Abstract
Dematin and adducin are actin-binding proteins located at the spectrin-actin junctions, also called the junctional complex, in the erythrocyte membrane. Here we propose a new model whereby dematin and adducin link the junctional complex to human erythrocyte plasma membrane. Using a combination of surface labeling, immunoprecipitation, and vesicle proteomics approaches, we have identified glucose transporter-1 as the receptor for dematin and adducin in the human erythrocyte membrane. This finding is the first description of a transmembrane protein that binds to dematin and adducin, thus providing a rationale for the attachment of the junctional complex to the lipid bilayer. Because homologues of dematin, adducin, and glucose transporter-1 exist in many non-erythroid cells, we propose that a conserved mechanism may exist that couples sugar and other related transporters to the actin cytoskeleton.
There is considerable interest in the elucidation of the mechanism that governs the linkage of elongated spectrin molecules to the erythrocyte plasma membrane (1–5). The mechanism by which the “head” region of the spectrin dimer, which participates in tetramer formation, binds to the membrane via ankyrin and band 3 has been well characterized (1–3); however, the mechanism that anchors the tail end of the spectrin dimer to the plasma membrane is not completely understood. This region, containing a short actin protofilament and the tail ends of spectrin polypeptides, is often referred to as the “spectrin-actin junction” or the “junctional complex” (4, 5). Protein 4.1R, dematin, and adducin have been directly visualized by immunogold electron microscopy as components of the junctional complex (6). Other proteins that are likely to be present at the junctional complex include tropomyosin, tropomodulin, p55, and calmodulin. Among the junctional complex components, the best characterized membrane-protein interaction involves protein 4.1R (5, 7). Protein 4.1R binds directly to p55 and glycophorin C, a transmembrane protein, and this ternary complex is believed to attach the junctional complex to the plasma membrane (4, 5, 8, 9). Our recent findings from the genetically altered mice suggest that other components of the junctional complex may also perform this crucial function (10, 11).
Dematin and adducin are two cytoskeletal proteins that could perform a function that is similar to the vertical interactions of protein 4.1R. These actin-binding phosphoproteins are located at the erythrocyte junctional complex. Individually they exert modest effects on erythrocyte shape and membrane stability, as revealed by their gene knock-out phenotypes in mice (12–14). Our more recent findings indicate that dematin and adducin serve redundant functions at the junctional complex, and mutant mice lacking both dematin-headpiece domain and β-adducin display severe defects in erythrocyte shape, membrane instability, and hemolysis (10). The presence of mis-shapen and fragmented erythrocytes correlated with their increased osmotic fragility and reduced life span in vivo (10). In addition, we demonstrated that purified dematin can bind to the trypsin-sensitive sites present on human erythrocyte inside-out vesicles in a saturable manner. These observations demonstrated that dematin and adducin together play an essential role in the maintenance of erythrocyte shape and membrane stability. Based on these findings, we have proposed a model where dematin and adducin could function as adaptors by linking the junctional complex to the membrane via a transmembrane receptor. To experimentally validate this model, we carried out a systematic search for the membrane receptor that could bind to dematin and adducin in the human erythrocyte membrane. Here we demonstrate that glucose transporter-1 (GLUT1)2 is one such receptor that binds to both dematin and β-adducin in the human erythrocyte plasma membrane. Our model predicts that GLUT1, a multispan transmembrane receptor, assembles a novel cytoskeletal complex with dematin and β-adducin at the tail ends of spectrin tetramers, and this ternary complex provides a new mechanism for the linkage of the junctional complex to human erythrocyte plasma membrane.
EXPERIMENTAL PROCEDURES
Proteomics of Erythrocyte Inside-out Vesicles (IOVs)—A comprehensive proteomics analysis was performed on highly purified alkali-stripped inside-out vesicles. Freshly collected human erythrocytes from an adult male were washed with PBS to remove the buffy coat, and sequentially passed through two columns containing a mixture of Sigma Cell type 50 and α-cellulose. Ghosts were prepared by hypotonic lysis, and loosely attached ghosts were recovered by tilting the tube to remove the red button. IOVs were prepared by incubation of ghosts in 50 volumes of the IOV buffer (0.1 mm sodium phosphate, pH 7.5, 0.2 mm dithiothreitol, and 0.1 mm EDTA) overnight at 4 °C. IOVs were recovered by centrifugation at 45,000 × g for 30 min and washed with the IOV-resuspension buffer (0.4 mm sodium phosphate, pH 7.5, 0.1 mm EDTA). Peripheral membrane proteins were stripped from the IOVs by incubation in an alkaline solution (0.5 mm EGTA, pH 11.0) at 37 °C for 1 h. The alkali-stripped IOVs were washed and separated from the right side-out vesicles (ROVs) by density gradient ultracentrifugation using a dextrose barrier solution (15, 16). Briefly, the IOVs were diluted 3-fold and gently layered on an equal volume of 4.5% (w/v) dextran solution. The gradient was centrifuged at 100,000 × g for 1 h at 4 °C. The IOVs remained at the top of the barrier solution, and the ROVs pelleted at the bottom. Purified-stripped IOVs were gently recovered, washed with the resuspension buffer, and dissolved in 0.3 mm sodium phosphate, pH 7.5, 0.1 mm dithiothreitol, 0.1 mm EDTA, and 20 mg/ml sucrose. The mouse erythrocyte IOVs were prepared using a similar approach except that the alkali-stripped IOVs were not further purified by density gradient ultracentrifugation. Purified IOVs were quick frozen in liquid nitrogen, and comprehensive proteomics was performed at the Wistar Institute Proteomics Facility in Philadelphia.
Briefly, the IOVs were solubilized in 5× SDS sample buffer and 20 mm tris(2-carboxyethyl)phosphine and run for 4 cm on a one-dimensional 4–12% polyacrylamide gel using a BisTris-based buffer system. Gels were stained using colloidal Coomassie solution, and one 16-μg lane was divided in slices for trypsin digestion. Gel slices were destained, reduced using 20 mm tris(2-carboxyethyl)phosphine, alkylated using 40 mm iodoacetamide, and digested using 20 μl of 0.02 μg/ml modified trypsin in ammonium bicarbonate buffer containing 30% acetonitrile for 18 h at 37 °C. After digestion, the supernatant was removed; peptides trapped in the gel were extracted with an additional aliquot of digestion buffer for 45 min, and both supernatants were pooled. Digests were subjected to nanobore capillary reverse-phase high pressure liquid chromatography, which was interfaced directly with MS/MS analysis on a ThermoFinnigan LTQ FT ICR mass spectrometer. MS/MS data were searched versus the NCBI nr data base using SEQUEST, and results were analyzed using Proteomics Browser. The data shown in supplemental Tables 1 and 2 were obtained from the entire gel lane.
Preparation of Ghosts and Membrane Skeletons—Ghosts were prepared by hypotonic lysis of washed erythrocytes in 5 mm phosphate buffer, pH 8.0, 0.1 mm EDTA, and 0.1 mm phenylmethylsulfonyl fluoride. Triton shells/skeletons were prepared by incubation of the ghosts with 1% Triton X-100 in 50 mm Tris-HCl, pH 7.5, 1.0 mm EDTA, 1.0 mm EGTA, 1.0 mm phenylmethylsulfonyl fluoride, and KCl either at 0.1 or 0.5 m for 60 min at 4 °C. After centrifugation at 55,000 × g for 60 min at 4 °C, the skeletal pellet was rinsed gently with PBS and dissolved in the sample buffer at 37 °C for 30 min.
