Abstract
Metrics derived from pedigrees are key to investigating several major issues in evolutionary biology, including the quantitative genetic architecture of traits, inbreeding depression, and the evolution of cooperation and inbreeding avoidance. There is merit in studying these issues in natural populations experiencing spatially and temporally variable environmental conditions, since these analyses may yield different results from laboratory studies and allow us to understand population responses to rapid environmental change. Partial pedigrees are now available for several natural populations which are the subject of long-term individual-based studies, and analyses using these pedigrees are leading to important insights. Accurate pedigree construction supported by molecular genetic data is now feasible across a wide range of taxa, and even where only imprecise pedigrees are available it is possible to estimate the consequences of imprecision for the questions of interest. In outbred diploid populations, the pedigree approach is superior to analyses based on marker-based pairwise estimators of coancestry.
Keywords: pedigree, parentage, microsatellite, heritability, inbreeding, kin selection
1. Introduction
The past few years have seen a dramatic increase in the use of multigenerational pedigrees of natural populations in evolutionary biology studies. In this review I outline the origins and benefits of this trend, summarize the available approaches for recovering pedigrees, discuss the consequences of imperfect information and show why recovered pedigrees are superior to proposed alternative approaches.
A pedigree is one of the simplest concepts in biology and probably one of the best understood biological concepts among non-scientists; after all, we each have a family tree, as do our pets and our farm animals. For more than a century, geneticists have recognized the value of pedigrees for studying the inheritance of polymorphisms, inbreeding depression and quantitative genetic variation. It has taken a great deal longer for wild pedigrees to be used—why?
Pedigree analysis within studies of individuals living in the wild has only been made possible by a series of developments. First, the intensive study of breeding success and other traits for all individuals of a species living in a particular area in the wild over several years, although initiated as early as 1936 (Richdale 1957), only became fashionable in the 1980s as ecologists recognized the value of individual life-history data for understanding population processes, and behavioural ecologists sought to measure the results of behavioural strategies in the currency of reproductive success (Clutton-Brock 1988). In many cases, measuring the reproductive success of individuals amounts to recording parentage, meaning pedigrees can be constructed. The first uses of pedigrees for socially monogamous birds (‘social pedigrees’) to investigate inbreeding (Bulmer 1973) and quantitative genetic variation (Boag & Grant 1978) followed soon after.
A second major contribution to modern wild pedigree analysis was made by the discovery of abundant, highly variable neutral genetic markers. The first breakthrough was multilocus DNA fingerprinting with minisatellites (Jeffreys et al. 1985a,b), which was rapidly applied to wild populations to assign parentage (Burke & Bruford 1987; Wetton et al. 1987). The second breakthrough was DNA profiling using microsatellites (Litt & Luty 1989; Tautz 1989; Weber & May 1989), which soon superceded DNA fingerprinting for wild population studies. When combined with appropriate statistical analysis (see §3 below), these techniques enable us to confirm suspected pedigree links, or infer parentage or sibship among groups of individuals, with far greater accuracy than is possible from behavioural data alone. Within virtually every social system observed in the field, a great variety of actual mating systems has been revealed. In socially monogamous birds, extra-pair paternity EPP rates range up to 55% across species and vary between populations within species (reviewed by Griffith et al. 2002). Among cooperative breeders, the dominant male in a meerkat (Suricata suricatta) group fathers, 60–80% of the offspring born in the group (Griffin et al. 2003) while in the superb fairy-wren (Malurus cyaneus) all group males together sire just 24% of offspring in the local nest (Mulder et al. 1994). In haplodiploid social hymenoptera, worker relatedness ranges from the often-predicted 0.75 right down to ˜ 0, depending on the number of queens and their number of mates (Avise 2004). Among polygynous breeders, harbour seals (Phoca vitulina) show remarkably low variance in male mating success (Coltman et al. 1998), while red deer (Cervus elaphus) show higher variance in mating success than behavioural data suggest (Pemberton et al. 1992). Soay sheep (Ovis aries) are so promiscuous that 74% of twins have different fathers (Pemberton et al. 1999).
A third cause of the recent increase in wild pedigree analyses is the increasing sophistication of statistical methods with which to conduct downstream analyses. For example, Keller (1998) was the first to conduct a comprehensive analysis of inbreeding depression in life-history components in a large wild pedigree (the social pedigree of Mandarte Island song sparrows, Melospiza melodia) including the estimation of lethal equivalents. In quantitative genetics, the application of the animal model with restricted maximum likelihood from animal breeding, which can deal with unbalanced, incomplete data and make efficient use of all the information available, is very recent (Kruuk et al. 2000; Milner et al. 2000; Kruuk 2004).
Finally, there is a growing realization that the evolutionary genetics of wild populations may not be well represented by laboratory population studies. Most obviously, wild populations have different histories of inbreeding and selection than laboratory populations. Possibly more important is the effect of temporal and spatial heterogeneity in environmental conditions. The demonstration that heritability (Wilson et al. 2006) and inbreeding depression (Keller et al. 2002) can vary systematically with temporal environmental change even within the same study population gives strong support to the view that many evolutionary genetic topics need to be addressed in the wild.
2. What do pedigrees offer?
Pedigrees of free-living populations allow us to estimate the coefficient of coancestry between two individuals x and y (fxy or Θxy, also called the coefficient of kinship or coefficient of consanguinity) which is the probability that two alleles (at the same locus) drawn at random (one from each individual) are identical by descent (Lynch & Walsh 1998). In turn, this allows us to estimate the coefficient of relatedness between two individuals (rxy) as 2fxy and the inbreeding coefficient of an individual (f) as the coefficient of coancestry of its parents. When constructing pedigrees of wild populations, researchers have to make the initial assumption that founders and immigrants are unrelated and non-inbred. Under these circumstances, in a diploid species, the coefficient of coancestry is 0.25 between a parent and offspring, their coefficient of relatedness is 0.5 and the offspring of a parent–offspring mating has an inbreeding coefficient of 0.25.
Between them, the coancestry, relatedness and inbreeding coefficients allow many questions across evolutionary genetics to be addressed. When estimating quantitative genetics parameters such as the heritability of a trait or the genetic correlation between two traits, 2fxy is the metric used to describe the genetic relationship between individuals (Lynch & Walsh 1998). Quantitative genetic analysis in natural populations is currently focused on two great questions: how to explain the maintenance of quantitative genetic variation even in traits that are under directional selection (Coltman et al. 2001; Foerster et al. 2007), and how to explain how natural populations respond to selection, including the frequent observation of stasis instead of predicted change (Merilä et al. 2001; Kruuk et al. 2002b, 2003; Wilson et al. 2006, 2007). In both cases, there appear to be several explanations with empirical support and it will take further research in multiple study systems to elucidate general patterns. Nor are these purely academic issues; they are extraordinarily relevant to understanding how natural populations will cope with climate change.
The coefficient of relatedness, 2fxy is also a key parameter in the kin selection theory for the evolution of cooperative behaviour (Hamilton 1964). Its use in natural populations has greatly illuminated our understanding of cooperation. Interestingly, one of the general effects of being able to estimate relatedness has been to emphasize alternative, direct benefit mechanisms which probably serve to maintain cooperative societies (Clutton-Brock 2002; Griffin & West 2002, 2003).
