Abstract
To explore the basis of metastasis, we compared the human breast cancer lines MCF-7 and MDA-MB453, which have low invasive ability, with their sublines MCF7-I4 and MDA-MB453-I4 with high invasive ability for gene expression and signaling pathways. We previously showed that the I4 lines had dramatically elevated levels of Twist compared with their parental lines. In this study, we observed significantly increased STAT3 Tyr705 phosphorylation, but not the STAT3 protein levels, in the I4 lines. Activation of STAT3 by interleukin-6 or expression of activated Src induced Twist expression at protein and mRNA levels. Inhibiting STAT3 by a small molecule inhibitor, JSI-124, STAT3 small hairpin RNAs, or dominant negative STAT3 resulted in significant reduction of Twist protein and mRNA expression. STAT3 directly bound to the second proximal STAT3-binding site on the human Twist promoter and activated its transcriptional activity. Inhibition of STAT3 reduced migration, invasion, and colony formation of the I4 cells. Ectopic expression of Twist significantly rescued those phenotypes. Ten normal and 46 tumor specimens of breast tissues were examined for activation of STAT3 and expression of Twist. There was a strong correlation between Tyr705 p-STAT3 and Twist level in the late stage tumor tissues. Our results indicate that activated STAT3 transcriptionally induces Twist, which plays an important role in promoting migration, invasion, and anchorage-independent growth. Together with our previous observation that Twist transcriptionally induces AKT2 to mediate Twist-promoted oncogenic functions, we conclude that STAT3, Twist, and AKT2 form a functional signaling axis to regulate pivotal oncogenic properties of cancer cells.
Twist, a highly conserved basic helix-loop-helix transcriptional factor, was previously shown to play a pivotal role in mesodermal, myoblast, and osteoblast differentiation (1-3). Mutational inactivation of the Twist gene resulted in Saethre-Chotzen syndrome, an autosomal dominant disorder characterized by premature fusion of the cranial sutures, skull deformations, limb abnormalities, and facial dysmorphisims (4). Recent studies have demonstrated that Twist also played a key role in the development and progression of human cancer (5).
Twist is frequently overexpressed in human rhabdomyosarcoma, gastric carcinoma, melanoma, breast cancer, prostate cancer, liver carcinoma, and glioma (5). The elevated Twist protein levels are associated with advanced tumor stage and poor prognosis in several types of cancer (6-8). Further, increased Twist in cancer cells has been shown to promote metastatic ability in vivo and induce epithelial to mesenchymal transition, cell survival, angiogenesis, and chemoresistance in vitro (9-14). In addition, ectopic expression of Twist in mouse embryonic fibroblasts promotes soft agar colony formation, indicating its role in malignant transformation (10). A number of downstream target genes of Twist have been identified and shown to mediate its function (10). Previous studies have demonstrated that epithelial to mesenchymal transition-associated molecules, such as E-cadherin and N-cadherin, are tightly regulated by Twist (9, 15). We have recently shown that AKT signaling is promoted by Twist through transcriptional up-regulation of AKT2 (16). However, the molecular mechanism for the up-regulation of Twist in cancer cells is less clear.
Signal transducer and activator of transcription 3 (STAT3)4 protein is a member of a family of latent cytoplasmic transcriptional factors that convey signals from the cell surface to the nucleus upon activation by cytokines and growth factors (17). Engagement of cell surface receptors by polypeptide ligands induces tyrosine phosphorylation of STAT3 protein by Janus kinases, growth factor receptor tyrosine kinases, and, in some cases, Src family tyrosine kinases. The phospho-STAT3 protein dimerizes and translocates to nucleus to regulate expression of the genes harboring STAT3-binding sites in their promoters (18). STAT3 has been shown to regulate genes that control fundamental biological processes including cell proliferation, survival, and development (18). Activated STAT3 was able to transform cells in vitro (19) and is required for cell transformation of a number of oncogenes (12, 19, 20). Numerous studies have detected constitutively active STAT3 in diverse human tumor specimens (21-24). Activated STAT3 in tumor cells participates in regulating expression of genes involved in controlling cell cycle progression, apoptosis, and angiogenesis (25).
We previously established a model system using Boyden chamber invasion assay to select highly invasive cells from a population of low invasive breast cancer lines and showed an increased level of Twist in the selected highly invasive cells (17). We further demonstrated that Twist transcriptionally induced AKT2 to mediate migration, invasion, and drug resistance (16). In the present study, we detected an elevated level of activated STAT3 in the highly invasive cells and demonstrated its role in transcriptional regulation of Twist. We showed that STAT3 bound to the Twist promoter and activated its transcriptional activity. Twist was able to rescue the oncogenic phenotypes resulting from inhibition of STAT3. Further, elevated p-STAT3 and Twist are highly correlated in breast cancer. Our finding demonstrates for the first time that Twist is a transcriptional target of STAT3. Together with our previous report (16), we conclude that STAT3, Twist, and AKT2 are functionally linked in a signaling pathway to regulate oncogenic properties of breast cancer cells.
EXPERIMENTAL PROCEDURES
Cell Lines, Transfection, and Tumor Specimens—The breast cancer cell lines MCF-7 and MDA-MB-453 and their derived highly invasive cell lines MCF-7-I4 and MDA-MB-453-I4 have been obtained as previously described (16). The cells were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal calf serum (FCS) and antibiotics. The cultures were maintained in a humidified incubator at 37 °C in the presence of 5% CO2. The transfections were done with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Anonymous primary human breast cancer specimens were procured in the H. Lee Moffitt Cancer Center under the institutional review board approved protocol. The tissues were snap frozen and stored at -70 °C.
Antibodies and Reagents—Antibodies against Tyr(P)705 STAT3 and STAT3 were purchased from Cell Signaling. Mono- and polyclonal anti-Twist antibodies were obtained from Abcam and homemade, respectively (17). Anti-tubulin antibody and growth factor reduced Matrigel were from Sigma and BD Biosciences, respectively. STAT3 inhibitor Cucurbitacin/JSI-124 was purchased from Calbiochem.