Surface Labeling and Immunoprecipitation Assay—To identify the dematin-binding receptor(s), co-immunoprecipitation of biotin-labeled erythrocyte membrane proteins was performed. Human erythrocytes (250-μl packed volume) were surface-labeled with 1.0 mg of NHS-biotin in 1.0 ml of PBS for 30 min at 4 °C. Unconjugated biotin was quenched with Tris-HCl solution, and labeled erythrocytes were extensively washed with Tris-buffered saline. Ghosts and IOVs were prepared for the co-immunoprecipitation experiments. IOVs (0.5 mg of total protein) were pre-cleared with 50 μl of a 50% suspension of protein-G-Sepharose beads in the immunoprecipitation (IP) buffer (20 mm Tris-HCl, pH 8.0, 137 mm NaCl, 1.0 mm MgCl2, 1.0 mm CaCl2, 1% Nonidet P-40, and 10% glycerol) for 1 h at 4 °C. IP was performed with 2.0 μg of anti-dematin monoclonal antibody, and protein-G-Sepharose-bound immune complex was eluted, blotted, and probed with the streptavidin-horseradish peroxidase to detect biotin-labeled bands.
Expression of Glucose Transporter-1 in Human Epithelial Cells—To evaluate the interactions of GLUT1 with dematin and adducin, transfections and co-IP assays were performed. For transient transfections, HEK293T cells (human embryonic kidney epithelial) were plated at a density of 2 × 106 cells/plate (10 cm). Plasmids for dematin (dematin 48-kDa/pcDNA3.1) and GLUT1 (GLUT1/pEGFP-C1) were isolated by the Qiagen maxiprep kit and transfected using Lipofectamine 2000 (Invitrogen). After 20 h of transfection, cells were rinsed briefly with PBS and harvested in 1.0 ml of IP buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.5% Nonidet P-40). After cell lysis at 4 °C for 30 min, the lysate was cleared by centrifugation at 15,000 × g for 20 min. For the IP assay, an anti-dematin monoclonal antibody (2.0 μg/sample) was added to the lysate and an anti-CD3 monoclonal antibody was used as a negative control. The immune complex was harvested using protein-G-Sepharose beads, and bound GLUT1 was detected using an anti-GLUT1 rabbit polyclonal antibody. Essentially the same strategy was used to test the binding of dematin with GLUT1cyto4, a chimeric construct, where the large middle cytoplasmic loop of GLUT1 was replaced by the same loop from GLUT4. To investigate the interactions of adducin with GLUT1 and GLUT1cyto4 in HEK293T cells, a FLAG-tagged β-adducin construct (human β-adducin/pCMV-FLAG) was made, and co-IP assays were performed using an anti-FLAG monoclonal antibody conjugated to the beads.
Endogenous Association of Dematin with GLUT1—For the endogenous dematin-GLUT1 complex, human erythrocyte ghosts (400 μl, 1.0 mg of total protein) were mixed with an IP buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 0.5% Nonidet P-40), and the lysate was pre-cleared with 50 μl of 50% protein-G-Sepharose beads for 1 h at 4 °C. Pre-cleared lysate was immunoprecipitated with either 2.5 μg of anti-dematin monoclonal antibody or an unrelated anti-CD3 monoclonal antibody. After incubation for 4.0 h at 4 °C, the immune complex was harvested using 50 μl of a 50% suspension of protein-G-Sepharose beads. To distinguish any signal in the Western blots from the IgG heavy chains, which migrate near the position of GLUT1, additional negative controls with monoclonal antibodies without the lysate were included.
In Vitro Interaction of Dematin with GLUT1—To demonstrate an interaction between GLUT1 and purified dematin, a pulldown assay was designed. Full-length GLUT1 was expressed in the HEK293T cells as described earlier. The GST fusion proteins containing either the full-length dematin or its core domain were expressed in bacteria and purified using glutathione-Sepharose affinity chromatography. The GLUT1-transfected cells were harvested and lysed in the IP buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, and 0.5% Nonidet P-40). The pre-cleared lysates were incubated with purified GST fusion proteins immobilized on the glutathione-Sepharose beads at 4 °C. The beads were recovered by centrifugation and washed, and the bound proteins were analyzed by Western blotting using an anti-GLUT1 polyclonal antibody.
RESULTS
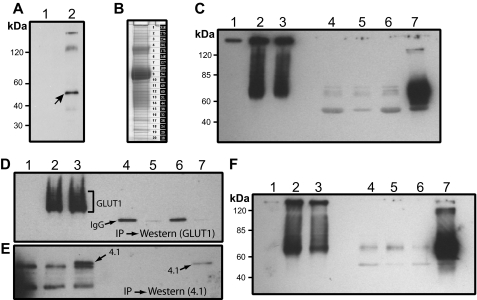
Dematin Binds to a Surface-labeled Protein in Human Erythrocytes—Recently, we reported that native dematin, purified from human erythrocyte ghosts, associated with trypsin-sensitive sites on the cytoplasmic surface of alkali-stripped inside-out vesicles (10). This observation suggested that dematin may bind to a transmembrane protein in the human erythrocyte membrane. To identify this putative receptor, we immunoprecipitated dematin from the surface-labeled human erythrocytes. Freshly collected human erythrocytes were surface-labeled using NHS-biotin. After the removal of free biotin and extensive surface blocking, the labeled erythrocytes were hypotonically ruptured to prepare ghosts and IOVs. Spectrin-actin-depleted IOVs were solubilized in the non-ionic detergent, and dematin was immunoprecipitated using a monoclonal antibody. Biotin-labeled surface membrane proteins that associated with the dematin immune complex were visualized by the streptavidin-peroxidase conjugate. Western blotting revealed specific co-precipitation of an ∼54-kDa protein in the dematin immune complex (Fig. 1A, lane 2, arrow). Two relatively weaker bands migrating in the range of 100–250 kDa were also observed, although the reproducibility of the very top band was not consistent in multiple experiments. These results suggested that dematin binds to an ∼54-kDa membrane protein in the human erythrocyte plasma membrane.
FIGURE 1.
Interaction of dematin with GLUT1 in transfected cells. A, surface labeling of red blood cell membrane proteins with NHS-biotin and co-immunoprecipitation. Lanes 1 and 2, inside-out vesicles with no antibody control or anti-dematin monoclonal antibody, respectively. B, Coomassie-stained gel of purified human IOVs used in the proteomics analysis. C, interaction of dematin and GLUT1 in transfected HEK293T cells. Lane 1, nontransfected cell lysate; lane 2, GLUT1-transfected cell lysate; lane 3, dematin- and GLUT1-transfected cell lysate; lanes 4 and 5, GLUT1-transfected lysate, co-IP with CD3 and dematin monoclonal antibodies, respectively; lanes 6 and 7, GLUT1- and dematin-transfected lysate, co-IP with CD3 and dematin monoclonal antibodies, respectively. Blot was probed with a polyclonal GLUT1 antibody. D, co-IP of protein 4.1 and GLUT1 in HEK293T cells. Lane 1, nontransfected cell lysate; lane 2, GLUT1-transfected cell lysate; lane 3, GLUT1 and protein 4.1R (with Xpress tag) co-transfected cell lysate; lanes 4 and 5, GLUT1-transfected cell lysate IP with CD3 or Xpress mAb, respectively; lanes 6 and 7, GLUT1 and protein 4.1R co-transfected cell lysate IP with CD3 or Xpress mAb, respectively. Blot was probed with a polyclonal GLUT1 antibody. E, same as D and the blot was probed with a polyclonal antibody against protein 4.1R. F, interaction of dematin and GLUT1cyto4 in transfected HEK293T cells. The experimental conditions were the same as in C except that the GLUT1cyto4 was transfected instead of GLUT1.