The coefficient of inbreeding is required for estimating inbreeding depression. Inbreeding depression is a near-universal feature of diploid organisms, but precise estimates of its magnitude based on pedigrees of individuals living in the wild are still uncommon (Keller & Waller 2002; Kruuk et al. 2002a). As a result, at present we are relatively ignorant of the extent and causes of observed variation in inbreeding depression between populations, how inbreeding depression varies across the lifespan, whether it is common for inbreeding depression to interact with environmental conditions (see Keller et al. 2002; Marr et al. 2006), and to what extent it contributes to change in population size (Keller & Waller 2002). Again, these issues are highly relevant to the future survival of threatened populations in the face of environmental change. The extent to which organisms avoid inbreeding is also of substantial evolutionary interest in its own right. Inbreeding avoidance appears to have driven the evolution of outcrossing mechanisms in plants and may have driven the evolution of sex-biased dispersal in vertebrates (Handley & Perrin 2007), but the extent to which animals also actively avoid incest through mate choice is unclear. Incest avoidance is clearly present in some cooperative breeders (Cockburn et al. 2003; Koenig & Haydock 2004), however in other social systems, choosing the correct null model to compare with observed behaviour is difficult (Pärt 1996). So far, two studies of non-social passerines using pedigree coancestry and realistic null models have found little evidence for a behavioural inbreeding avoidance strategy (Keller & Arcese 1998; Hansson et al. 2007).
3. Pedigree construction approaches
(a) Field observation
Pedigrees are formed from the accumulation of parent–offspring or sib–sib links. Field observations are the key starting point for pedigree construction, since they often supply hypotheses for genealogy, and if the hypotheses are correct, they make genetic analysis more powerful. For example, knowing the identity and having genotype information for a mother greatly increases the power of paternity analysis. In many birds and mammals, multiple offspring, likely to be sibs, are reared in the same place, a nest or burrow, where they can be marked and sampled. In many birds, parental care is indicative of parentage, though should not be assumed without some molecular analysis (see above). In most mammals, pregnancy and lactation provide excellent maternal information. In some species, additional information can be obtained through modest intervention; in their study of side-blotched lizards (Uta stansburiana), Sinervo and co-authors bring gravid females into the laboratory briefly for the egg-laying period, hatch the eggs in captivity, and then sample and release the offspring at the mother's capture site, giving perfect maternal information with minimal influence on reproductive success (Sinervo & Zamudio 2001). At the very least, field observations of marked individuals are useful in determining which candidate parents were in the study area during the mating and parturition periods.
(b) Markers for parentage and sibship inference
For inference of family relationships, microsatellites have been the marker of choice for several years (Parker et al. 1998; Jones & Ardren 2003). No other marker type combines the following desirable features: single locus information, codominance, high variability due to many alleles at low frequency, potential for high throughput through automation and short DNA fragments amenable to analysis of forensic samples obtained from wild populations. Identifying microsatellite markers for novel species is through de novo discovery or by taking advantage of their cross-species utility (Barbará et al. 2007). In recent years, centrally funded facilities and commercial companies specializing in finding microsatellites have arisen, so that obtaining loci for parentage analysis is now often a matter of time and money. However, it would be wrong not mention some difficulties. In some taxa, microsatellites are hard to find or insufficiently polymorphic for the task at hand. Genotyping is error-prone, mutations occur and an appreciable proportion of loci has segregating null alleles (Dakin & Avise 2004), all of which can cause false parentage exclusion. A technical issue affecting long-term studies is that microsatellite allele sizes change, and not necessarily in a consistent fashion, between detection platforms (J. M. Pemberton 1999, personal observation).
In future, single nucleotide polymorphisms (SNPs) may be commonly used in pedigree reconstruction in natural populations. Although individually much less informative than microsatellites, they exist in large numbers and scoring is potentially less error-prone than with microsatellites. As a result, discriminatory power for both identifying individuals and parents is potentially very high (Anderson & Garza 2006). Panels of SNPs have now been developed for farm animals and humans and studies confirm their usefulness compared with standard microsatellite panels (e.g. Phillips et al. 2007; Rohrer et al. 2007); the development of SNP panels for well-established long-term natural study populations seems probable in the near future.
(c) Parentage assignment
Parentage analysis using genetic markers requires careful statistical analysis. There is a substantial literature, an array of freeware computer programs and a recent review on the subject (Jones & Ardren 2003). In brief, the simplest approach is to use exclusion with associated exclusion probabilities, calculated from allele frequencies, to provide statistical support. However, few studies of natural populations use an exclusionary approach, since candidate sampling is almost never complete, marker panels are not always powerful enough to exclude all but one candidate and genotyping error can easily cause false exclusion of a true parent. Instead, most workers adopt a likelihood approach, which makes better use of candidate genotype information as well as using allele frequencies. Specifically, among those candidates not excluded at a locus, an individual that is homozygous for a required allele is twice as likely to be the true parent as an individual that is heterozygous for the required allele. The nine freeware programs comprehensively reviewed and tabulated by Jones & Arden (2003) take different approaches to dealing with the range of complexities encountered in wild populations such as the existence of large numbers of candidates (of one, both or even unknown sexes), some or all of which may not be sampled or even enumerated; mutations, genotyping errors and null alleles; insufficiently informative marker data; relatives among the candidates; and the assessment of statistical confidence.
There have been some advances in parentage analysis outwith and since the Jones & Ardren (2003) review of parentage inference methods. The authors omitted mention of the first full probability, Bayesian, approach to parentage analysis in the absence of any parental information (Emery et al. 2001) which is presented as the program Parentage, available at www.mas.ncl.ac.uk/∼nijw/. Duchesne et al. (2005) present Pasos, available at www.bio.ulaval.ca/louisbernatchez/, an open-system (i.e. allows for unsampled candidates) stable mate for their previous program, Papa. A useful feature of Pasos is that it explicitly estimates the number of unsampled candidates. Cervus (Marshall et al. 1998) has proved one of the most popular programs, but Kalinowski et al. (2007) point out an error in the way its likelihood equations accounted for genotyping error. This has been corrected in Cervus v. 3.0, which can now also conduct simultaneous analysis of maternity and paternity and is available from a new website www.fieldgenetics.com.
An interesting recent advance concerns the direct incorporation of field information into parentage analysis. In principle, it makes efficient use of the data, and reduces certain biases, to incorporate information about candidates, for example, spatial proximity, into the same analysis as the genetic marker information. Hadfield et al. (2006) took this approach in the case of the Seychelles warbler (Acrocephalus sechellensis), in which microsatellite variation is low, helpers at the nest of both sexes, which are relatives of the dominant pair, are potential parents and there are extra-territory fertilizations. This Bayesian approach (available as MasterBayes at http://www.R-project.org) found several different extra-group paternity assignments compared with previous methods. In the future, using developments of this approach, it will be possible to estimate quantitative genetic parameters or inbreeding depression at the same time as the pedigree (Hadfield et al. 2006).
The development of such a diversity of parentage inference programs is a reflection of the diversity of problems encountered during parentage analysis in natural populations. However, by far the most common problems in parentage analysis are that candidate parents are poorly sampled and the amount of marker information available is marginal for confident resolution of parentage links, even when the true parents are sampled (Marshall et al. 1998; Jones & Ardren 2003), suggesting that we should never skimp on sampling effort, the number of loci screened and the accuracy with which the loci are screened.