Cloning and Construction of the Human Twist Promoter—Full-length human Twist promoter was generated by nested PCR using genomic DNA isolated from the human melanoma cell line FEMX. The first PCR primers used for the Twist promoter were 5′-GCGTATCCAAGCATTTGGAATTGGGG-3′ (forward) and 5′-CTCTCGAGCGGCGACGCGTGGCCTC-3′ (reverse). Full-length Twist promoter and its subsequent truncation mutants were created by a second round of PCR using primers: Full forward, 5′-CCGGGTACCCTTTCAAGGTCACAATGCGGAGCC-3′; Truncation 1 (-745) forward, 5′-CCGGGTACCAGCGTCAGACTGGGTCGTTGTAGAGG-3′; Truncation 2 (-451) forward, 5′-CCGGGTACCGAGATGAGACATCACCCACTGTGTAG-3′; Truncation 3 (-186) forward, 5′-CCGGGTACCTTTGGGAGGACGAATTGTTAGACC-3′; Truncation 4 (-105) forward, 5′-CCGGGTACCACTTCGAAAAGTCCCTCCTCCTC-3′; and Common reverse, 5′-ATACTCGAGTGGGCGAGAGCTGCAGACTTGG-3′. The PCR products were digested with KpnI/XhoI and subcloned into pGL3-Luciferase vector. The final constructs were confirmed by DNA sequencing. Myc-tagged Twist plasmid was previously described (16).
Western Blot—Western blot analysis was performed as described previously (12). Briefly, the cells were lysed with radioimmune precipitation assay buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 5 μg/ml aprotinin, 5 μg/ml leupeptin, and 1 mm phenylarsine oxide), and the total lysate was separated by SDS-PAGE and immunoblotted with appropriate antibodies as indicated in the figure legends.
RNA Isolation and Reverse Transcription (RT)-PCR Analysis—Total RNA was extracted using TRIzol (Invitrogen) and was further purified with RNeasy purification kit (Qiagen). The first strand cDNA synthesis was carried out with SuperScript II reverse transcriptase (Invitrogen) using 2 μg of total RNA and poly(dT) primer. PCR was performed with gene specific primers for glyceraldehyde-3-phosphate dehydrogenase (forward, 5′-GACCCCTTCATTGACCTCAAC-3′; reverse, 5′-CTTCTCCATGGTGGTGAAGAC-3′) as an internal control and Twist (forward, 5′-GGAGTCCGCAGTCTTACGAG-3′; reverse, 5′-TCTGGAGGACCTGGTAGAGG-3′).
Cell Migration and Invasion Assay—Invasion assays were performed in a Boyden chamber with polyethylene terephthalate filter inserts for 24-well plates containing 0.8-μm pores (Falcon 3097 and 3504) as described before (17). The filters were coated on ice with 100 μl of growth factor reduced Matrigel at 0.5-0.8 mg/ml protein. 1 × 104 cells were plated in 300 μl of 0.1% bovine serum albumin-DMEM into the upper chamber. The lower chamber was filled with 500 μl of 10% FCS-DMEM. After culturing for 6-12 h, noninvaded cells in the inserts were removed with cotton swabs. The invaded cells on the underside were treated with a fixative/staining solution (0.1% crystal violet, 1% formalin, 20% ethanol) for visualization. The invaded cells at underside of the membrane were photographed. Migration assay was performed using Boyden chamber similarly except without coating of Matrigel and 6-8 h of migration time allowed. Migrated/invaded cells were stained, photographed digitally, and quantified using ImageJ software. Statistical analysis was performed using a two-sample t test assuming equal variance, and p values were calculated based on a two-tailed test.
Soft Agar Colony Formation Assay—The assay was as described before (12, 17). 1 × 105 cells were suspended in 10% FCS-DMEM containing 0.4% agar. The cells were then placed into a dish containing a hard agar base composed of 10% FCS-DMEM and 0.75% agar. The cultures were returned to the incubator and fed every 2 days with 500 μl of normal growth medium. Photomicrographs of colonies were taken 2-3 weeks later and quantified by ImageJ software.
Luciferase Reporter Assay—The cells were seeded in a 24-well plate and transfected with Twist-Luc reporter plasmid, pRenilla-luciferase plasmid, and additional plasmids as described in the figure legend. The amount of DNA in each transfection was kept constant by the addition of empty vector, pCMV-Tag3B. Thirty-six hours post transfection, the cell lysates were prepared, and firefly luciferase and Renilla luciferase were assayed according to the manufacturer's protocol (Promega). The same amount of cell extract was used for the assay, and the firefly luciferase activity was normalized to that of Renilla luciferase. Luciferase activity was expressed as relative light units or fold change. Each experiment was repeated three times in triplicate.
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assay was performed essentially as previously described with modifications (26). Briefly, soluble chromatin was prepared from a total of 2 × 107 asynchronously growing MDA-MB-453I4 cells that were transfected with STAT3 and v-Src. The chromatin solution was diluted 10-fold with ChIP dilution buffer (1.1% Triton X-100, 1.2 mm EDTA, 167 mm NaCl, 16.7 mm Tris-HCl, pH 8.1, 0.01% SDS, plus protease and phosphatase inhibitors), and precleared with protein A beads blocked with 2 μg of sheared salmon sperm DNA and preimmune serum. The precleared chromatin solution was divided and utilized in immunoprecipitation assays with either an anti-STAT3 antibody or an anti-GFP antibody. Following multiple washes, the antibody-protein-DNA complex was eluted from the beads by resuspending the pellets in 1% SDS, 0.1 m NaHCO3 at room temperature for 20 min. After reversal cross-link incubation at 67 °C, protein and RNA were removed by incubation with 10 μg of proteinase K and 10 μg of RNase A at 42 °C for 3 h. Purified DNA was subjected to PCR with primers specific for the STAT3-binding sites upstream of the transcriptional start site. The sequences of the PCR primers used are: proximal forward, 5′-GCCAGGTCGTTTTTGAATGG-3′, and reverse, 5′-CGTGCAGGCGGAAAGTTTGG-3′; distal forward, 5′-TGCCTTTCCCATGGACTGGG-3′, and reverse, 5′-GAGTTCCAAAGGCCAAACCG-3′. The proximal and distal PCR products contain the three proximal and two distal STAT3-binding sites, respectively.