Identification of the Dematin-binding Protein by Vesicle Proteomics—Because the gel mobility of ∼54-kDa transmembrane protein is near the location where the immunoglobulin heavy chain and the endogenous dematin migrate, it was technically difficult to identify this protein in the dematin immunoprecipitates by direct sequencing of the gel band. Therefore, we performed a comprehensive proteomics of human erythrocyte IOVs with the expectation that this approach might reveal the identities of candidate ∼54-kDa membrane proteins that could potentially bind to dematin. A critical requirement for the success of this approach was the isolation of highly purified membrane vesicles free of peripheral membrane proteins. To accomplish this goal, we first carried out a rigorous removal of leukocytes and platelets from freshly isolated human erythrocytes. After the removal of spectrin and actin from erythrocyte ghosts by multiple low ionic strength extractions, the IOVs were stripped of nearly all peripheral membrane proteins by extraction with the pH 11 solution (17). Finally, the alkali-stripped IOVs were biochemically purified from the ROVs by density gradient centrifugation. The polypeptides of purified IOVs were separated by gel electrophoresis, and bands were sequentially excised for analysis by mass spectrometry (Fig. 1B) as described under “Experimental Procedures.”
The proteomics data, shown in supplemental Table 1, represent 60 relatively abundant proteins identified in the alkali-stripped IOVs of human erythrocytes. Although the proteins are arranged according to the abundance of total MS/MS spectra observed divided by mass or normalized spectral counts (data not shown), the protein ranking is a rough estimate of relative protein abundance. This is because some peptides derived from membrane proteins will be impossible to detect by the methods used here, as they are either too hydrophobic or are heavily glycosylated. Therefore, in principle, some membrane proteins may be missed using this approach. The absence of peptides from proteins generally associated with platelets and leukocytes further confirms the purity of human erythrocyte IOVs used in this study. There are several noteworthy features of the proteomics analysis of human erythrocyte IOVs shown in supplemental Table 1. For example, the stripped IOVs still retain a subpopulation of peripheral membrane proteins such as band 4.2, p55, ankyrin, β-hemoglobin, and the RAB family of small GTPases. Because this subpopulation of peripheral membrane proteins resisted stringent membrane dissociation at pH 11, we speculate that their tight membrane association presumably originates from their lipid modifications in vivo or some as yet unknown covalent modification that anchors them so tightly to the cytoplasmic face of the plasma membrane. This speculation is consistent with the known lipid modifications of protein 4.2, p55, ankyrin, and small GTPases (18–20). Interestingly, ubiquitin was present in all gel bands, and based on the spectral counts, it appears that ubiquitin is an abundant polypeptide in the mature human erythrocyte membrane IOVs. The reason for the tight ubiquitin association with membrane proteins in the terminally differentiated erythrocytes is not clear at this stage, although it is now known that mono-ubiquitination is associated with signaling rather than degradation pathways.
Several recent studies have reported detailed proteomics of the membrane and cytosolic fractions of human erythrocytes (21, 22). Overall, our proteomic data are consistent with these studies. However, we believe that the proteomic data shown in this study constitute the first report where alkali-stripped IOVs were biochemically purified from ROVs for mass spectrometry. Because the contamination of ROVs in the conventional preparations of IOVs generally contributes to the presence of trapped peripheral membrane proteins, our results provide the most rigorous identification of the transmembrane and tightly associated lipid-modified peripheral membrane proteins in human erythrocytes. Importantly, the vesicle proteomic data revealed at least five proteins in the vicinity of the 54-kDa region of human IOVs. These proteins include the solute carrier family 2 (glucose transporter-1), solute carrier family 29 (nucleoside transporter-1 or ENT1), Scianna blood group antigen, solute carrier family 43 (member 3), and the Rh protein (supplemental Table 1). Based on the known properties of these proteins, and some initial observations on the obesity phenotype of dematin headpiece knock-out mice,3 GLUT1 emerged as a likely candidate receptor suitable for binding to dematin and adducin in human erythrocytes.
Dematin Binds to GLUT1—To evaluate the biochemical interaction between dematin and GLUT1, a transfection and co-immunoprecipitation-based approach was used. Full-length dematin (48-kDa subunit) and full-length GLUT1 were transiently transfected into the human embryonic kidney (HEK293T) epithelial cells, and dematin was immunoprecipitated using a monoclonal antibody. An unrelated monoclonal antibody against the CD3 antigen was used as a negative control. The dematin immune complex was examined for the presence of GLUT1 by Western blotting using a polyclonal antibody against GLUT1. The expression of transfected GLUT1 alone (Fig. 1C, lane 2) and upon co-transfection with dematin (Fig. 1C, lane 3) was confirmed by the presence of a diffuse band, and higher molecular mass polypeptides, presumably because of its glycosylation and oligomerization in the epithelial cells. In the transfected cells with GLUT1 alone, immunoprecipitation with either the anti-CD3 or anti-dematin monoclonal antibody did not show any GLUT1 signal, indicating the absence of any significant amount of endogenous dematin in the HEK293T cells (Fig. 1C, lanes 4 and 5, respectively). In contrast, the immunoprecipitation of dematin from cells that were co-transfected with GLUT1 and dematin showed a significant amount of GLUT1 in the dematin immune complex (Fig. 1C, lane 7). As expected, the anti-CD3 immune complex was negative for the presence of GLUT1 (Fig. 1C, lane 6). These results suggest a specific biochemical interaction between dematin and GLUT1 under these conditions. To further demonstrate the specificity of dematin-GLUT1 interaction, we transfected an Xpress-tagged protein 4.1R construct and GLUT1 in HEK293T cells. We have previously shown that this 4.1R construct expresses both 80- and 135-kDa isoforms of protein 4.1R (23). Co-immunoprecipitation of protein 4.1 using an Xpress monoclonal, under the same conditions as used for the dematin-GLUT1 interaction, did not immunoprecipitate GLUT1 (Fig. 1, D and E).
The membrane topology model of GLUT1 predicts several protein segments facing the cytosolic compartment. These segments include a 12-amino acid N-terminal domain, a major 64-amino acid cytoplasmic loop, a 42-amino acid C-terminal domain, and four short internal loops of 8–10 amino acids (24). Because this topology model is conserved among class I glucose transporters and other related transporters, we investigated whether the interaction of dematin with GLUT1 was sensitive to the specificity of the 64-amino acid cytoplasmic loop of GLUT1. A chimeric full-length GLUT1 construct where the 64-amino acid cytoplasmic loop of GLUT1 was replaced with a similar loop from the insulin-sensitive GLUT4 transporter, which is expressed predominantly in the muscle and adipocytes, was tested for binding to dematin. This chimeric protein, termed GLUT1cyto4, bound dematin as efficiently as GLUT1 (Fig. 1F, lane 7). This result suggests that dematin interacts either with the 64-amino acid cytoplasmic loops of both GLUT1 and GLUT4 or it binds to GLUT1 on a site other than the 64-amino acid cytoplasmic loop. To distinguish this possibility, we engineered a His-tagged dematin construct (48-kDa subunit) and the GST-tagged 64-amino acid cytoplasmic loop of GLUT1 as fusion proteins. Pulldown assays established that recombinant dematin can directly bind to the 64-amino acid cytoplasmic loop fusion protein of GLUT1 (data reviewed but not shown). To quantify this interaction, we developed a sandwich-type Biosensor assay. An antibody against the His tag was immobilized to the CM5 Biosensor chip, and the His-tagged dematin was conjugated to the chip surface. The GST-loop fusion protein of GLUT1 was passed over the immobilized dematin, and its binding affinity was estimated to be ∼145 nm. The GST was used as a negative control. Future studies designed to quantify the interaction of full-length GLUT1 with limited truncations of both N- and C-terminal segments, their chimeras, and in vitro mutagenesis of the intracellular loop would be required to precisely determine the molecular basis of dematin-GLUT1 interaction in human erythrocytes.