(d) Sibship reconstruction
Another approach to partial pedigree construction using genotype data is to recover full and half sibships from samples of individuals. Here, methods have developed rapidly over the last few years (Blouin 2003). Butler et al. (2004) tested four algorithms for full sibship reconstruction ranging from an exclusionary approach through methods using MCMC to maximize the likelihood of partitions between sibships, and showed that they varied in accuracy depending on the structure of the data in terms of family size, and that all were sensitive to genotyping error. The approach of reconstructing sibships using MCMC laid out by Thomas & Hill (2002) has been developed further, particularly to deal with genotyping error, by Wang (2004) and is available as the program Colony from www.zoo.cam.ac.uk/ioz/software.htm. Although some downstream analyses can be carried out with sibship data, they do not of themselves allow, for example, the analysis of inbreeding depression, and the challenge now is to combine sibship inference with parentage analysis to construct more complete pedigrees. One possible approach was demonstrated in a study of bighorn sheep (Ovis canadensis) in which candidate sires were only partially sampled. Coltman et al. (2005) genotyped offspring without identifiable sires at 32 loci and used Colony to infer 38 half sib clusters among 167 offspring. This information substantially increased the number of pedigree links available for quantitative genetic analysis.
(e) Pedigree reconstruction without field data
With enough polymorphic markers, it should be possible to reconstruct a pedigree of a sample of individuals without the need for any field information. Methods using simulated annealing algorithms have already been proposed and tested by Almudevar (2003) and Fernandez & Toro (2006) and this field seems likely to expand greatly as marker information increases for natural populations. However, from the perspective of downstream analyses, ecological information about individuals will nearly always be useful. For example, information on year of birth often explains trait variation and is usefully fitted as an additional effect in quantitative genetic analyses.
4. Pedigree quality
Pedigrees for wild populations vary in depth, accuracy, size, completeness and structure, and a fast-growing literature describes the effect of this variation on the results obtained from evolutionary genetic analyses. This is useful both from the perspective of those planning to try and recover pedigrees for wild populations and for those analysing pedigrees for which there is no prospect of retrospective pedigree improvement, because the individuals have died or dispersed without sampling.
(a) Pedigree depth
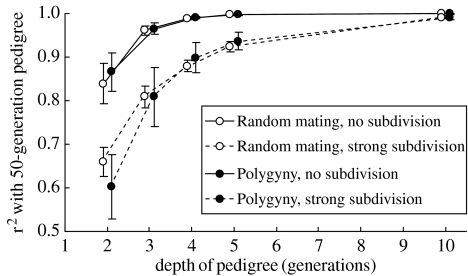
Coancestry and relatedness are greatest between members of the same or adjacent generations (e.g. parent–offspring), and the inbreeding coefficient of an individual is greatest when close relatives mate. This is good news for studies of wild populations, for it means that it is not necessary to have great depth of pedigree, in terms of generations, to capture most of the variance in these parameters. This point was made very clear by Balloux et al. (2004) who investigated the correlation between f calculated over generations 2, 3, …, 10 and f calculated over 50 generations in simulated populations covering four example vertebrate breeding systems and population structures. Within just five generations, 90% of the variance in 50-generation f is captured, regardless of population detail, and some simulated structures reached this figure far sooner (figure 1).
Figure 1.
Correlation between inbreeding coefficients calculated using pedigrees 2, 3, 4, 5 and 10 generations deep and inbreeding coefficients calculated using pedigrees 50 generations deep, reproduced with permission from Balloux et al. (2004). Two breeding systems (polygyny and random mating) and two population structures (400 individuals with no structure and 400 individuals divided into 20 populations of 20 individuals) were simulated.
(b) Pedigree accuracy
The accuracy of pedigree links is a major concern for all studies. In general, errors might be expected to result in downward-biased and less precise estimates of heritability (Kruuk 2004) and this has indeed been observed, for example, in Darwin's finches (Geospiza fortis) using parent–offspring regression (Keller et al. 2001). However, Charmantier & Réale (2005) examined the effect of extra-pair paternities in simulated and real pedigrees of a socially monogamous bird species and showed that, provided the number of families studied is sufficient, animal model heritability estimates are surprisingly robust to EPP rates up to 20%. This finding is also good news for those using molecular parentage analysis with marginal power (see above). Nevertheless, for small sample sizes or highly heritable traits, heritability and other quantitative genetic parameters will be downward biased as the accuracy of pedigree links declines, and systematic patterns in pedigree errors, such as misassignment of paternity to spatially closest males, could cause environmental covariance to be misinterpreted as genetic covariance.
Similarly, estimates of inbreeding depression will be imprecise when pedigree links are inaccurate. Inbreeding coefficients calculated from the social pedigree suggest that Mandarte Island song sparrows experience substantial inbreeding depression in several traits (Keller et al. 1994; Keller 1998). A microsatellite analysis of four cohorts of chicks showed that due to EPPs, 28% of paternal links in the social pedigree are wrong (O'Connor et al. 2006). Using this information, Marr et al. (Amy B. Marr, Louis C. Dallaire and Lukas F. Keller 2007, personal communication) estimated inbreeding depression in the population with increasing proportions of paternity error (28%, the existing social pedigree, to 100%) and then extrapolated to a predicted inbreeding depression if there was no paternity error. For two of the traits studied, inbreeding depression was predicted to be significantly higher when pedigree errors were zero, suggesting that estimates of inbreeding depression emerging from this study to date are conservative.
(c) Pedigree structure
More subtle issues surround the actual pattern of pedigree links in time and space. Polygynous species yield pedigrees which are good for estimating maternal and shared environment effects, since paternal half sibs have different mothers who may range in habitats of different qualities (Kruuk & Hadfield 2007). Long-lived and/or iteroparous species lend themselves to studies of ontogenetic effects and genetic×environment interactions (Wilson et al. 2005, 2006, 2007). Adding newly collected trait data for recent cohorts to a large pedigree of a short generation time bird (great tit, Parus major) and a smaller pedigree of a more iteroparous long-lived bird (mute swan, Cygnus olor) had contrasting effects (Quinn et al. 2006). In general, quantitative genetic parameters were estimated with greater precision in the great tit pedigree, presumably because sample sizes were greater and first and second degree relatives with measured traits were more likely to occur in adjacent sampling years.
A general difficulty is that owing to the variety of pedigrees and genetic architectures observed, it is hard to determine how powerful a pedigree is for measuring specific parameters and the extent to which errors or gaps in pedigree links will affect results. To address this issue for quantitative genetic studies, Morrissey et al. (2007) suggest a framework in which an empirically acquired pedigree and a user-supplied quantitative genetic architecture for traits can both be manipulated (e.g. wrong pedigree links can be created), and then used in animal models, to discover just how robust results obtained with real trait data are likely to be. A computer package, Pedantics, is available at http://wildevolution.biology.ed.ac.uk/awilson/pedantics.html for this purpose.
5. Alternatives to pedigrees and insights arising from them
The existence of extensive microsatellite genotype data for free-living populations, often combined with information about traits, including behaviour, for the individuals involved, has led to alternative non-pedigree-based approaches to parameter estimation in studies of quantitative genetics, inbreeding and cooperation. These approaches use genotype data as a proxy for the coancestry and inbreeding coefficients outlined above and have great attraction since they avoid the laborious process of parentage analysis and the time required for generations to pass.