Electrophoretic Mobility Shift Assay (EMSA)—MCF7-I4 cells were transfected with FLAG-tagged STAT3. Following 48 h of incubation, nuclear extract was prepared as previously described (21). Normalized extracts containing ∼5 μg of protein were incubated with 32P-radiolabeled double-stranded oligonucleotide probe. The oligonucleotides containing the putative STAT3-binding sites in the Twist promoter and their derivatives are as follows: (-107, e.g. the second STAT3-binding site) wild type, 5′-GCGGAAACTTTCCTATAAAACTTCG-3′; mutant, 5′-GCGGAAACTggCCTATAAAACTTCG-3′ (-96, e.g. the first STAT3 binding) wild type, 5′-CTATAAAACTTCGAAAAGTCCCTCC-3′. Protein-DNA complexes were resolved by nondenaturing polyacrylamide gel electrophoresis and visualized by autoradiography.
Short Hairpin RNA Construction—The shRNA duplexes were constructed based on the sequences obtained from the Sigma MISSION shRNA Library of the RNAi Consortium. RNA targeting regions with a hairpin sequence (TTCAAGAGA) was cloned into pSIREN-RetroQ-linker at BamHI and EcoRI sites (a generous gift from Dr. Domenico Tortorella, Mount Sinai School of Medicine). Sequences are as shown: shSTAT3-1, 5′-gatccGCGGATCATAAGGTCAGGAGATTTCAAGAGAATCTCCTGACCTTATGATCCGTTTTTTg-3′; shSTAT3-2, (5′-gatccGCTGACCAACAATCCCAAGAATTCAAGAGATTCTTGGGATTGTTGGTCAGCTTTTTTg-3′); shSTAT3-3, 5′-gatccGCTGAAATCATCATGGGCTATTTCAAGAGAATAGCCCATGATGATTTCAGCTTTTTTg-3′; shSTAT3-4, 5′-gatccGCACAATCTACGAAGAATCAATTCAAGAGATTGATTCTTCGTAGATTGTGCTTTTTTg-3′; and shSTAT3-5, 5′-gatccGCAAAGAATCACATGCCACTTTTCAAGAGAAAGTGGCATGTGATTCTTTGCTTTTTTg-3′. A pool of these pSIREN-RetroQ-shSTATs and a control pSIREN-RetroQ-shRNA that specifically targets GFP protein were used to transfect MCF-7-I4 and MDA-MB-453-I4 cells. Stable STAT3 knockdown pool cells were obtained by puromycin selection.
Western Analysis of Normal and Tumor Tissues—For total protein lysate preparation, an appropriate piece of the tissues was cut and minced, an aliquot of radioimmune precipitation assay buffer was added, and the tissues suspension was subjected to tissue homogenization. The clear lysates were recovered after centrifugation. An equivalent amount of protein from each sample was analyzed by Western blotting.
Immunohistochemistry—Immunohistochemistry was performed as previously described (17), anti-Twist (Santa Cruz), and anti-phospho-STAT3-Tyr705 antibodies described above were used for the staining. Briefly, following paraffin section rehydration, the samples were treated with solution containing 0.3% hydrogen peroxide for 30 min to block endogenous peroxidase activity. After antigen retrieval in citrate buffer, the sections were incubated with the primary antibody (1:100 in phosphate-buffered saline, 1% bovine serum albumin overnight at 4 °C) and then with biotinylated secondary antibody (Vector Laboratories, Burlingame, CA). The signal was amplified by avidin-biotin complex formation and developed with diaminobenzidine followed by counterstaining with hematoxylin, dehydrated in alcohol and xylene, and mounted.
RESULTS
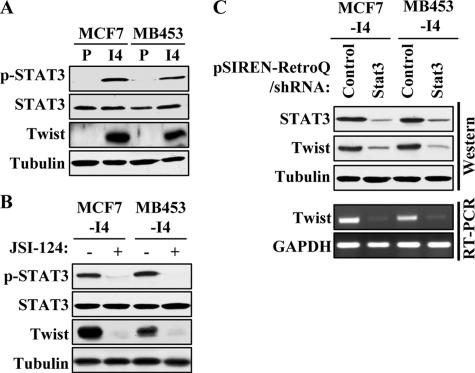
STAT3 Is Activated in the Selected Invasive I4 Cells, and Blocking STAT3 Activation Reduces Twist Expression at Protein and mRNA Levels—To better understand the molecular mechanism of breast cancer metastasis, we previously established highly invasive lines MCF-7-I4 and MDA-MB-453-I4 by four-round selection of MCF-7 and MDA-MB-453 using Boyden chamber invasion assay and showed that AKT2 and Twist were elevated in the selected invasive cells compared with the parental cells. Further, we demonstrated that Twist bound to the AKT2 promoter and induced AKT2 expression (16). However, the basis for up-regulation of Twist in I4 cells is unknown. Frequent activation of STAT3 in late stage and high grade breast cancer prompted us to examine whether STAT3 is activated in the I4 cells. Immunoblotting analysis revealed that the levels of Tyr(P)705 STAT3, a residue reflecting activation upon its phosphorylation, were increased in both MCF-7-I4 and MDA-MB-453-I4 cells, whereas the total levels of STAT3 did not change as compared with their parental cells (Fig. 1A). Because both STAT3 and Twist contributed to tumor progression, we considered the possibility that STAT3 and Twist could be functionally linked. To test this possibility, MCF-7-I4 and MDA-MB-453-I4 cells were treated with the small molecule inhibitor of STAT3, JSI-124 (27). Following treatment with JSI-124 for 24 h, we observed markedly reduced phospho-STAT3 levels and correspondingly decreased Twist protein expression (Fig. 1B), suggesting that Twist protein level is affected by the status of STAT3 activation. To further demonstrate the regulation of Twist by STAT3, we performed shRNA-mediated knockdown of STAT3 in the I4 cells. MCF-7-I4 and MDA-MB453-I4 cells were treated with either a control pSIREN-RetroQ-shRNA or a pool of pSIREN-RetroQ-shRNAs targeting five different regions of STAT3. As shown in Fig. 1C, the level of Twist protein decreased concurrently with STAT3 knockdown. The STAT3-knocked down I4 cells showed a dramatic reduction of Twist mRNA as measured by semi-quantitative RT-PCR. Those data led us to hypothesize that Twist is a downstream target gene of STAT3.