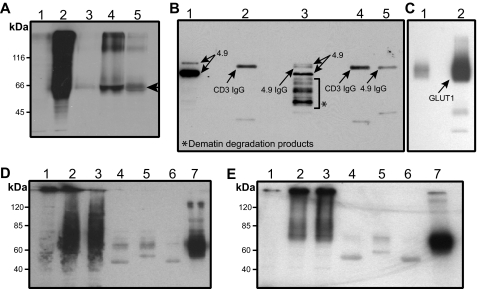
To further map the GLUT1-binding site within dematin, a pulldown assay was designed. Full-length dematin and its core domain were expressed in bacteria and purified as GST fusion proteins. It is noteworthy that the core domain of dematin contains a PEST sequence, and therefore the recombinant core domain degrades rapidly under in vitro expression conditions (11). Full-length GLUT1 was expressed in HEK293T cells, and the lysate was incubated with purified GST fusion proteins of dematin-immobilized on the glutathione-Sepharose beads. Western blotting showed that GLUT1 binds to both full-length and the core domain of dematin (Fig. 2A, lanes 4 and 5, respectively). The core domain, however, generated a relatively weaker signal suggesting that the full-length dematin polypeptide provides a more robust binding surface for GLUT1 under these conditions.
FIGURE 2.
GLUT1 interactions with dematin and β-adducin. A, in vitro interaction of dematin with GLUT1. Lane 1, nontransfected HEK293T cell lysate; lane 2, GLUT1-transfected cell lysate; lanes 3–5, GLUT1 pulldown (arrowhead); lane 3, GST alone; lane 4, dematin full-length; and lane 5, dematin core domain. Blot was probed with an anti-GLUT1 polyclonal antibody. B, dematin Western blot of endogenous co-IP of dematin and GLUT1 from human erythrocyte ghosts. Lane 1, pre-cleared lysate; lane 2, co-IP with an unrelated monoclonal antibody CD3; lane 3, co-IP with anti-dematin monoclonal antibody; and lanes 4 and 5, no ghosts lysate controls showing signals from unrelated and dematin monoclonal antibodies. It is noteworthy that dematin is highly prone to degradation, particularly in the presence of non-ionic detergents, because of the presence of a PEST motif. The degradation products, as seen in lane 3, have been sequenced to confirm their origin from the dematin protein. C, GLUT1 Western blot of endogenous co-IP of dematin and GLUT1 from human erythrocyte ghosts. Lanes 1 and 2 correspond to lanes 2 and 3 of dematin blot in B. D, interaction of FLAG-tagged β-adducin and GLUT1 in transfected HEK293T cells. Lane 1, nontransfected cell lysate; lane 2, GLUT1-transfected cell lysate; lane 3, β-adducin and GLUT1-transfected cell lysate; lanes 4 and 5, GLUT1-transfected lysate, co-IP with anti-CD3 and FLAG antibodies, respectively; and lanes 6 and 7, GLUT1- and β-adducin-transfected lysate, co-IP with CD3 and FLAG antibodies, respectively. Blot was probed with an anti-GLUT1 polyclonal antibody. E, interaction of FLAG-tagged β-adducin with GLUT1cyto4 in transfected HEK293T cells. The experimental conditions were the same as in D except that GLUT1cyto4 was transfected instead of GLUT1.
To demonstrate an endogenous interaction between dematin and GLUT1, the dematin-GLUT1 complex was immunoprecipitated from human erythrocyte ghosts. It is noteworthy that the immunoprecipitation of erythrocyte membrane proteins is often difficult, generally requiring a large amount of the antibody and thus generating excessive nonspecific background signals. With this limitation in perspective, we optimized the conditions to immunoprecipitate the dematin-GLUT1 complex from the detergent-solubilized ghosts using an anti-dematin monoclonal antibody. To distinguish the GLUT1 signal from the IgG heavy chain, which migrates at a similar position as GLUT1, two additional controls without lysates were included. The anti-CD3 monoclonal antibody was used as a negative control. Western blots of GLUT1 and dematin were probed using an anti-GLUT1 polyclonal antibody and an affinity-purified anti-dematin polyclonal antibody. The representative blots are shown in Fig. 2B (dematin blot) and Fig. 2C (GLUT1 blot). The presence of GLUT1 in the dematin immunoprecipitate (Fig. 2C, lane 2) suggests the existence of dematin-GLUT1 complex in the human erythrocyte plasma membrane. Using monoclonal antibodies against ankyrin and stomatin, we immunoprecipitated ankyrin and stomatin from human erythrocyte ghosts to establish the specificity of endogenous dematin-GLUT1 interaction. Western blotting indicated the absence of GLUT1 in the ankyrin immunoprecipitate, whereas a significant amount of GLUT1 was detected in the stomatin immunoprecipitates (data reviewed but not shown). It is noteworthy here that the absence of stomatin in the dematin immunoprecipitate (data reviewed but not shown) suggests that either a separate population of GLUT1-stomatin complex exists in the human erythrocyte membrane or the detection of stomatin in the dematin immunoprecipitate is not feasible under these conditions. Moreover, published studies have shown that band 3 and actin are not associated with the co-immunoprecipitates of stomatin and GLUT1 (25).
Adducin Also Binds to GLUT1—To investigate whether GLUT1 can independently interact with adducin, we generated a FLAG-tagged construct of full-length human β-adducin for expression in the HEK293T epithelial cells. The GLUT1 and β-adducin constructs were co-expressed in the HEK293T cells, and β-adducin was immunoprecipitated using an anti-FLAG antibody. The anti-CD3 antibody served as a negative control. When β-adducin was immunoprecipitated from the cells overexpressing GLUT1 alone, there was no precipitation of GLUT1 with either the anti-FLAG or the anti-CD3 antibody (Fig. 2D, lanes 4 and 5, respectively). In contrast, when β-adducin was immunoprecipitated from cells that were co-transfected with β-adducin and GLUT1 constructs, a significant amount of GLUT1 was co-precipitated with β-adducin (Fig. 2D, lane 7). Immunoprecipitation of β-adducin also pulled down the chimeric GLUT1cyto4 protein where the large cytoplasmic loop of GLUT1 has been replaced with that of GLUT4 (Fig. 2E, lane 7). Together, these results suggest a biochemical interaction between β-adducin and GLUT1 under these conditions. As the main thrust of this study was on the dematin-GLUT1 interaction, we have not yet examined the existence of the endogenous adducin-GLUT1 complex in the human erythrocytes membranes as well as direct binding of β-adducin with the 64-amino acid cytoplasmic loop of GLUT1.