(a) Coancestry and relatedness
Conceptually, the sharing of marker alleles between two individuals, after taking account of population allele frequencies, yields an estimate of coancestry. Many different marker-based estimators of pairwise coancestry have been derived over recent years including method-of-moments estimators, maximum-likelihood estimators, two-gene estimators, four-gene estimators and different approaches to allele frequency correction (Queller & Goodnight 1989; Ritland 1996a; Van de Casteele et al. 2001; Thomas 2005; Oliehoek et al. 2006).
Pairwise coancestry estimators based on marker data have been widely used in the behavioural ecology literature in studies of cooperation. Less commonly, they have been used to investigate inbreeding avoidance behaviour (e.g. Reusch et al. 2001). Furthermore, it is in principal possible to use them (with phenotypic data) to infer quantitative genetic parameters without the need to resolve pedigrees (Ritland 1996b, 2000). Heritability estimates using this method were predicted from the start to be highly dependent on the variance in relatedness, and indeed it turns out that heritabilities calculated for outbred vertebrates are erratically different from animal model estimates applied to pedigree data for the same sample (Thomas et al. 2002; Wilson et al. 2003; Coltman 2005; Garant & Kruuk 2005; Frentiu 2008). Furthermore, in the context of quantitative genetics and inbreeding depression, the lack of information on precise ancestry is a great disadvantage, for it prevents study of additional and often important sources of variance such as maternal effects.
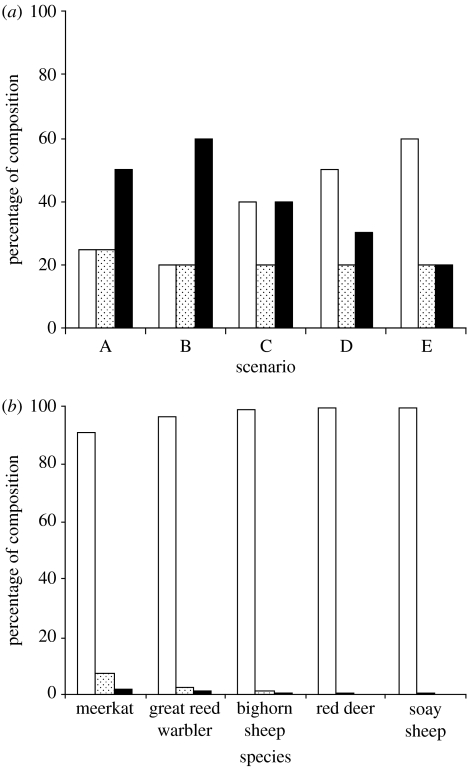
Closer inspection of pairwise marker-based coancestry estimators has shown that at least for outbred vertebrates, they are rather imprecise. Mean and variance in coancestry in real study populations is far lower than has typically been assumed for testing the average performance of coancestry estimators (e.g. compare Van de Casteele et al. (2001) with Csilléry et al. (2006); figure 2), and the low precision with which just a few loci can capture this variance merely adds to the difficulties. Pairwise, marker-based coancestry estimators should therefore be used with care in evolutionary studies: they are at their best when applied in scenarios with high variance in pedigree relatedness (e.g. within some haplodiploid hymenopteran colonies or selected samples of individuals likely to show high variance in coancestry). In all other scenarios, including tests of cooperative behaviour, it is questionable how powerful tests using pairwise relatedness really are.
Figure 2.
Comparison of hypothetical and observed distributions of relatedness in outbred vertebrate populations. (a) Percentage of pairs of individuals which, if drawn at random from a population, would fall into different relatedness categories as used by Van de Casteele et al. (2001) in a study of the average performance of marker-based relatedness estimators. White bars, r=0; stippled bars, r=0.25; black bars, r=0.5. The five different scenarios for relatedness structure suggested by the authors are shown. Note that parent–offspring and full sib categories used by the authors have been collapsed into a single category here. (b) The same information for five wild pedigrees analysed by Csilléry et al. (2006). White bars, r=0; stippled bars, r=0.25; black bars, r=0.5 (as above). All species previously identified in text except great reed warbler (Acrocephalus arundinaceus). For simplicity, these figures were derived by restricting analysis to two-generation deep pedigrees; relaxing this restriction adds additional classes of relatedness but does not alter the view that the overwhelming majority of randomly drawn pairs have r∼0.
(b) Inbreeding coefficients
Inbred individuals should be more homozygous than outbred individuals after correcting for population allele frequencies, and again a variety of marker-based estimators of individual inbreeding coefficients have been proposed (Hill et al. 1995; Ritland 1996a; Coulson et al. 1998; Coltman et al. 1999; Amos et al. 2001). Despite a probable publication bias, there is a certain consistency to findings of a positive correlation between standardized heterozygosity and fitness in natural populations (Coltman & Slate 2003).
The idea that heterozygosity or inbreeding coefficient estimated from a few marker loci has precision for measuring inbreeding depression in normally outbred diploids has recently been eroded from several directions. The observed correlation between pedigree inbreeding coefficients and marker-based estimates of inbreeding is often low despite good data (Markert et al. 2004; Slate et al. 2004; Overall et al. 2005; Rodriguez-Ramilo et al. 2007) and this is largely because the mean and variance of inbreeding coefficients are both low in those natural populations so far studied (Slate et al. 2004). Similar conclusions were reached by a simulation study (Balloux et al. 2004). As for coancestry, so for inbreeding coefficients: marker-based estimators of inbreeding are at their most useful when inbreeding and variance of inbreeding is high, as for example in selfing plants.
An alternative explanation for heterozygosity–fitness correlations is therefore required. Many individual-based studies which have published such correlations work with small, introduced or expanding populations in which linkage disequilibrium may extend over large distances (Hansson et al. 2001; Hansson 2004). Also, sometimes the correlation is driven by a subset of markers (Slate & Pemberton 2002). One suggestion is therefore that the correlations are due to associative overdominance, that is, alleles at fitness loci which are in linkage disequilibrium with the screened microsatellites, an idea also known as the ‘local effects hypothesis’ (Hansson & Westerberg 2002).
In conclusion, it is theoretically possible to estimate levels of coancestry and inbreeding from marker data. In practice, this approach is imprecise in several wild populations of outbred organisms studied so far. Greater precision is obtained by using marker data to determine parentage and sibships, yielding a pedigree from which coefficients of coancestry can be calculated and with which fixed effects and a range of variance components can be appropriately assessed.
6. The future
Wild pedigrees form a crucial part of a rich seam of data from individual-based projects, analyses of which are likely to stretch for years into the future. There is a correct pedigree for every individual in a population and it is well worth striving to ascertain that pedigree since the information allows better downstream analysis in every way. The main way to resolve pedigrees well is to sample individuals as completely as possible and to use a sufficiently informative panel of markers. Where retrospective social pedigrees cannot be corrected through molecular genetics, or populations are just too large for detailed molecular genetic analysis of all individuals to be practical, the tools are now available to allow estimation of bias and imprecision using pedigree error rates obtained from testing a sample of individuals.
The majority of wild pedigrees analysed to date are for small birds or large mammals, reflecting the relative ease with which individuals can be studied in these groups. In the future, it is to be hoped that the ingenuity of fieldworkers and the power of molecular genetics can greatly expand the taxonomic range of studies to enable exploration of patterns of quantitative genetic variation and inbreeding depression across a wider range of life histories and breeding systems than currently available.
