/jbc.M414196200. [DOI] [PubMed] [Google Scholar]
- John, M., Schmidt, J., Kneifel, H. Iodine–maltosaccharide complexes: Relation between chain-length and colour. Carbohydr. Res. 1983;119:254–257. [Google Scholar]
- Kumar, S., Nussinov, R. Salt bridge stability in monomeric proteins. J. Mol. Biol. 1999;293:1241–1255. doi: 10.1006/jmbi.1999.3218. [DOI] [PubMed] [Google Scholar]
- Lehmann, M., Wyss, M. Engineering proteins for thermostability: The use of sequence alignments versus rational design and directed evolution. Curr. Opin. Biotechnol. 2001;12:371–375. doi: 10.1016/s0958-1669(00)00229-9. [DOI] [PubMed] [Google Scholar]
- Makhatadze, G.I., Loladze, V.V., Ermolenko, D.N., Chen, X., Thomas, S.T. Contribution of surface salt bridges to protein stability: Guidelines for protein engineering. J. Mol. Biol. 2003;327:1135–1148. doi: 10.1016/s0022-2836(03)00233-x. [DOI] [PubMed] [Google Scholar]
- Miyazaki, J., Nakaya, S., Suzuki, T., Tamakoshi, M., Oshima, T., Yamagishi, A. Ancestral residues stabilizing 3-isopropylmalate dehydrogenase of an extreme thermophile: Experimental evidence supporting the thermophilic common ancestor hypothesis. J. Biochem. 2001;129:777–782. doi: 10.1093/oxfordjournals.jbchem.a002919. [DOI] [PubMed] [Google Scholar]
- Mondon, P., Souyris, N., Douchy, L., Crozet, F., Bouayadi, K., Kharrat, H. Method for generation of human hyperdiversified antibody fragment library. Biotechnol. J. 2007;2:76–82. doi: 10.1002/biot.200600205. [DOI] [PubMed] [Google Scholar]
- Pearlman, D.A., Case, D.A., Caldwell, J.W., Ross, W.S., Cheatham, T.E., DeBolt, S., Ferguson, D., Seibel, G., Kollman, P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 1995;91:1–41. [Google Scholar]
- Perl, D., Mueller, U., Heinemann, U., Schmid, F.X. Two exposed amino acid residues confer thermostability on a cold shock protein. Nat. Struct. Biol. 2000;7:380–383. doi: 10.1038/75151. [DOI] [PubMed] [Google Scholar]
- Petrounia, I.P., Arnold, F.H. Designed evolution of enzymatic properties. Curr. Opin. Biotechnol. 2000;11:325–330. doi: 10.1016/s0958-1669(00)00107-5. [DOI] [PubMed] [Google Scholar]
- Pizzut-Serin, S., Potocki-Véronèse, G., van der Veen, B.A., Albenne, C., Monsan, P., Remaud-Simeon, M. Characterisation of a novel amylosucrase from Deinococcus radiodurans . FEBS Lett. 2005;579:1405–1410. doi: 10.1016/j.febslet.2004.12.097. [DOI] [PubMed] [Google Scholar]
- Potocki de Montalk, G., Remaud-Simeon, M., Willemot, R.M., Planchot, V., Monsan, P. Sequence analysis of the gene encoding amylosucrase from Neisseria polysaccharea and characterization of the recombinant enzyme. J. Bacteriol. 1999;181:375–381. doi: 10.1128/jb.181.2.375-381.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potocki de Montalk, G., Remaud-Simeon, M., Willemot, R.M., Sarcabal, P., Planchot, V., Monsan, P. Amylosucrase from Neisseria polysaccharea: Novel catalytic properties. FEBS Lett. 2000;471:219–223. doi: 10.1016/s0014-5793(00)01406-x. [DOI] [PubMed] [Google Scholar]
- Potocki-Véronèse, G., Putaux, J.L., Dupeyre, D., Albenne, C., Remaud-Simeon, M., Monsan, P., Buleon, A. Amylose synthesized in vitro by amylosucrase: Morphology, structure, and properties. Biomacromolecules. 2005;6:1000–1011. doi: 10.1021/bm049326g. [DOI] [PubMed] [Google Scholar]
- Reetz, M.T., Carballeira, J.D., Vogel, A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew. Chem. Int. Ed. Engl. 2006;45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]
- Sarcabal, P., Remaud-Simeon, M., Willemot, R., Potocki de Montalk, G., Svensson, B., Monsan, P. Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 2000;474:33–37. doi: 10.1016/s0014-5793(00)01567-2. [DOI] [PubMed] [Google Scholar]
- Skov, L.K., Mirza, O., Henriksen, A., De Montalk, G.P., Remaud-Simeon, M., Sarcabal, P., Willemot, R.M., Monsan, P., Gajhede, M. Amylosucrase, a glucan-synthesizing enzyme from the α-amylase family. J. Biol. Chem. 2001;276:25273–25278. doi: 10.1074/jbc.M010998200. [DOI] [PubMed] [Google Scholar]
- van den Burg, B., Eijsink, V.G. Selection of mutations for increased protein stability. Curr. Opin. Biotechnol. 2002;13:333–337. doi: 10.1016/s0958-1669(02)00325-7. [DOI] [PubMed] [Google Scholar]
- van der Veen, B.A., Potocki-Véronèse, G., Albenne, C., Joucla, G., Monsan, P., Remaud-Simeon, M. Combinatorial engineering to enhance amylosucrase performance: Construction, selection, and screening of variant libraries for increased activity. FEBS Lett. 2004;560:91–97. doi: 10.1016/S0014-5793(04)00077-8. [DOI] [PubMed] [Google Scholar]
- van der Veen, B.A., Skov, L.K., Potocki-Véronèse, G., Gajhede, M., Monsan, P., Remaud-Simeon, M. Increased amylosucrase activity and specificity, and identification of regions important for activity, specificity and stability through molecular evolution. FEBS J. 2006;273:673–681. doi: 10.1111/j.1742-4658.2005.05076.x. [DOI] [PubMed] [Google Scholar]
- Vieille, C., Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001;65:1–43. doi: 10.1128/MMBR.65.1.1-43.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, L., Kurek, I., English, J., Keenan, R. Laboratory-directed protein evolution. Microbiol. Mol. Biol. Rev. 2005;69:373–392. doi: 10.1128/MMBR.69.3.373-392.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]