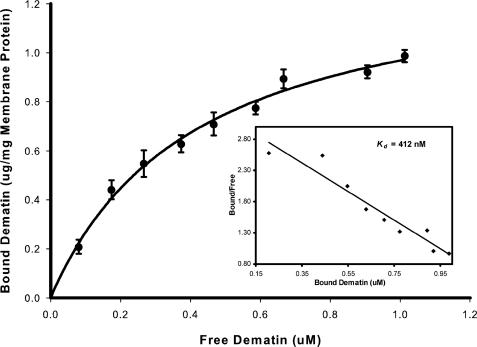
Dematin Binds to Erythrocyte Membrane Receptor with High Affinity—We have recently shown that dematin binds to human erythrocyte membrane vesicles via the trypsin-sensitive binding sites (10). To quantify this interaction, we measured the binding of purified dematin, isolated from human erythrocytes, to alkali-stripped erythrocyte membrane vesicles. Binding of native dematin to membrane vesicles was quantified by a sandwich-based immunoassay as described previously (26). Dematin bound to the stripped membrane vesicles with a Kd of 412 nm (Fig. 3). Scatchard analysis of the binding data indicates a single class of dematin-binding sites located on the cytoplasmic face of membrane vesicles, with 1.4 μg of purified dematin bound per mg of stripped vesicle membrane protein at saturation (Fig. 3). With the assumption that GLUT1 harbors the single class of dematin-binding sites on the human erythrocyte membrane vesicles, and the known abundance of GLUT1 constituting 2–6% of total erythrocyte membrane protein mass (24, 27), our binding measurements suggest a stoichiometric association of dematin trimer with a GLUT1 dimer or tetramer in the human erythrocyte plasma membrane. We have also examined the direct interaction of recombinant β-adducin with human erythrocyte membrane, and we found a saturable binding with the alkali-stripped membrane vesicles (data reviewed but not shown). The β-adducin-membrane binding could not be quantified because of the unavailability of a sensitive quantitative assay for β-adducin. Together, these results indicate that both dematin and adducin can bind to the cytoplasmic surface of human erythrocyte membrane that is devoid of most peripheral membrane proteins.
FIGURE 3.
Binding of dematin to erythrocyte membrane vesicles. Highly purified dematin isolated from human erythrocyte membranes was incubated with alkali-stripped membrane vesicles as described under “Experimental Procedures.” Binding of dematin to membrane vesicles was quantified by a sandwich ELISA as described before (10). Binding data were analyzed using the SigmaPlot software. The inset shows the results of the Scatchard analysis of the binding data yielding a Kd of 412 nm for the interaction of trimeric dematin to the stripped membrane vesicles.
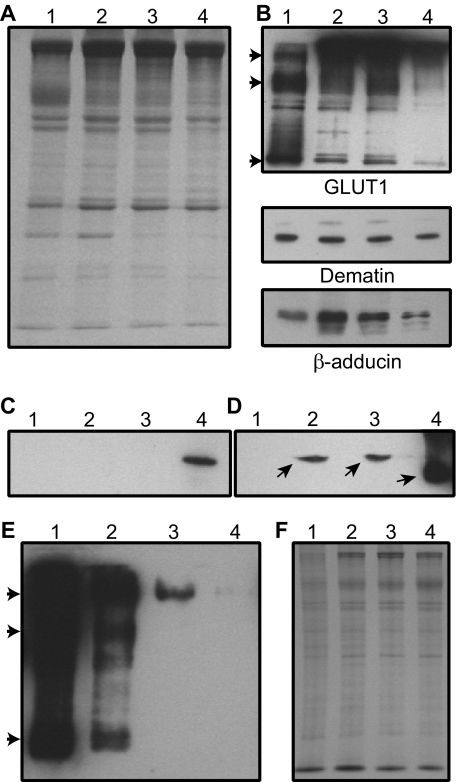
Co-fractionation of Human Erythrocyte Dematin, Adducin, and GLUT1—To determine whether dematin, adducin, and GLUT1 co-distribute during membrane fractionation, human erythrocyte ghosts were solubilized by Triton X-100 in the presence and absence of salt (Fig. 4A, Coomassie stain). Western blotting indicated that a significant amount of GLUT1 remains associated with the membrane skeletons prepared under low ionic strength conditions (Fig. 4B, lane 2). Interestingly, we found that GLUT1 tends to oligomerize into dimer and tetramer forms under these conditions (Fig. 4B, arrowheads). It appears that the oligomeric forms of GLUT1 increase when the membranes are extracted with the Triton X-100 to isolate the membrane skeletons (Fig. 4B). The relative amount of GLUT1 associated with the skeletons remained unchanged when the membrane skeletons were prepared in the presence of 0.1 m KCl (Fig. 4B, lane 3). In fact, the presence of GLUT1 was still detectable when the membrane skeletons were prepared in the presence of 0.5 m KCl (Fig. 4B, lane 4). It is to be noted that the protein loading of ghosts for GLUT1 was reduced by 5-fold (Fig. 4B, lane 1) to minimize the overexposure of the GLUT1 signal in the Western blots. These results suggest that GLUT1, dematin, and adducin form a ternary complex that resists biochemical dissociation upon detergent extraction of the human erythrocyte ghosts. Although it is difficult to precisely quantify the amount of various species of GLUT1 associated with the skeletons, a rough estimate based on the densitometric analysis of bands suggests ∼80% retention of GLUT1 in the membrane skeletons as compared with human erythrocyte ghosts. Because 5-fold less protein was loaded in the ghosts lane (Fig. 4B, lane 1), this amount corresponds to ∼15% retention of GLUT1 in the membrane skeletons. With a conservative estimate of ∼400,000 copies of GLUT1 in each erythrocyte, it would predict ∼30,000 copies of dimeric GLUT1 associated with the membrane skeletons. Together, these observations suggest that there is a sufficient number of copies of GLUT1 to hold the remaining adducin and dematin in the human erythrocyte membrane skeletons.
FIGURE 4.
Distribution of GLUT1, dematin, and β-adducin in human erythrocyte membrane skeletons. A, Coomassie gel shows ghosts (lane 1), skeletons (lane 2), skeletons prepared with 0.1 m KCl (lane 3), and skeletons prepared with 0.5 m KCl (lane 4). B, same material was blotted using antibodies against GLUT1 (monomer, dimer, and tetramer forms of GLUT1 are indicated by arrowheads), dematin, and β-adducin. Proteins were loaded in the same relative proportion as shown in A, except for GLUT1 blot (lane 1) where 5-fold less protein was loaded. C and D, Western blot analysis of GLUT1 in mouse erythrocytes. C, adult mouse erythrocyte ghosts were probed for GLUT1. WT (lane 1), dematin-headpiece knockout (lane 2), β-adducin knockout (lane 3), and DAKO (lane 4). D, WT (lane 1), DAKO (lane 2), WT anemic (lane 3), WT 1-day-old (lane 4). Protein loading was 100-fold less in lane 4. GLUT1 position is indicated by arrows. E, Western blot analysis of GLUT1 in peripheral blood of 1-day-old (lane 1), 5-day-old (lane 2), 10-day-old (lane 3), and 15-day-old (lane 4) mouse pups. Arrowheads indicate the positions of GLUT1 monomer, dimer, and tetramer. F, protein loading in E was in similar proportion as shown in the Coomassie Blue-stained gel.