Only some rather general applications of wild pedigrees have been outlined above, but there are more potential topics for investigation in the future. Given enough markers, wild pedigrees can be used to construct linkage maps for study pedigrees, which can then be used to map polymorphisms and quantitative trait loci (addressed elsewhere in this volume (Slate 2008)) and analyse the rate of decay of linkage disequilibrium with genetic distance. No authors have yet investigated dominance variance in a pedigree for a natural population and it is not yet clear whether any wild pedigree structures lend themselves to such analysis. Much further investigation into the effects of imperfect pedigree information can be expected, including the assumptions that founders and immigrants are outbred and unrelated.
Acknowledgments
Thanks to two referees, both the editors and Tristan Marshall for their comments on the MS.
Footnotes
One contribution of 18 to a Special Issue ‘Evolutionary dynamics of wild populations’.
References
- Almudevar A. A simulated annealing algorithm for maximum likelihood pedigree reconstruction. Theor. Popul. Biol. 2003;63:63–75. doi: 10.1016/s0040-5809(02)00048-5. doi:10.1016/S0040-5809(02)00048-5 [DOI] [PubMed] [Google Scholar]
- Amos W, Wilmer J.W, Fullard K, Burg T.M, Croxall J.P, Bloch D, Coulson T. The influence of parental relatedness on reproductive success. Proc. R. Soc. B. 2001;268:2021–2027. doi: 10.1098/rspb.2001.1751. doi:10.1098/rspb.2001.1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson E.C, Garza J.C. The power of single-nucleotide polymorphisms for large-scale parentage inference. Genetics. 2006;172:2567–2582. doi: 10.1534/genetics.105.048074. doi:10.1534/genetics.105.048074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avise J.C. Sinauer; Sunderland, MA: 2004. Molecular markers, natural history and evolution. [Google Scholar]
- Balloux F, Amos W, Coulson T. Does heterozygosity estimate inbreeding in real populations? Mol. Ecol. 2004;13:3021–3031. doi: 10.1111/j.1365-294X.2004.02318.x. doi:10.1111/j.1365-294X.2004.02318.x [DOI] [PubMed] [Google Scholar]
- Barbará A, Palma-Silva C, Paggi G.M, Bered F, Fay M.F, Lexer C. Cross-species transfer of nuclear microsatellite markers: potenital and limitations. Mol. Ecol. 2007;17:3759–3767. doi: 10.1111/j.1365-294X.2007.03439.x. doi:10.1111/j.1365-294X.2007.03439.x [DOI] [PubMed] [Google Scholar]
- Blouin M.S. DNA-based methods for pedigree reconstruction and kinship analysis in natural populations. Trends Ecol. Evol. 2003;18:503–511. doi:10.1016/S0169-5347(03)00225-8 [Google Scholar]
- Boag P.T, Grant P.R. Heritability of external morphology in Darwin finches. Nature. 1978;274:793–794. doi:10.1038/274793a0 [Google Scholar]
- Bulmer M.G. Inbreeding in great-tit. Heredity. 1973;30:313–325. doi: 10.1038/hdy.1973.41. [DOI] [PubMed] [Google Scholar]
- Burke T, Bruford M.W. DNA fingerprinting in birds. Nature. 1987;327:149–152. doi: 10.1038/327149a0. doi:10.1038/327149a0 [DOI] [PubMed] [Google Scholar]
- Butler K, Field C, Herbinger C.M, Smith B.R. Accuracy, efficiency and robustness of four algorithms allowing full sibship reconstruction from DNA marker data. Mol. Ecol. 2004;13:1589–1600. doi: 10.1111/j.1365-294X.2004.02152.x. doi:10.1111/j.1365-294X.2004.02152.x [DOI] [PubMed] [Google Scholar]
- Charmantier A, Réale D. How do misassigned paternities affect the estimation of heritability in the wild? Mol. Ecol. 2005;14:2839–2850. doi: 10.1111/j.1365-294X.2005.02619.x. doi:10.1111/j.1365-294X.2005.02619.x [DOI] [PubMed] [Google Scholar]
- Clutton-Brock T.H, editor. Reproductive success. Studies of individual variation in contrasting breeding systems. University of Chicago press; Chicago, IL: 1988. [Google Scholar]
- Clutton-Brock T.H. Behavioral ecology—breeding together: kin selection and mutualism in cooperative vertebrates. Science. 2002;296:69–72. doi: 10.1126/science.296.5565.69. doi:10.1126/science.296.5565.69 [DOI] [PubMed] [Google Scholar]
- Cockburn A, Osmond H.L, Mulder R.A, Green D.J, Double M.C. Divorce, dispersal and incest avoidance in the cooperatively breeding superb fairy-wren Malurus cyaneus. J. Anim. Ecol. 2003;72:189–202. doi:10.1046/j.1365-2656.2003.00694.x [Google Scholar]
- Coltman D.W. Testing marker-based estimates of heritability in the wild. Mol. Ecol. 2005;14:2593–2599. doi: 10.1111/j.1365-294X.2005.02600.x. doi:10.1111/j.1365-294X.2005.02600.x [DOI] [PubMed] [Google Scholar]
- Coltman D.W, Slate J. Microsatellite measures of inbreeding: a meta-analysis. Evolution. 2003;57:971–983. doi: 10.1111/j.0014-3820.2003.tb00309.x. [DOI] [PubMed] [Google Scholar]
- Coltman D.W, Bowen W.D, Wright J.M. Male mating success in an aquatically mating pinniped, the harbour seal (Phoca vitulina), assessed by microsatellite DNA markers. Mol. Ecol. 1998;7:627–638. doi: 10.1046/j.1365-294x.1998.00373.x. doi:10.1046/j.1365-294x.1998.00373.x [DOI] [PubMed] [Google Scholar]
- Coltman D.W, Pilkington J.G, Smith J.A, Pemberton J.M. Parasite-mediated selection against inbred Soay sheep in a free-living, island population. Evolution. 1999;53:1259–1267. doi: 10.1111/j.1558-5646.1999.tb04538.x. doi:10.2307/2640828 [DOI] [PubMed] [Google Scholar]
- Coltman D.W, Pilkington J, Kruuk L.E.B, Wilson K, Pemberton J.M. Positive genetic correlation between parasite resistance and body size in a free-living ungulate population. Evolution. 2001;55:2116–2125. doi: 10.1111/j.0014-3820.2001.tb01326.x. [DOI] [PubMed] [Google Scholar]
- Coltman D.W, O'Donoghue P, Hogg J.T, Festa-Bianchet M. Selection and genetic (co)variance in bighorn sheep. Evolution. 2005;59:1372–1382. doi:10.1111/j.0014-3820.2005.tb01786.x [PubMed] [Google Scholar]
- Coulson T.N, Pemberton J.M, Albon S.D, Beaumont M, Marshall T.C, Slate J, Guinness F.E, Clutton-Brock T.H. Microsatellites reveal heterosis in red deer. Proc. R. Soc. B. 1998;265:489–495. doi: 10.1098/rspb.1998.0321. doi:10.1098/rspb.1998.0321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csilléry K, Johnson T, Beraldi D, Clutton-Brock T, Coltman D, Hansson B, Spong G, Pemberton J.M. Performance of marker-based relatedness estimators in natural populations of outbred vertebrates. Genetics. 2006;173:2091–2101. doi: 10.1534/genetics.106.057331. doi:10.1534/genetics.106.057331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakin E.E, Avise J.C. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–509. doi: 10.1038/sj.hdy.6800545. doi:10.1038/sj.hdy.6800545 [DOI] [PubMed] [Google Scholar]