FIGURE 1.
Phospho-STAT3 and Twist levels are elevated in I4 cells. A, Western blot analysis. Parental (P) and I4 of MCF-7 and MDA-MB-453 cells were grown at 90% confluence, lysed, and immunoblotted with phospho-STAT3-Tyr705 (top panel), STAT3 (second panel), Twist (third panel), or tubulin (bottom panel) antibodies. B, MCF-7-I4 and MDA-MB-453-I4 cells were treated with 0.5 μm JSI-124 for 24 h and then lysed, and the total lysates were separated by SDS-PAGE and immunoblotted with the indicated antibodies. C, the I4 cells were treated with a pool of five pSIREN-RetroQ/shRNAs targeting the human STAT3 sequence or the control shRNA (GFP). After 72 h of incubation, protein and total RNA were isolated from the cells. Equal amounts of protein lysates were separated on SDS-PAGE and blotted with the indicated antibodies (top three panels). RNA was subjected to semi-quantitative RT-PCR analysis for the expression of Twist. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control (bottom panel).
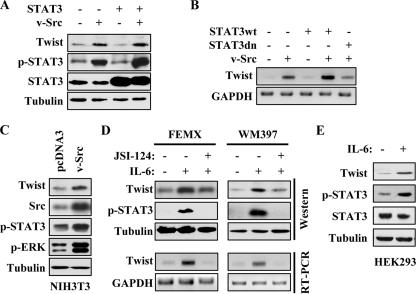
Activation of STAT3 Increases Twist Expression—To test our hypothesis, we ectopically expressed STAT3 and/or v-Src, which is known to phosphorylate and activate STAT3 (20, 28), in MDA-MB-453 cells and monitored for Twist expression. Overexpression of wild type STAT3 alone did not yield an increase in Twist protein levels; however, expression of Src alone or co-expression of v-Src and STAT3 led to a significant increase in phospho-STAT3 and Twist levels, with the latter having a slightly higher level (Fig. 2A). The mRNA level of Twist was examined by semi-quantitative RT-PCR. Ectopic expression of v-Src alone increased Twist mRNA, and a further increase was detected when additional STAT3 was provided. The effect of v-Src on Twist levels could be reversed by the expression of a dominant negative STAT3 mutant unable to bind DNA (29) (Fig. 2B), suggesting that this was a STAT3-dependent phenomenon. We next asked whether this relationship could be observed under persistent activation of STAT3. To address this, we examined Twist protein levels in v-Src stably transformed NIH3T3 line and its normal counterpart, pcDNA3-NIH3T3. As shown in Fig. 2C, NIH3T3-Src cells had elevated phopho-STAT3 and Twist levels as compared with those of the pcDNA3-NIH3T3 cells. ERK phosphorylation was used as an independent assessment of v-Src activity, and as expected, a significantly higher level of p-ERK was detected in NIH3T3-Src cells.
FIGURE 2.
Activation of STAT3 induces Twist expression. A, Western blot analysis. MDA-MB-453 cells were transiently transfected with v-Src or/and STAT3. Following 48 h of incubation, the cell lysates were prepared and analyzed by Western blot with the indicated antibodies. B, semi-quantitative RT-PCR. MDA-MB-453 cells were transiently transfected with the indicated plasmids. Total RNA was isolated after 48 h of transfection, and RT-PCR was performed as described under “Experimental Procedures.” C, v-Src stably transformed NIH3T3 and pcDNA3-transfected NIH3T3 cells were grown to 90% confluence. Whole cell extracts were isolated and immunoblotted with indicated antibodies. D, FEMX and WM397 melanoma cells were treated with IL-6 (20 ng/ml) with or without 0.5 μm STAT3 inhibitor JSI-124 for 24 h. Whole cell lysates and total RNA were isolated. Western blot analysis was performed with the indicated antibodies. Semi-quantitative RT-PCR was carried out for the expression of Twist and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). E, HEK293 cells were treated with IL-6 for 24 h, and the cell lysates were immunoblotted with the indicated antibodies.
Besides the nonreceptor Src family protein tyrosine kinase, STAT3 can also be activated by cytokine signaling. IL-6 has been shown to be a potent STAT3 activation signal (30). To further demonstrate the regulation of Twist by STAT3 signaling, we treated two melanoma cell lines, FEMX and WM397, and HEK293 cells with IL-6 alone or with IL-6 plus STAT3 inhibitor JSI-124. IL-6 treatment resulted in increased p-STAT3 as well as Twist at protein and mRNA levels, and the induction was inhibited in the presence of JSI-124 (Fig. 2, D and E). Together, our results indicate that Twist expression is regulated by STAT3, suggesting that up-regulation of Twist in the I4 cells and certain tumor cells could be due to the activation of STAT3.
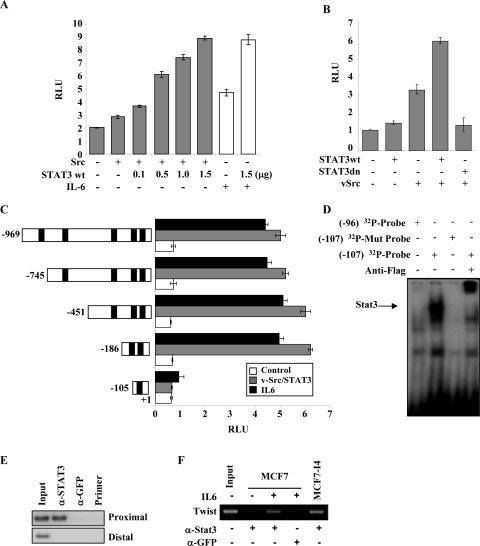
STAT3 Directly Binds to and Regulates the Twist Promoter—By examining and comparing the sequences of human and mouse Twist genomic structure, we identified the putative human Twist promoter and found five putative sites conforming to the consensus STAT3-binding sequences TT(N4-6)AA (Fig. 3) (17). Subsequently, we cloned the human Twist promoter into the pGL3-Luciferase vector. A dual luciferase reporter assay was performed to determine whether activated STAT3 was able to transactivate the human Twist promoter. Co-expression of v-Src and increasing amounts of STAT3 led to a dosage-dependent induction of Twist promoter activity, whereas v-Src alone only slightly enhanced the Twist promoter activity. Moreover, the addition of IL-6 induced the promoter activity, which was further enhanced by STAT3 (Fig. 4A). As expected from v-Src-mediated activation of STAT3, co-expression of v-Src and STAT3 led to a 6-fold increase of Twist promoter activity, which was inhibited to the basal level upon introduction of dominant negative STAT3 (Fig. 4B). These results raised the possibility that activated STAT3 may regulate Twist promoter via those recognition sites.