GLUT1 Is Not Detectable in Adult Mouse Erythrocytes— Originally, we planned to use the dematin and adducin single and double knock-out mice to investigate the effects of dematin and adducin deficiency on the retention of GLUT1 in the erythrocyte ghosts. To our surprise, Western blotting failed to detect any GLUT1 in the wild type adult mouse erythrocytes (Fig. 4C, lane 1). Interestingly, the presence of GLUT1 was detectable in the erythrocyte ghosts of dematin and adducin double knock-out (DAKO) mice, which contain ∼40% reticulocytes (Fig. 4C, lane 4) (10). This finding suggested that GLUT1 may be expressed in mouse erythroblasts and reticulocytes, but it is absent in mature erythrocytes. To test this possibility, wild type mice were made anemic by repeated bleedings as confirmed by significant reticulocytosis (∼30%). Alternatively, peripheral blood was collected from the newborn mouse pups, which contain ∼20% reticulocytes. Ghosts were prepared from the peripheral blood erythrocytes of adult anemic and newborn pups and were examined for the presence of GLUT1 by Western blotting. Again, the presence of GLUT1 was not detectable in the adult mouse erythrocytes (Fig. 4D, lane 1), but GLUT1 was present in the DAKO ghosts (Fig. 4D, lane 2), adult anemic mouse ghosts (Fig. 4D, lane 3), and in the ghosts of 1-day-old pups (Fig. 4D, lane 4). The amount of total protein loaded for the 1-day-old mouse pups is 100-fold less (Fig. 4D, lane 4) as compared with the other samples, indicating a substantially higher amount of GLUT1 in the newborn mouse red blood cells. To investigate this observation further, we examined the expression of erythroid GLUT1 as a function of neonatal mouse age (Fig. 4, E and F). Western blotting confirmed robust expression of GLUT1 in the peripheral blood of a 1-day-old mouse (Fig. 4E, lane 1). The erythroid GLUT1 rapidly down-regulated with mouse age at day 5 (Fig. 4E, lane 2) and day 10 (Fig. 4E, lane 3) and essentially disappeared by day 15 (Fig. 4E, lane 4) after birth. Interestingly, all species of erythroid GLUT1, including the monomer, dimer, and tetramer forms, begin to disappear with mouse age (Fig. 4E, arrowheads). In fact, the only detectable species of erythroid GLUT1 present at day 10 is the residual GLUT1 tetramer (Fig. 4E, lane 3). Together, these results suggest that GLUT1 is present in the mouse erythroid precursors and reticulocytes, rapidly down-regulates with mouse age, and is completely eliminated from the red blood cells of adult mice.
The absence of GLUT1 in the adult mouse erythrocytes was further confirmed by the vesicle proteomics of mouse IOVs (supplemental Table 2). This approach was similar to the analysis of human erythrocyte IOVs except that the biochemical separation of IOVs from ROVs by density gradient centrifugation was not performed. The identity of 102 relatively abundant proteins in the adult mouse erythrocyte membrane vesicles is shown in supplemental Table 2. The absence of GLUT1 is consistent with our Western blotting results indicating that GLUT1 is not detectable in the adult mouse erythrocytes. Interestingly, the mouse proteomics list includes a number of transporters that conform to the predicted membrane topology of GLUT1. Whether one or more of these transporters may substitute for GLUT1 in the adult mouse erythrocytes and link the junctional complex to the plasma membrane via dematin and adducin remains to be investigated in future studies.
DISCUSSION
The core of the erythrocyte membrane skeleton consists primarily of spectrin, actin, protein 4.1, and dematin. It is now well recognized that the spectrin-actin junctions, also called the junctional complex, are essential for the maintenance of erythrocyte shape, membrane stability, and mechanical properties. A precise understanding of the junctional complex composition, organization, and the mode of its linkage to the plasma membrane is therefore of fundamental importance to the erythrocyte biology. A better understanding of the junctional complex is also of considerable significance in non-erythroid cells because the homologues of junctional complex proteins exist in a variety of other cells. The current model of the red cell membrane suggests that the spectrin-actin junctions are linked to the plasma membrane via the protein 4.1 and glycophorin C. In this study, we provide the first evidence that dematin and adducin can independently bind to GLUT1, and we propose that a macromolecular complex between dematin, adducin, and GLUT1 provides a new mechanism for the vertical linkage of the spectrin-actin junctions to the plasma membrane in human erythrocytes. Evidence accumulated in the past 2 decades in fact suggests that alternate mechanisms may exist that can link the spectrin-actin junctions to the erythrocyte plasma membrane independent of the protein 4.1-glycophorin C bridge (28–35). Genetic evidence from human anemia mutations also supports the existence of additional vertical tethers that could link the spectrin-actin junctions to the membrane (29, 36).
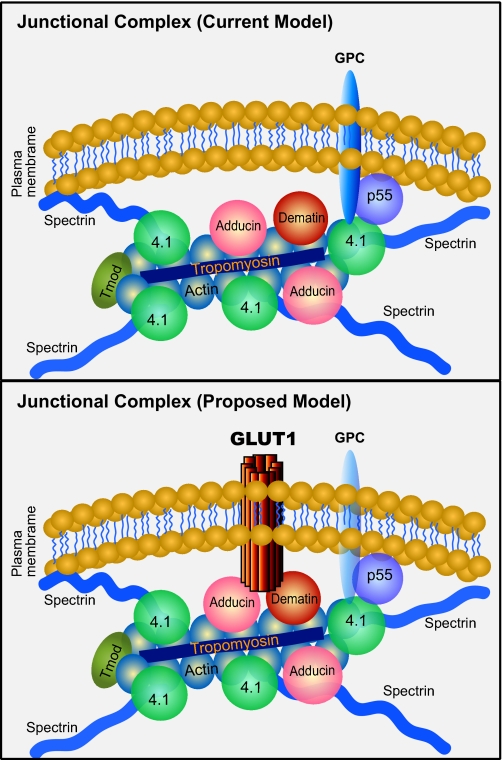
Several lines of evidence suggest that the biochemical properties of dematin are consistent with its role as an adaptor protein. (a) When spectrin and actin are removed from the erythrocyte IOVs, the bulk of dematin remains associated with these vesicles. (b) In reconstitution experiments, dematin binds to the IOVs by a specific and saturable interaction that is trypsin-sensitive. (c) Dematin has been visualized at the junctional complex by immunogold electron microscopy. (d) Dematin is an excellent substrate of multiple protein kinases. Given dematin's stoichiometry, actin binding properties, and phosphorylation-dependent actin cross-linking function, it is an ideal cytoskeletal protein to serve as a molecular bridge between the spectrin-actin junctions and the plasma membrane. However, a critical missing link was the identification of the transmembrane receptor that could link dematin to the lipid bilayer. In this study, we demonstrate that both dematin and adducin can independently bind to GLUT1, a major transmembrane protein in the human erythrocyte membrane. We propose that the macromolecular complex formed between GLUT1, dematin, and adducin anchors the spectrin-actin junctions to the membrane in human erythrocytes (Fig. 5).
FIGURE 5.
Proposed model of the erythrocyte junctional complex. Our proposed model predicts a major role of GLUT1 interactions with dematin and adducin in stabilizing the vertical linkage of the junctional complex. In principle, the GLUT1 complex with dematin and adducin can be further strengthened by putative interactions of other transmembrane proteins with GLUT1 and their subsequent interactions with components of the junctional complex. It remains to be established whether dematin and adducin interact with the same or separate GLUT1 molecules that in turn participate in dimer or tetramer formation in the plasma membrane.