- Duchesne P, Castric T, Bernatchez L. Pasos (parental allocation of singles in open systems): a computer program for individual parental allocation with missing parents. Mol. Ecol. Notes. 2005;5:701–704. doi:10.1111/j.1471-8286.2005.01001.x [Google Scholar]
- Emery A.M, Wilson I.J, Craig S, Boyle P.R, Noble L.R. Assignment of paternity groups without access to parental genotypes: multiple mating and developmental plasticity in squid. Mol. Ecol. 2001;10:1265–1278. doi: 10.1046/j.1365-294x.2001.01258.x. doi:10.1046/j.1365-294X.2001.01258.x [DOI] [PubMed] [Google Scholar]
- Fernandez J, Toro M.A. A new method to estimate relatedness from molecular markers. Mol. Ecol. 2006;15:1657–1667. doi: 10.1111/j.1365-294X.2006.02873.x. doi:10.1111/j.1365-294X.2006.02873.x [DOI] [PubMed] [Google Scholar]
- Foerster K, Coulson T, Sheldon B.C, Pemberton J.M, Clutton-Brock T.H, Kruuk L.E.B. Sexually antagonistic genetic variation for fitness in red deer. Nature. 2007;447:1107–1110. doi: 10.1038/nature05912. doi:10.1038/nature05912 [DOI] [PubMed] [Google Scholar]
- Frentiu F.D, Clegg S.M, Chittock J, Burke T, Blows M.W, Owens I.P.F. Pedigree-free animal-models: the relatedness matrix reloaded. Proc. R. Soc. B. 2008;275:639–647. doi: 10.1098/rspb.2007.1032. doi:10.1098/rspb.2007.1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garant D, Kruuk L.E.B. How to use molecular marker data to measure evolutionary parameters in wild populations. Mol. Ecol. 2005;14:1843–1859. doi: 10.1111/j.1365-294X.2005.02561.x. doi:10.1111/j.1365-294X.2005.02561.x [DOI] [PubMed] [Google Scholar]
- Griffin A.S, West S.A. Kin selection: fact and fiction. Trends Ecol. Evol. 2002;17:15–21. doi:10.1016/S0169-5347(01)02355-2 [Google Scholar]
- Griffin A.S, West S.A. Kin discrimination and the benefit of helping in cooperatively breeding vertebrates. Science. 2003;302:634–636. doi: 10.1126/science.1089402. doi:10.1126/science.1089402 [DOI] [PubMed] [Google Scholar]
- Griffin A.S, Pemberton J.M, Brotherton P.N.M, McIlrath G, Gaynor D, Kansky R, O'Riain J, Clutton-Brock T.H. A genetic analysis of breeding success in the cooperative meerkat (Suricata suricatta) Behav. Ecol. 2003;14:472–480. doi:10.1093/beheco/arg040 [Google Scholar]
- Griffith S.C, Owens I.P.F, Thuman K.A. Extra pair paternity in birds: a review of interspecific variation and adaptive function. Mol. Ecol. 2002;11:2195–2212. doi: 10.1046/j.1365-294x.2002.01613.x. doi:10.1046/j.1365-294X.2002.01613.x [DOI] [PubMed] [Google Scholar]
- Hadfield J.D, Richardson D.S, Burke T. Towards unbiased parentage assignment: combining genetic, behavioural and spatial data in a Bayesian framework. Mol. Ecol. 2006;15:3715–3730. doi: 10.1111/j.1365-294X.2006.03050.x. doi:10.1111/j.1365-294X.2006.03050.x [DOI] [PubMed] [Google Scholar]
- Hamilton W.D. Genetical evolution of social behaviour I. J. Theor. Biol. 1964;7:1–16. doi: 10.1016/0022-5193(64)90038-4. doi:10.1016/0022-5193(64)90038-4 [DOI] [PubMed] [Google Scholar]
- Handley L.J.L, Perrin N. Advances in our understanding of mammalian sex-biased dispersal. Mol. Ecol. 2007;16:1559–1578. doi: 10.1111/j.1365-294X.2006.03152.x. doi:10.1111/j.1365-294X.2006.03152.x [DOI] [PubMed] [Google Scholar]
- Hansson B. Marker-based relatedness predicts egg-hatching failure in great reed warblers. Conserv. Genet. 2004;5:339–348. doi:10.1023/B:COGE.0000031143.51063.cb [Google Scholar]
- Hansson B, Westerberg L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002;11:2467–2474. doi: 10.1046/j.1365-294x.2002.01644.x. doi:10.1046/j.1365-294X.2002.01644.x [DOI] [PubMed] [Google Scholar]
- Hansson B, Bensch S, Hasselquist D, Akesson M. Microsatellite diversity predicts recruitment of sibling great reed warblers. Proc. R. Soc. B. 2001;268:1287–1291. doi: 10.1098/rspb.2001.1640. doi:10.1098/rspb.2001.1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B, Jack L, Christians J.K, Pemberton J.M, Akesson M, Westerdahl H, Bensch S, Hasselquist D. No evidence for inbreeding avoidance in a great reed warbler population. Behav. Ecol. 2007;18:157–164. doi:10.1093/beheco/arl062 [Google Scholar]
- Hill W.G, Babiker H.A, Ranford-Cartwright L.C, Walliker D. Estimation of inbreeding coefficients from genotypic data on multiple alleles, and application to estimation of clonality in malaria parasites. Genet. Res. 1995;65:53–61. doi: 10.1017/s0016672300033000. [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J, Wilson V, Thein S.L. Hypervariable minisatellite regions in human DNA. Nature. 1985a;314:67–73. doi: 10.1038/314067a0. doi:10.1038/314067a0 [DOI] [PubMed] [Google Scholar]
- Jeffreys A.J, Wilson V, Thein S.L. Individual-specific ‘fingerprints’ of human DNA. Nature. 1985b;316:76–79. doi: 10.1038/316076a0. doi:10.1038/316076a0 [DOI] [PubMed] [Google Scholar]
- Jones A.G, Ardren W.R. Methods of parentage analysis in natural populations. Mol. Ecol. 2003;12:2511–2523. doi: 10.1046/j.1365-294x.2003.01928.x. doi:10.1046/j.1365-294X.2003.01928.x [DOI] [PubMed] [Google Scholar]
- Kalinowski S.T, Taper M.L, Marshall T.C. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. doi:10.1111/j.1365-294X.2007.03089.x [DOI] [PubMed] [Google Scholar]
- Keller L.F. Inbreeding and its fitness effects in an insular population of song sparrows (Melospiza melodia) Evolution. 1998;52:240–250. doi: 10.1111/j.1558-5646.1998.tb05157.x. doi:10.2307/2410939 [DOI] [PubMed] [Google Scholar]
- Keller L.F, Arcese P. No evidence for inbreeding avoidance in a natural population of song sparrows (Melospiza melodia) Am. Nat. 1998;152:380–392. doi: 10.1086/286176. doi:10.1086/286176 [DOI] [PubMed] [Google Scholar]
- Keller L.F, Waller D.M. Inbreeding effects in wild populations. Trends Ecol. Evol. 2002;17:230–241. doi:10.1016/S0169-5347(02)02489-8 [Google Scholar]