FIGURE 3.
Human and mouse Twist promoter. Human and mouse Twist promoter sequences were obtained from NCBI MapViewer. Putative STAT3-binding sites are shown. The TATA box is denoted according to Wang et al. (31). The capital letters denote the 5′ region of Twist encoding the mRNA transcript.
FIGURE 4.
STAT3 binds to and transactivates human Twist promoter. A and B, luciferase reporter assay. HEK293 cells were transfected with pGL3-Twist-Luc (firefly) and Renilla luciferase as well as the indicated plasmids or stimulated with IL-6. Following 36 h of culture and treatment with or without IL-6, luciferase activity was measured and normalized to Renilla luciferase activity. The relative luciferase activity was expressed in arbitrary units in a scale of 1 to 10. The results are the means ± S.E. of three independent experiments performed in triplicate. C, a series of deletion mutants of the Twist promoter (left) were introduced into HEK293 cells together with (gray bars) or without (white and black bars) v-Src and STAT3. After 36 h of incubation in a medium with (black bars) or without (white and gray bars) IL-6, the cells were subjected to luciferase reporter assay as described above. D, EMSA. Equal amounts of 32P-labeled oligonucleotides were incubated with nuclear extract prepared from FLAG-STAT3-transfected MCF7-I4 cells. Supershift was examined by incubation of the reactions with anti-FLAG antibody. E and F, ChIP assay. MDA-MB-453 cells were transfected with STAT3 and v-Src (E), and MCF7 and MCF7-I4 cells were treated with or without IL-6 as indicated (F). Twenty-four hours post-transfection or treatment, chromatin immunoprecipitation assay was carried out using anti-STAT3 antibody or irrelevant anti-GFP antibody as a negative control. The protein-chromatin immunoprecipitates were subjected to PCR analysis as described under “Experimental Procedures.” Input lane, total genomic DNA used as control for the PCR reaction. Lane 4 of E is a PCR control, and the reaction does not contain DNA. wt, wild type.
Because the Twist promoter contains five putative STAT3-binding sites, we next defined which binding site(s) was/were responsive to STAT3. Serial truncations of the human Twist promoter were created based on the location of the STAT3-binding sites. Luciferase assay showed that the second STAT3-binding motif on the Twist promoter proximal to the transcriptional starting site is responsible for the full STAT3-mediated activation of the promoter (Fig. 4C). In addition, it appeared to have a minor repressive element between -745 and -451 bp of the promoter (Figs. 3 and 4). To determine whether STAT3 could bind the Twist promoter in vitro and in intact cells, we performed EMSA and ChIP assays. EMSA experiments showed that the second but not the first STAT3-binding site directly bound to STAT3 (Fig. 4D). A ChIP assay was carried out using two sets of primers that covered the five candidate STAT3-binding sites. The first set of primers covered the proximal three STAT3-binding sites, whereas the second set of primers covered the distal two STAT3-binding sites. As shown in Fig. 4E, the proximal primers yielded Twist promoter DNA in the chromatin immunoprecipitated with an anti-STAT3 antibody. By contrast, primers corresponding to 5′ region containing the two distal STAT3 sites of the Twist promoter did not detect promoter DNA in the anti-STAT3 immunoprecipitates (Fig. 4E). This result was further confirmed in MCF7 cells that were treated with or without IL-6 (Fig. 4F and data not shown). Because the second proximal STAT3 site mediated the maximal Twist promoter activity, whereas the first and third sites did not facilitate the promoter activity (Fig. 4C), our data suggest that the second proximal site is the major STAT3 recognition site. This site is 75 bases upstream of the TATA box previously mapped by Wang et al. (31). Taken together, our data provide evidence that STAT3 directly binds the Twist promoter and activates its transcription.
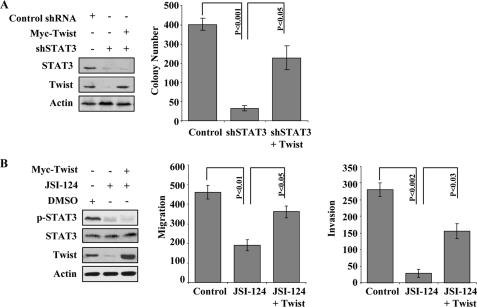
Twist Rescues STAT3-mediated Colony Formation, Migration, and Invasion upon Knockdown of STAT3—Having explored the transcriptional regulatory mechanism of STAT3-mediated Twist expression, we proceeded to address the biological significance of the STAT3-Twist connection in the regulation of anchorage-independent growth, migration, and invasion. The stable knockdown of STAT3 in MDA-MB-453-I4 cells, selected with puromycin, led to a dramatic decrease in colony formation ability, which was partially rescued by the co-expression of cytomegalovirus promoter-driven Twist (Fig. 5A). This supports the notion that Twist is downstream of STAT3 and could mediate STAT3 oncogenic function. In addition, we assessed the effect of STAT3-Twist axis on cell migration and invasion. MDA-MB-453-I4 cells were treated with the STAT3 inhibitor JSI-124 or vehicle Me2SO control and then examined for migration and invasion. Inhibition of STAT3 activation significantly attenuated both migration and invasion abilities of the I4 cells, which again were partially but significantly rescued by Twist (Fig. 5B). Although inhibition of STAT3 also deceased cell survival and proliferation moderately (supplemental Fig. S1), this should not have significant impact on our cell migration and invasion experiments, because the same number of treated and untreated cells was used right before the Boyden chamber assays. These data suggest that the STAT3-Twist axis is an important pathway in regulating invasion and anchorage-independent growth and that STAT3 exerts its function, at least in part, through Twist.