A surprising finding in our study was the complete absence of GLUT1 in the adult mouse erythrocytes (Fig. 4), particularly because the glucose transport has been documented in the adult mouse erythrocytes (37). Our Western blotting and proteomics data unequivocally demonstrate that GLUT1 is present in mouse reticulocytes, but it is absent in adult mouse erythrocytes in sharp contrast to adult human erythrocytes. Although the significance of this observation is not yet clear, it appears that dematin and adducin may use an alternate receptor in adult mouse erythrocytes to anchor the junctional complex to the lipid bilayer. Future identification of this receptor will allow testing of the hypothesis that the same receptor may bind to dematin and adducin in the adult erythrocytes of several other species, such as pig, rabbit, sheep, horse, and cat, where the glucose transport activity or GLUT1 appears to be absent (38–40).
Glucose is a universal energy-producing substrate, and the regulation of its transport activity across the plasma membrane is of fundamental importance to cellular homeostasis. The GLUT family consists of 14 members that regulate sugar transport in a tissue-specific manner (41, 42). Among these transporters, the GLUT1 protein is ubiquitous and regulates basal glucose transport either alone or in combination with other transporters in the erythroid, endothelial, epithelial, and neuronal cells (43, 44). The GLUT1 protein, the founding member of the GLUT family (45), is the most abundant glucose transporter found in human erythrocytes and constitutes 2–6% of the membrane protein mass (24, 27). The unusually high abundance of GLUT1 in human erythrocytes, and its rapid glucose transport kinetics that far exceeds the physiological sugar requirement of an erythrocyte, has raised the possibility that GLUT1 and other related transporters may in fact perform a structural rather than the transport function in adult erythrocytes of many species. Our findings, as reported in this study, provide the first evidence in support of this hypothesis.
Besides erythrocytes, the GLUT1 protein is an abundant sugar transporter in endothelial cells of the blood-brain barrier. Because both dematin and adducin are expressed in the neuronal and endothelial cells, it would be interesting to determine whether the dematin-adducin-GLUT1 bridge is also preserved in other tissues. Defects in both GLUT1 and adducin have been shown to alter brain functions (46–48). We have not examined the neurological status of the dematin headpiece domain knock-out mice as yet. It is known that genetic inactivation of mouse GLUT1 results in embryonic lethality suggesting a critical role of this transporter in the cellular physiology of multiple tissues (49, 50). As our results indicate the significance of the dematin-adducin-GLUT1 bridge in the maintenance of erythrocyte membrane stability, a possibility now exists that this novel complex could also have significant functional implications in the regulation of membrane stability and glucose homeostasis in many non-erythroid cells. As dematin and adducin are also abundant in the adipocytes and muscle cells, the interactions of dematin and adducin with GLUT1 or GLUT4 could play a functional role in the intracellular trafficking of glucose transporters in non-erythroid cells. Indeed, our preliminary studies on dematin and GLUT4 show an insulin-dependent co-localization of these proteins in a distinct population of membrane vesicles in mouse adipocytes.3
The GLUT1 transporter-binding protein, GLUT1CBP, and stomatin are two factors that are known to regulate GLUT1 activity in non-erythroid cells (25, 51–53). Stomatin interacts with GLUT1 in erythrocytes (25), but its effect on the transport activity of erythrocyte GLUT1 is not known (25, 53). Because dematin, adducin, and GLUT1 are widely expressed, the proposed dematin-adducin-GLUT1 interactions may constitute the first cytoskeletal complex for GLUT1 thus providing both mechanical and biochemical functions in a variety of non-erythroid cells. Finally, our findings also raise the possibility that alterations in dematin and adducin structure and function may contribute to the overall phenotype of GLUT1 deficiency in patients with the GLUT1 deficiency syndrome (48, 54, 55).
Supplementary Material
Acknowledgments
We are grateful to Dr. Ronald Dubreuil of the University of Illinois for the invaluable advice and suggestions throughout the course of this study. We also thank Drs. John Quigley and Richard J. Labotka of the University of Illinois at Chicago for their helpful suggestions regarding the nature of membrane transporters in red cells and hematology issues that considerably improved this study. We are thankful to Dr. Hsin-Yao Tang for assistance with performing the liquid chromatography-MS/MS analyses. We thank Dr. Rainer Prohaska of the Medical University of Vienna, Austria and Dr. Clive Palfrey of the University of Chicago for their generous gifts of stomatin and adducin antibodies, respectively. We also thank Brendan Quinn, Shafi Kuchay, Xuerong Li, and other members of our laboratory for general discussions and help with many aspects of this study. Finally, we are thankful to Dena Inempolidis and Deanna Rybak for their careful reading and assistance during the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants HL051445 (to A. H. C.) and HL38794 (to D. W. S.). This work was also supported by the American Heart Association, Southeast Affiliate, Award 0555389B (to B. C. R.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1 and 2.
Footnotes
The abbreviations used are: GLUT1, glucose transporter-1; WT, wild type; DAKO, dematin-adducin double knockout; PBS, phosphate-buffered saline; IOVs, inside-out vesicles; NHS, N-hydroxysuccinimide; IP, immunoprecipitation; MS/MS, tandem mass spectrometry; BisTris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; ROV, right side-out vesicle; GST, glutathione S-transferase.
A. H. Chishti, unpublished data.
References
- 1.Lux, S. E., and Palek, J. (1995) in BLOOD: Principles and Practice of Hematology (Handin, R. I., Lux, S. E., Thomas P., and Stossel, T. P., eds) pp. 1701-1718, J. B. Lippincott, Philadelphia
- 2.Luna, E. J., and Hitt, A. L. (1992) Science 258 955-964 [DOI] [PubMed] [Google Scholar]
- 3.Chasis, J. A., and Mohandas, N. (1992) Blood 80 1869-1879 [PubMed] [Google Scholar]
- 4.Bennett, V. (1989) Biochim. Biophys. Acta 988 107-121 [DOI] [PubMed] [Google Scholar]
- 5.Gilligan, D. M., and Bennett, V. (1993) Semin. Hematol. 30 74-83 [PubMed] [Google Scholar]
- 6.Derick, L. H., Liu, S. C., Chishti, A. H., and Palek, J. (1992) Eur. J. Cell Biol. 57 317-320 [PubMed] [Google Scholar]
- 7.Conboy, J. G. (1993) Semin. Hematol. 30 58-73 [PubMed] [Google Scholar]