- Keller L.F, Arcese P, Smith J.N.M, Hochachka W.M, Stearns S.C. Selection against inbred song sparrows during a natural population bottleneck. Nature. 1994;372:356–357. doi: 10.1038/372356a0. doi:10.1038/372356a0 [DOI] [PubMed] [Google Scholar]
- Keller L.F, Grant P.R, Grant B.R, Petren K. Heritability of morphological traits in Darwin's finches: misidentified paternity and maternal effects. Heredity. 2001;87:325–336. doi: 10.1046/j.1365-2540.2001.00900.x. doi:10.1046/j.1365-2540.2001.00900.x [DOI] [PubMed] [Google Scholar]
- Keller L.F, Grant P.R, Grant B.R, Petren K. Environmental conditions affect the magnitude of inbreeding depression in survival of Darwin's finches. Evolution. 2002;56:1229–1239. doi: 10.1111/j.0014-3820.2002.tb01434.x. doi:10.1111/j.0014-3820.2002.tb01434.x [DOI] [PubMed] [Google Scholar]
- Koenig W.D, Haydock J. Incest and incest avoidance. In: Koenig W, Dickinson J, editors. Ecology and evolution of cooperative breeding in brids. Cambridge University Press; Cambridge, UK: 2004. pp. 142–156. [Google Scholar]
- Kruuk L.E.B. Estimating genetic parameters in natural populations using the ‘animal model’. Phil. Trans. R. Soc. B. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. doi:10.1098/rstb.2003.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L.E.B, Hadfield J.D. How to separate genetic and environmental causes of similarity between relatives. J. Evol. Biol. 2007;20:1890–1903. doi: 10.1111/j.1420-9101.2007.01377.x. doi:10.1111/j.1420-9101.2007.01377.x [DOI] [PubMed] [Google Scholar]
- Kruuk L.E.B, Clutton-Brock T.H, Slate J, Pemberton J.M, Brotherstone S, Guinness F.E. Heritability of fitness in a wild mammal population. Proc. Natl Acad. Sci. USA. 2000;97:698–703. doi: 10.1073/pnas.97.2.698. doi:10.1073/pnas.97.2.698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L.E.B, Sheldon B.C, Merila J. Severe inbreeding depression in collared flycatchers (Ficedula albicollis) Proc. R. Soc. B. 2002a;269:1581–1589. doi: 10.1098/rspb.2002.2049. doi:10.1098/rspb.2002.2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruuk L.E.B, Slate J, Pemberton J.M, Brotherstone S, Guinness F, Clutton-Brock T. Antler size in red deer: heritability and selection but no evolution. Evolution. 2002b;56:1683–1695. doi: 10.1111/j.0014-3820.2002.tb01480.x. doi:10.1111/j.0014-3820.2002.tb01480.x [DOI] [PubMed] [Google Scholar]
- Kruuk L.E.B, Merila J, Sheldon B.C. When environmental variation short-circuits natural selection. Trends Ecol. Evol. 2003;18:207–209. doi:10.1016/S0169-5347(03)00073-9 [Google Scholar]
- Litt M, Luty J.A. A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am. J. Hum. Genet. 1989;44:397–401. [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. Sinauer; Sunderland, MA: 1998. Genetics and analysis of quantitative traits. [Google Scholar]
- Markert J.A, Grant P.R, Grant B.R, Keller L.F, Coombs J.L, Petren K. Neutral locus heterozygosity, inbreeding, and survival in Darwin's ground finches (Geospiza fortis and G. scandens) Heredity. 2004;92:306–315. doi: 10.1038/sj.hdy.6800409. doi:10.1038/sj.hdy.6800409 [DOI] [PubMed] [Google Scholar]
- Marr A.B, Arcese P, Hochachka W.M, Reid J.M, Keller L.F. Interactive effects of environmental stress and inbreeding on reproductive traits in a wild bird population. J. Anim. Ecol. 2006;75:1406–1415. doi: 10.1111/j.1365-2656.2006.01165.x. doi:10.1111/j.1365-2656.2006.01165.x [DOI] [PubMed] [Google Scholar]
- Marshall T.C, Slate J, Kruuk L.E.B, Pemberton J.M. Statistical confidence for likelihood-based paternity inference in natural populations. Mol. Ecol. 1998;7:639–655. doi: 10.1046/j.1365-294x.1998.00374.x. doi:10.1046/j.1365-294x.1998.00374.x [DOI] [PubMed] [Google Scholar]
- Merilä J, Sheldon B.C, Kruuk L.E.B. Explaining statis: microevolutionary studies in natural populations. Genetica. 2001;112–113:199–222. doi:10.1023/A:1013391806317 [PubMed] [Google Scholar]
- Milner J.M, Pemberton J.M, Brotherstone S, Albon S.D. Estimating variance components and heritabilities in the wild: a case study using the ‘animal model’ approach. J. Evol. Biol. 2000;13:804–813. doi:10.1046/j.1420-9101.2000.00222.x [Google Scholar]
- Morrissey M.B, Wilson A.J, Pemberton J.M, Ferguson M. A framework for power and sensitivity analyses for quantitative genetic studies of natural populations, and case studies in Soay sheep (Ovis aries) J. Evol. Biol. 2007;20:2309–2321. doi: 10.1111/j.1420-9101.2007.01412.x. doi:10.1111/j.1420-9101.2007.01412.x [DOI] [PubMed] [Google Scholar]
- Mulder R.A, Dunn P.O, Cockburn A, Lazenbycohen K.A, Howell M.J. Helpers liberate female fairy-wrens from constraints on extra-pair mate choice. Proc. R. Soc. B. 1994;255:223–229. doi:10.1098/rspb.1994.0032 [Google Scholar]
- O'Connor K.D, Marr A.B, Arcese P, Keller L.F, Jeffery K.J, Bruford M.W. Extra-pair fertilization and effective population size in the song sparrow Melospiza melodia. J. Avian Biol. 2006;37:572–578. doi:10.1111/j.2006.0908-8857.03681.x [Google Scholar]
- Oliehoek P.A, Windig J.J, van Arendonk J.A.M, Bijma P. Estimating relatedness between individuals in general populations with a focus on their use in conservation programs. Genetics. 2006;173:483–496. doi: 10.1534/genetics.105.049940. doi:10.1534/genetics.105.049940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overall A.D.J, Byrne K.A, Pilkington J.G, Pemberton J.M. Heterozygosity, inbreeding and neonatal traits in Soay sheep on St Kilda. Mol. Ecol. 2005;14:3383–3393. doi: 10.1111/j.1365-294X.2005.02682.x. doi:10.1111/j.1365-294X.2005.02682.x [DOI] [PubMed] [Google Scholar]
- Parker P.G, Snow A.A, Schug M.D, Booton G.C, Fuerst P.A. What molecules can tell us about populations: choosing and using a molecular marker. Ecology. 1998;79:361–382. [Google Scholar]
- Pärt T. Problems with testing inbreeding avoidance: the case of the collared flycatcher. Evolution. 1996;50:1625–1630. doi: 10.1111/j.1558-5646.1996.tb03934.x. doi:10.2307/2410898 [DOI] [PubMed] [Google Scholar]
- Pemberton J.M, Albon S.D, Guinness F.E, Clutton-Brock T.H, Dover G.A. Behavioral estimates of male mating success tested by DNA fingerprinting in a polygynous mammal. Behav. Ecol. 1992;3:66–75. doi:10.1093/beheco/3.1.66 [Google Scholar]