FIGURE 5.
Twist mediates the effects of STAT3 on colony formation, migration, and invasion. A, colony assays were performed using 1 × 105 cells/6-cm dish. Stable STAT3 knockdown MDA-MB-453-I4 cells were fed with 500 μl of 10% FCS-DMEM every 2 days. Photomicrographs were taken after 14 days. The colonies were quantified by ImageJ software (National Institutes of Health) from digitized images from three independent experiments. The average number of colonies with respective S.E. were calculated and presented. The left panels show the expression of knockdown of STAT3 and transfected Twist. B, migration and invasion assay. MDA-MB453-I4 cells were transfected with pCMV vector or pCMV-Twist and treated with 0.5 μm JSI-124 as indicated. Twenty-four hours post-treatment, 1 × 105 cells each were seeded onto the migration/invasion chamber for the respective assays as described under “Experimental Procedures.” After 6 h, the migrated/invaded cells were quantified. The experiments were repeated three times in triplicate. The p values for comparisons are indicated. The immunoblots show the inhibition of p-STAT3 and the expression of transfected Twist (left panels).
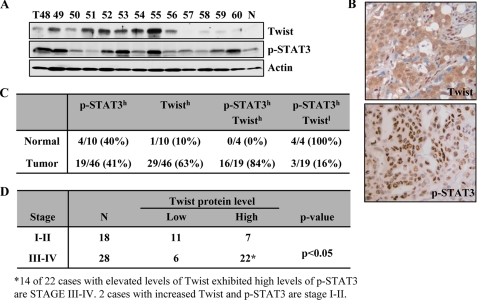
Overexpression of Twist Correlates with STAT3 Activation in Late Stage of Breast Cancer—Having observed that STAT3 mediated Twist expression in established cancer lines, we asked whether this connection occurs in tumors. We examined 46 primary breast carcinomas and 10 normal control specimens for phospho-STAT3 and Twist expression by Western blot and immunohistochemistry (Fig. 6, A and B). Of the 46 breast tumors, 19 had elevated phospho-STAT3 levels, and 29 showed overexpression of Twist. Of the tumors with elevated phospho-STAT3, 16 (84%) also had elevated Twist levels, whereas 3 (16%) had low Twist levels (Fig. 6A). Overexpression of phospho-STAT3 and Twist are located specifically to the nucleus of the cancer cells and not to that of the surrounding stromal cells (Fig. 6B). By contrast, four “normal” samples, which have benign fibrocystic changes, had elevated phospho-STAT3 levels, and only one had moderate level of Twist expression (Fig. 6C). Notably, whereas elevated levels of Twist were not significantly different between the tumor grades, increased Twist expression was closely associated with late stage of breast tumors (Fig. 6D). Further, co-overexpression of Twist and p-STAT3 was predominantly detected in late stage tumors (14 out of 22 cases) (Fig. 6D). These data further support the findings of biochemical and functional links between STAT3 and Twist.
FIGURE 6.
Overexpression of Twist correlates with activated STAT3 in late stage breast cancer. A, Western blot analysis. Representative tumor and normal breast tissue lysates were analyzed by Western blot with the indicated antibodies. Intensity of Twist, phospho-STAT3-Tyr705, and actin were quantified via ImageJ software (National Institutes of Health) and normalized to actin. The overexpression of Twist and activated STAT3 in tumor samples was scored based on the average values of the normal tissues from three independent experiments. B, representative photomicrographs of sections of breast tumor tissue from the same patient stained with anti-phospho-STAT3-Tyr705 and -Twist antibodies. C, summary of the immunoblotting and immunohistochemistry staining results. D, association between expression of Twist and/or p-STAT3 with breast cancer stages.
DISCUSSION
Using our selected invasive breast cancer line model (16), we demonstrated a direct link between activated STAT3 and Twist, two key transcriptional factors that are associated with breast cancer progression. The correlation of STAT3 and Twist levels has been observed before; however, the specific mechanisms of how Twist level was regulated by STAT3 were not clear (32). Our study indicates that constitutively active STAT3 is a causal factor for up-regulation of Twist at the transcriptional level. Our data demonstrate that Twist is a target gene of STAT3, and STAT3-Twist signaling plays an important role in regulating migration/invasion and anchorage independent growth of breast cancer cells. Our data (supplemental Fig. S1) showed that transfection with STAT3-shRNA resulted in about 20% inhibition of growth at 48 h; thus both inhibition of STAT3-promoted proliferation and anchorage-independent survival may contribute to the observed 90% inhibition of colony formation upon treatment with STAT3-shRNA (Fig. 5). Our study provided the evidence showing that Twist is transcriptionally regulated by STAT3 and mediates STAT3 function. Identification of Twist as a transcriptional target and mapping of the STAT3-binding site on Twist promoter is in agreement with that observed by Lo et al. (33). Together with our previous report that Twist transcriptionally activates AKT2, STAT3/Twist/AKT2 forms a signaling axis involved in breast cancer progression and metastasis. This also at least in part explains the frequent co-expression of these molecules in cancer cells.
A number of reports have demonstrated that Twist is frequently overexpressed in a variety of types of human malignancy and whose alteration is associated with late stage and high grade tumors. However, no evidence shows alterations of Twist at the DNA level during the tumor progression. In the present study, we showed frequent co-existence of activated STAT3 and overexpression of Twist in human primary breast carcinoma. Strong association of phospho-STAT3 and elevated Twist expression in breast tumors underscored the clinical significance of the STAT3-Twist signaling axis. It is very likely that activated STAT3 is one of the major transcriptional factors that contribute to up-regulation of Twist in other human primary tumors. Another known transcriptional regulator of Twist is NF-κB (14); however, its role in Twist induction in cancer is unclear.