- 8.Marfatia, S. M., Lue, R. A., Branton, D., and Chishti, A. H. (1995) J. Biol. Chem. 270 715-719 [DOI] [PubMed] [Google Scholar]
- 9.Marfatia, S. M., Lue, R. A., Branton, D., and Chishti, A. H. (1994) J. Biol. Chem. 269 8631-8634 [PubMed] [Google Scholar]
- 10.Chen, H., Khan, A. A., Liu, F., Gilligan, D. M., Peters, L. L., Messick, J., Haschek-Hock, W. M., Li, X., Ostafin, A. E., and Chishti, A. H. (2007) J. Biol. Chem. 282 4124-4135 [DOI] [PubMed] [Google Scholar]
- 11.Azim, A. C., Knoll, J. H., Beggs, A. H., and Chishti, A. H. (1995) J. Biol. Chem. 270 17407-17413 [DOI] [PubMed] [Google Scholar]
- 12.Gilligan, D. M., Lozovatsky, L., Gwynn, B., Brugnara, C., Mohandas, N., and Peters, L. L. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 10717-10722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muro, A. F., Marro, M. L., Gajovic, S., Porro, F., Luzzatto, L., and Baralle, F. E. (2000) Blood 95 3978-3985 [PubMed] [Google Scholar]
- 14.Khanna, R., Chang, S. H., Andrabi, S., Azam, M., Kim, A., Rivera, A., Brugnara, C., Low, P. S., Liu, S. C., and Chishti, A. H. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 6637-6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jarvis, S. M. (1988) Biochem. J. 249 383-389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steck, T. L., and Kant, J. A. (1974) Methods Enzymol. 31 172-180 [DOI] [PubMed] [Google Scholar]
- 17.Boukharov, A. A., and Cohen, C. M. (1998) Biochem. J. 330 1391-1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das, A. K., Bhattacharya, R., Kundu, M., Chakrabarti, P., and Basu, J. (1994) Eur. J. Biochem. 224 575-580 [DOI] [PubMed] [Google Scholar]
- 19.Chishti, A. H. (1998) Curr. Opin. Hematol. 5 116-121 [DOI] [PubMed] [Google Scholar]
- 20.Staufenbiel, M., and Lazarides, E. (1986) Proc. Natl. Acad. Sci. U. S. A. 83 318-322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kakhniashvili, D. G., Bulla, L. A., Jr., and Goodman, S. R. (2004) Mol. Cell. Proteomics 3 501-509 [DOI] [PubMed] [Google Scholar]
- 22.Pasini, E. M., Kirkegaard, M., Mortensen, P., Lutz, H. U., Thomas, A. W., and Mann, M. (2006) Blood 108 791-801 [DOI] [PubMed] [Google Scholar]
- 23.Huang, S., Lichtenauer, U. D., Pack, S., Wang, C., Kim, A. C., Lutchman, M., Koch, C. A., Torres-Cruz, J., Huang, S. C., Benz, E. J., Jr., Christiansen, H., Dockhorn-Dworniczak, B., Poremba, C., Vortmeyer, A. O., Chishti, A. H., and Zhuang, Z. (2001) Eur. J. Clin. Investig. 31 907-914 [DOI] [PubMed] [Google Scholar]
- 24.Hruz, P. W., and Mueckler, M. M. (2001) Mol. Membr. Biol. 18 183-193 [DOI] [PubMed] [Google Scholar]
- 25.Zhang, J. Z., Hayashi, H., Ebina, Y., Prohaska, R., and Ismail-Beigi, F. (1999) Arch. Biochem. Biophys. 372 173-178 [DOI] [PubMed] [Google Scholar]
- 26.Chishti, A., Levin, A., and Branton, D. (1988) Nature 334 718-721 [DOI] [PubMed] [Google Scholar]
- 27.Klepper, J., Garcia-Alvarez, M., O'Driscoll, K. R., Parides, M. K., Wang, D., Ho, Y. Y., and De Vivo, D. C. (1999) J. Clin. Lab. Anal. 13 116-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohanian, V., Wolfe, L. C., John, K. M., Pinder, J. C., Lux, S. E., and Gratzer, W. B. (1984) Biochemistry 23 4416-4420 [DOI] [PubMed] [Google Scholar]
- 29.Pinder, J. C., Chung, A., Reid, M. E., and Gratzer, W. B. (1993) Blood 82 3482-3488 [PubMed] [Google Scholar]
- 30.Takakuwa, Y., Tchernia, G., Rossi, M., Benabadji, M., and Mohandas, N. (1986) J. Clin. Investig. 78 80-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takakuwa, Y., Ishibashi, T., and Mohandas, N. (1990) Biorheology 27 357-365 [DOI] [PubMed] [Google Scholar]
- 32.Discher, D., Parra, M., Conboy, J. G., and Mohandas, N. (1993) J. Biol. Chem. 268 7186-7195 [PubMed] [Google Scholar]
- 33.Chang, S. H., and Low, P. S. (2001) J. Biol. Chem. 276 22223-22230 [DOI] [PubMed] [Google Scholar]
- 34.Discher, D. E., Winardi, R., Schischmanoff, P. O., Parra, M., Conboy, J. G., and Mohandas, N. (1995) J. Cell Biol. 130 897-907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hemming, N. J., Anstee, D. J., Mawby, W. J., Reid, M. E., and Tanner, M. J. (1994) Biochem. J. 299 191-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winardi, R., Reid, M., Conboy, J., and Mohandas, N. (1993) Blood 81 2799-2803 [PubMed] [Google Scholar]
- 37.Izumo, A., Tanabe, K., Kato, M., Doi, S., Maekawa, K., and Takada, S. (1989) Parasitology 98 371-379 [DOI] [PubMed] [Google Scholar]
- 38.Widdas, W. F. (1955) J. Physiol. (Lond.) 127 318-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laris, P. C. (1958) J. Cell. Physiol. 51 273-307 [DOI] [PubMed] [Google Scholar]
- 40.Kondo, T., and Beutler, E. (1980) J. Clin. Investig. 65 1-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Macheda, M. L., Rogers, S., and Best, J. D. (2005) J. Cell. Physiol. 202 654-662 [DOI] [PubMed] [Google Scholar]
- 42.Mueckler, M. (1994) Eur. J. Biochem. 219 713-725 [DOI] [PubMed] [Google Scholar]
- 43.Baldwin, S. A. (1993) Biochim. Biophys. Acta 1154 17-49 [DOI] [PubMed] [Google Scholar]
- 44.Gould, G. W., and Holman, G. D. (1993) Biochem. J. 295 329-341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mueckler, M., Caruso, C., Baldwin, S. A., Panico, M., Blench, I., Morris, H. R., Allard, W. J., Lienhard, G. E., and Lodish, H. F. (1985) Science 229 941-945 [DOI] [PubMed] [Google Scholar]
- 46.Rabenstein, R. L., Addy, N. A., Caldarone, B. J., Asaka, Y., Gruenbaum, L. M., Peters, L. L., Gilligan, D. M., Fitzsimonds, R. M., and Picciotto, M. R. (2005) J. Neurosci. 25 2138-2145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klepper, J., Salas-Burgos, A., Gertsen, E., and Fischbarg, J. (2005) Biochemistry 44 12621-12626 [DOI] [PubMed] [Google Scholar]
- 48.Pascual, J. M., Lecumberri, B., Wang, D., Yang, R., Engelstad, K., and De Vivo, D. C. (2004) Rev. Neurol. (Paris) 38 860-864 [PubMed] [Google Scholar]
- 49.Heilig, C., Brosius, F., Siu, B., Concepcion, L., Mortensen, R., Heilig, K., Zhu, M., Weldon, R., Wu, G., and Conner, D. (2003) Am. J. Pathol. 163 1873-1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, D., Pascual, J. M., Yang, H., Engelstad, K., Mao, X., Cheng, J., Yoo, J., Noebels, J. L., and De Vivo, D. C. (2006) Hum. Mol. Genet. 15 1169-1179 [DOI] [PubMed] [Google Scholar]
- 51.Bunn, R. C., Jensen, M. A., and Reed, B. C. (1999) Mol. Biol. Cell 10 819-832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Reed, B. C., Cefalu, C., Bellaire, B. H., Cardelli, J. A., Louis, T., Salamon, J., Bloecher, M. A., and Bunn, R. C. (2005) Mol. Biol. Cell 16 4183-4201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, J. Z., Abbud, W., Prohaska, R., and Ismail-Beigi, F. (2001) Am. J. Physiol. 280 C1277-C1283 [DOI] [PubMed] [Google Scholar]
- 54.Klepper, J., and Voit, T. (2002) Eur. J. Pediatr. 161 295-304 [DOI] [PubMed] [Google Scholar]
- 55.Klepper, J., and Leiendecker, B. (2007) Dev. Med. Child Neurol. 49 707-716 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.