- Pemberton J.M, Coltman D.W, Smith J.A, Pilkington J.G. Molecular analysis of a promiscuous, fluctuating mating system. Biol. J. Linn. Soc. 1999;68:289–301. doi:10.1006/bijl.1999.0342 [Google Scholar]
- Phillips C, et al. Evaluaton of the Genplex SNP typing system and a 49plex forensic marker panel. Foren. Sci. Int. Genet. 2007;1:180–185. doi: 10.1016/j.fsigen.2007.02.007. doi:10.1016/j.fsigen.2007.02.007 [DOI] [PubMed] [Google Scholar]
- Queller D.C, Goodnight K.F. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. doi:10.2307/2409206 [DOI] [PubMed] [Google Scholar]
- Quinn J.L, Charmantier A, Garant D, Sheldon B.C. Data depth, data completeness, and their influence on quantitative genetic estimation in two contrasting bird populations. J. Evol. Biol. 2006;19:994–1002. doi: 10.1111/j.1420-9101.2006.01081.x. doi:10.1111/j.1420-9101.2006.01081.x [DOI] [PubMed] [Google Scholar]
- Reusch T.B.H, Haberli M.A, Aeschlimann P.B, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300–302. doi: 10.1038/35104547. doi:10.1038/35104547 [DOI] [PubMed] [Google Scholar]
- Richdale L.E. Clarendon press; Oxford, UK: 1957. A population study of penguins. [Google Scholar]
- Ritland K. Estimators for pairwise relatedness and individual inbreeding coefficients. Genet. Res. 1996a;67:175–185. [Google Scholar]
- Ritland K. Marker-based method for inferences about quantitative inheritance in natural populations. Evolution. 1996b;50:1062–1073. doi: 10.1111/j.1558-5646.1996.tb02347.x. doi:10.2307/2410647 [DOI] [PubMed] [Google Scholar]
- Ritland K. Marker-inferred relatedness as a tool for detesting heritability in nature. Mol. Ecol. 2000;9:1195–1204. doi: 10.1046/j.1365-294x.2000.00971.x. doi:10.1046/j.1365-294x.2000.00971.x [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ramilo S.T, Toro M.A, Caballero A, Fernandez J. The accuracy of a heritability estimator using molecular information. Conserv. Genet. 2007;8:1189–1198. doi:10.1007/s10592-006-9273-z [Google Scholar]
- Rohrer G.A, Freking B.A, Nonneman D. Single nucleotide polymorphisms for pig identification and parentage exclusion. Anim. Genet. 2007;38:253–258. doi: 10.1111/j.1365-2052.2007.01593.x. doi:10.1111/j.1365-2052.2007.01593.x [DOI] [PubMed] [Google Scholar]
- Sinervo A, Zamudio K.R. The evolution of alternative reproductive strategies: fitness differential, heritability, and genetic correlation between the sexes. J. Hered. 2001;92:198–205. doi: 10.1093/jhered/92.2.198. doi:10.1093/jhered/92.2.198 [DOI] [PubMed] [Google Scholar]
- Slate J. Robustness of linkage maps in natural populations: a simulation study. Proc. R. Soc. B. 2008;275:695–702. doi: 10.1098/rspb.2007.0948. doi:10.1098/rspb.2007.0948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slate J, Pemberton J.M. Comparing molecular measures for detecting inbreeding depression. J. Evol. Biol. 2002;15:20–31. doi:10.1046/j.1420-9101.2002.00373.x [Google Scholar]
- Slate J, David P, Dodds K.G, Veenvliet B.A, Glass B.C, Broad T.E, McEwan J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: theoretical expectations and empirical data. Heredity. 2004;93:255–265. doi: 10.1038/sj.hdy.6800485. doi:10.1038/sj.hdy.6800485 [DOI] [PubMed] [Google Scholar]
- Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989;17:6463–6471. doi: 10.1093/nar/17.16.6463. doi:10.1093/nar/17.16.6463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.C. The estimation of genetic relationships using molecular markers and their efficiency in estimating heritability in natural populations. Phil. Trans. R. Soc. B. 2005;360:1457–1467. doi: 10.1098/rstb.2005.1675. doi:10.1098/rstb.2005.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.C, Hill W.G. Sibship reconstruction in hierarchical population structures using Markov chain Monte Carlo techniques. Genet. Res. 2002;79:227–234. doi: 10.1017/s0016672302005669. doi:10.1017/S0016672302005669 [DOI] [PubMed] [Google Scholar]
- Thomas S.C, Coltman D.W, Pemberton J.M. The use of marker-based relationship information to estimate the heritability of body weight in a natural population: a cautionary tale. J. Evol. Biol. 2002;15:92–99. doi:10.1046/j.1420-9101.2002.00372.x [Google Scholar]
- Van de Casteele T, Galbusera P, Matthysen E. A comparison of microsatellite-based pairwise relatedness estimators. Mol. Ecol. 2001;10:1539–1549. doi: 10.1046/j.1365-294x.2001.01288.x. doi:10.1046/j.1365-294X.2001.01288.x [DOI] [PubMed] [Google Scholar]
- Wang J.L. Sibship reconstruction from genetic data with typing errors. Genetics. 2004;166:1963–1979. doi: 10.1534/genetics.166.4.1963. doi:10.1534/genetics.166.4.1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J.L, May P.E. Abundant class of human DNA polymorphisms which can be typed using the polymerase chain reaction. Am. J. Hum. Genet. 1989;44:388–396. [PMC free article] [PubMed] [Google Scholar]
- Wetton J.H, Carter R.E, Parkin D.T, Walters D. Demographic study of a wild house sparrow population by DNA fingerprinting. Nature. 1987;327:147–149. doi: 10.1038/327147a0. doi:10.1038/327147a0 [DOI] [PubMed] [Google Scholar]
- Wilson A.J, McDonald G, Moghadam H.K, Herbinger C.M, Ferguson M.M. Marker-assisted estimation of quantitative genetic parameters in rainbow trout, Oncorhynchus mykiss. Genet. Res. 2003;81:145–156. doi: 10.1017/s0016672302006055. doi:10.1017/S0016672302006055 [DOI] [PubMed] [Google Scholar]
- Wilson A.J, Kruuk L.E.B, Coltman D.W. Ontogenetic patterns in heritable variation for body size: using random regression models in a wild ungulate population. Am. Nat. 2005;166:E177–E192. doi: 10.1086/497441. doi:10.1086/497441 [DOI] [PubMed] [Google Scholar]
- Wilson A.J, Pemberton J.M, Pilkington J.G, Coltman D.W, Mifsud D.V, Clutton-Brock T.H, Kruuk L.E.B. Environmental coupling of selection and heritability limits evolution. PLoS Biol. 2006;4:1270–1275. doi: 10.1371/journal.pbio.0040216. doi:10.1371/journal.pbio.0040216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A.J, Pemberton J.M, Pilkington J.G, Clutton-Brock T.H, Coltman D.W, Kruuk L.E.B. Quantitative genetics of growth and cryptic evolution of body size in an island population. Evol. Ecol. 2007;21:337–356. doi:10.1007/s10682-006-9106-z [Google Scholar]