The importance of STAT3 in oncogenesis has been well established by its constitutive phosphorylation in the majority of human neoplasms and its capacity to induce cell transformation (19, 34). STAT3 and its downstream targets are implicated in all processes of tumorigenesis from initiation to progression to invasion (9, 17, 35). The last step, invasion, is of vital importance because the greatest challenge in cancer therapy lies in the prevention of the spread of cancer cells. Increasing evidence suggests that STAT3 is intimately involved in regulating cell motility, migration, and invasiveness in both physiological and pathological situations. For example, during normal zebrafish gastrulation STAT3 controls cell migration via up-regulation of the LIV-1 gene (36, 37), a zinc transporter required for Snail nuclear localization (38). In terms of cell migration, STAT3-deficient murine keratinocytes displayed deficiency in cell migration as assessed via wound assay, in part because of abnormal p130CAS phosphorylation (39). Finally, in different carcinomas, activated STAT3 is able to potentiate the metastatic spread. For example, in melanoma-derived cell lines, activated STAT3 transactivated matrix metalloproteinase 2 gene (MMP-2) (40). In bladder cancer cells, STAT3 activation induced genes encoding MMP-1 and MMP-10, leading to increased migration and invasion in vivo (41). STAT3 also mediated interleukin-6 (IL-6)-mediated T cell migration (42). In MCF-7 breast cancer cells, oncostatin M-mediated STAT3 activation resulted in morphological alterations and increased migration (43). In addition to p130CAS and MMP proteins, we showed in the present study that Twist is a downstream transcriptional target of STAT3 and mediates its action in cell migration and invasion.
Previously, we have shown that Twist transcriptionally regulated AKT2 and that AKT2 mediated Twist function in cancer cell migration, invasion, and survival (16). Based on our current observations, we believe that STAT3 may regulate AKT2 expression through Twist. Additionally, a previous report has demonstrated that STAT3 transcriptionally modulated AKT1 expression (44). These observations suggest that STAT3 may promote cell survival via AKT1 and enhance tumor invasion via Twist and AKT2. Pharmacological agents that inhibit STAT3 or its downstream mediators Twist and AKT2 may lead to inhibition of both tumor progression and invasion underscoring the significance of the STAT3-Twist-AKT2 axis as an attractive target for cancer therapy.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants CA29339 (to L.-H. W.) and CA107078 (to J. Q. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Footnotes
The abbreviations used are: STAT, signal transducers and activators of transcription; IL, interleukin; shRNA, small hairpin RNA; DMEM, Dulbecco's modified Eagle's medium; FCS, fetal calf serum; RT, reverse transcription; ChIP, chromatin immunoprecipitation; EMSA, electrophoretic mobility shift assay; GFP, green fluorescent protein; ERK, extracellular signal-regulated kinase; MMP, matrix metalloproteinase.
References
- 1.Bialek, P., Kern, B., Yang, X., Schrock, M., Sosic, D., Hong, N., Wu, H., Yu, K., Ornitz, D. M., Olson, E. N., Justice, M. J., and Karsenty, G. (2004) Dev. Cell 6 423-435 [DOI] [PubMed] [Google Scholar]
- 2.Baylies, M. K., and Bate, M. (1996) Science 272 1481-1484 [DOI] [PubMed] [Google Scholar]
- 3.Castanon, I., Von Stetina, S., Kass, J., and Baylies, M. K. (2001) Development 128 3145-3159 [DOI] [PubMed] [Google Scholar]
- 4.El Ghouzzi, V., Legeai-Mallet, L., Aresta, S., Benoist, C., Munnich, A., de Gunzburg, J., and Bonaventure, J. (2000) Hum. Mol. Genet. 9 813-819 [DOI] [PubMed] [Google Scholar]
- 5.Puisieux, A., Valsesia-Wittmann, S., and Ansieau, S. (2006) Br. J. Cancer 94 13-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, Z., Xie, D., Li, X., Wong, Y. C., Xin, D., Guan, X. Y., Chua, C. W., Leung, S. C., Na, Y., and Wang, X. (2007) Hum. Pathol. 38 598-606 [DOI] [PubMed] [Google Scholar]
- 7.Ohuchida, K., Mizumoto, K., Ohhashi, S., Yamaguchi, H., Konomi, H., Nagai, E., Yamaguchi, K., Tsuneyoshi, M., and Tanaka, M. (2007) Int. J. Cancer 120 1634-1640 [DOI] [PubMed] [Google Scholar]
- 8.Lee, T. K., Poon, R. T., Yuen, A. P., Ling, M. T., Kwok, W. K., Wang, X. H., Wong, Y. C., Guan, X. Y., Man, K., Chau, K. L., and Fan, S. T. (2006) Clin. Cancer Res. 12 5369-5376 [DOI] [PubMed] [Google Scholar]
- 9.Yang, J., Mani, S. A., Donaher, J. L., Ramaswamy, S., Itzykson, R. A., Come, C., Savagner, P., Gitelman, I., Richardson, A., and Weinberg, R. A. (2004) Cell 117 927-939 [DOI] [PubMed] [Google Scholar]
- 10.Maestro, R., Dei Tos, A. P., Hamamori, Y., Krasnokutsky, S., Sartorelli, V., Kedes, L., Doglioni, C., Beach, D. H., and Hannon, G. J. (1999) Genes Dev. 13 2207-2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mironchik, Y., Winnard, P. T., Jr., Vesuna, F., Kato, Y., Wildes, F., Pathak, A. P., Kominsky, S., Artemov, D., Bhujwalla, Z., Van Diest, P., Burger, H., Glackin, C., and Raman, V. (2005) Cancer Res. 65 10801-10809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zong, C. S., Zeng, L., Jiang, Y., Sadowski, H. B., and Wang, L. H. (1998) J. Biol. Chem. 273 28065-28072 [DOI] [PubMed] [Google Scholar]
- 13.Yuen, H. F., Chan, Y. P., Wong, M. L., Kwok, W. K., Chan, K. K., Lee, P. Y., Srivastava, G., Law, S. Y., Wong, Y. C., Wang, X., and Chan, K. W. (2006) J. Clin. Pathol. 60 510-514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pham, C. G., Bubici, C., Zazzeroni, F., Knabb, J. R., Papa, S., Kuntzen, C., and Franzoso, G. (2007) Mol. Cell Biol. 27 3920-3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alexander, N. R., Tran, N. L., Rekapally, H., Summers, C. E., Glackin, C., and Heimark, R. L. (2006) Cancer Res. 66 3365-3369 [DOI] [PubMed] [Google Scholar]
- 16.Cheng, G. Z., Chan, J., Wang, Q., Zhang, W., Sun, C. D., and Wang, L. H. (2007) Cancer Res. 67 1979-1987 [DOI] [PubMed] [Google Scholar]
- 17.Haura, E. B., Turkson, J., and Jove, R. (2005) Nat. Clin. Pract. Oncol. 2 315-324 [DOI] [PubMed] [Google Scholar]
- 18.Levy, D. E., and Darnell, J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3 651-662 [DOI] [PubMed] [Google Scholar]
- 19.Bromberg, J. F., Horvath, C. M., Besser, D., Lathem, W. W., and Darnell, J. E., Jr. (1998) Mol. Cell Biol. 18 2553-2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu, C. L., Meyer, D. J., Campbell, G. S., Larner, A. C., Carter-Su, C., Schwartz, J., and Jove, R. (1995) Science 269 81-83 [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, T. J., and John, S. (2005) Immunology 114 301-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedranzini, L., Leitch, A., and Bromberg, J. (2004) J. Clin. Investig. 114 619-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernandes, A., Hamburger, A. W., and Gerwin, B. I. (1999) Int. J. Cancer 83 564-570 [DOI] [PubMed] [Google Scholar]
- 24.Garcia, R., Yu, C. L., Hudnall, A., Catlett, R., Nelson, K. L., Smithgall, T., Fujita, D. J., Ethier, S. P., and Jove, R. (1997) Cell Growth Differ. 8 1267-1276 [PubMed] [Google Scholar]
- 25.Calo, V., Migliavacca, M., Bazan, V., Macaluso, M., Buscemi, M., Gebbia, N., and Russo, A. (2003) J. Cell Physiol. 197 157-168 [DOI] [PubMed] [Google Scholar]
- 26.Sierra, J., Villagra, A., Paredes, R., Cruzat, F., Gutierrez, S., Javed, A., Arriagada, G., Olate, J., Imschenetzky, M., Van Wijnen, A. J., Lian, J. B., Stein, G. S., Stein, J. L., and Montecino, M. (2003) Mol. Cell Biol. 23 3339-3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blaskovich, M. A., Sun, J., Cantor, A., Turkson, J., Jove, R., and Sebti, S. M. (2003) Cancer Res. 63 1270-1279 [PubMed] [Google Scholar]
- 28.Cao, X., Tay, A., Guy, G. R., and Tan, Y. H. (1996) Mol. Cell Biol. 16 1595-1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath, C. M., Wen, Z., and Darnell, J. E., Jr. (1995) Genes Dev. 9 984-994 [DOI] [PubMed] [Google Scholar]
- 30.Wegenka, U. M., Buschmann, J., Lutticken, C., Heinrich, P. C., and Horn, F. (1993) Mol. Cell Biol. 13 276-288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S. M., Coljee, V. W., Pignolo, R. J., Rotenberg, M. O., Cristofalo, V. J., and Sierra, F. (1997) Gene (Amst.) 187 83-92 [PubMed] [Google Scholar]
- 32.Ling, X., and Arlinghaus, R. B. (2005) Cancer Res. 65 2532-2536 [DOI] [PubMed] [Google Scholar]
- 33.Lo, H. W., Hsu, S. C., Xia, W., Cao, X., Shih, J. Y., Wei, Y., Abbruzzese, J. L., Hortobagyi, G. N., and Hung, M. C. (2007) Cancer Res. 67 9066-9076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spiekermann, K., Pau, M., Schwab, R., Schmieja, K., Franzrahe, S., and Hiddemann, W. (2002) Exp. Hematol. 30 262-271 [DOI] [PubMed] [Google Scholar]
- 35.Buettner, R., Mora, L. B., and Jove, R. (2002) Clin. Cancer Res. 8 945-954 [PubMed] [Google Scholar]
- 36.Yamashita, S., Miyagi, C., Carmany-Rampey, A., Shimizu, T., Fujii, R., Schier, A. F., and Hirano, T. (2002) Dev. Cell 2 363-375 [DOI] [PubMed] [Google Scholar]
- 37.Yamashita, S., Miyagi, C., Fukada, T., Kagara, N., Che, Y. S., and Hirano, T. (2004) Nature 429 298-302 [DOI] [PubMed] [Google Scholar]
- 38.Cano, A., Perez-Moreno, M. A., Rodrigo, I., Locascio, A., Blanco, M. J., del Barrio, M. G., Portillo, F., and Nieto, M. A. (2000) Nat. Cell Biol. 2 76-83 [DOI] [PubMed] [Google Scholar]
- 39.Kira, M., Sano, S., Takagi, S., Yoshikawa, K., Takeda, J., and Itami, S. (2002) J. Biol. Chem. 277 12931-12936 [DOI] [PubMed] [Google Scholar]
- 40.Xie, T. X., Wei, D., Liu, M., Gao, A. C., Ali-Osman, F., Sawaya, R., and Huang, S. (2004) Oncogene 23 3550-3560 [DOI] [PubMed] [Google Scholar]
- 41.Itoh, M., Murata, T., Suzuki, T., Shindoh, M., Nakajima, K., Imai, K., and Yoshida, K. (2006) Oncogene 25 1195-1204 [DOI] [PubMed] [Google Scholar]
- 42.McLoughlin, R. M., Jenkins, B. J., Grail, D., Williams, A. S., Fielding, C. A., Parker, C. R., Ernst, M., Topley, N., and Jones, S. A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 9589-9594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, F., Li, C., Halfter, H., and Liu, J. (2003) Oncogene 22 894-905 [DOI] [PubMed] [Google Scholar]
- 44.Park, K. J., Krishnan, V., O'Malley, B. W., Yamamoto, Y., and Gaynor, R. B. (2005) Mol. Cell 18 71-82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.