Abstract
Monocyte chemotactic protein-1 (MCP-1) has been recognized as an angiogenic chemokine. The molecular mechanism of MCP-1-mediated angiogenesis remains unknown. We recently identified a novel transcription factor, designated MCP-1-induced protein (MCPIP), in human monocytes after treatment with MCP-1. We investigated whether MCP-1-induced angiogenesis is mediated via MCPIP. Treatment of human umbilical vein endothelial cells (HUVECs) with MCP-1 induced expression of MCPIP and capillary-like tube formation. Knockdown of MCPIP by small interfering RNA (siRNA) suppressed MCP-1-induced angiogenesis-related gene VEGF and HIF-1α expression as well as tube formation. Transfection of HUVECs with an MCPIP expression vector induced angiogenesis-related genes and tube formation. Chromatin immunoprecipitation analysis revealed that cadherin (cdh) 12 and cdh19 are in vivo targets of MCPIP. Transfection of HUVECs with MCPIP expression vector activated the expression of cdh12 and cdh19 genes. Knockdown of cdh12 or cdh19 expression markedly inhibited MCPIP-induced capillary-like tube formation. Moreover, knockdown of MCPIP also significantly suppressed MCP-1-induced cdh12 and cdh19 gene expression. Our data strongly suggest that MCP-1-induced angiogenesis is mediated via MCPIP, at least in part through transcriptional activation of cdh12 and cdh19.
Angiogenesis, the formation of new blood vessels from pre-existing vessels in adult tissue, is a key process involved in inflammatory diseases such as diabetes, ischemic heart, and limb diseases and tumor growth (1, 2). Although the critical initiating event for the generation of new blood vessels has been attributed to the production of growth factors, recruitment of monocytes has been suggested to be important in the angiogenic cascade (3, 4). Accumulation of leukocytes at the inflammatory sites is regulated by chemotactic small molecular weight proteins called chemokines. Monocyte chemotactic protein-1 (MCP-1),2 a key CC chemokine responsible for trafficking and activation of monocytes/macrophages through its receptor CCR2, has been implicated in inflammation and angiogenesis (5, 6). Administration of exogenous MCP-1 has been shown to increase monocyte/macrophage recruitment, collateral vessel formation, and blood flow to the ischemic tissue in hindlimb models of ischemia (6–8). By drilling tunnels through myocardial tissue, monocytes/macrophages were reported to increase angiogenesis in ischemic myocardium (9). MCP-1 can also directly act on endothelial cells (ECs) to induce angiogenesis (10, 11). However, the mechanisms by which MCP-1 mediates these effects on angiogenesis are unclear.
We recently identified a novel transcription factor, designated MCP-1-induced protein (MCPIP), which was originally found in human monocytes after treatment with MCP-1 and is proapoptotic (12). MCP-1 induces this transcription factor, which in turn up-regulates members of the apoptotic gene family that have been linked to angiogenesis and vascular remodeling (13–15). Therefore, it appeared possible that MCP-1-induced angiogenesis might be mediated by transcription factor MCPIP. Here, we report that MCP-1 treatment of human umbilical vein endothelial cells (HUVECs) resulted in induction of MCPIP and that expression of MCPIP enhanced endothelial cell apoptosis, proliferation, migration, and expression of angiogenesis-related genes, resulting in capillary-like tube formation. All of these angiogenic effects of MCP-1 and expression of MCPIP were inhibited by MCPIP-specific small interfering RNA (siRNA). The chromatin immunoprecipitation assay revealed that cadherin (cdh)12 and cdh19 were in vivo targets of MCPIP. Knockdown of MCPIP expression significantly reduced transcript levels of cdh12 and cdh19. Moreover, knockdown of either cdh12 or cdh19 expression inhibited MCPIP-induced capillary-like tube formation. These results strongly suggest that MCP-1-induced angiogenesis is mediated via induction of MCPIP, the newly discovered transcription factor, at least in part through transcriptional activation of cdh12 and cdh19 that have not previously been implicated in angiogenesis.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions—The HUVECs (Clonetics) were grown in endothelial cell basal medium supplemented with hydrocortisone (1 μg/ml), bovine brain extract (12 μg/ml), gentamicin (50 μg/ml), amphotericin B (50 ng/ml), epidermal growth factor (10 ng/ml), and 2% fetal bovine serum (EGM Single Quots, Clonetics) as recommended by the manufacturer. HUVECs were used between passages 4 and 8. The cell line HEK293 was grown in Dulbecco modified Eagle's medium supplemented with 10% fetal calf serum. All cells were maintained at 37 °C in 5% CO2.
Plasmid Construction and Transfection—The human MCPIP cDNA encoding the full-length MCPIP (GenBank™ accession number: AY920403) was cloned into BamH1 and EcoR1 sites of a pEGFP/N1 vector to generate the GFP-MCPIP fusion protein as described previously (12). Transfection of MCPIP plasmid in HUVECs was performed using Lipofectamine PLUS Reagents (Invitrogen) with a transfection efficiency of about 60–70%, as determined by the green fluorescence.
Knockdown with siRNA—HUVECs, fourth generation, were cultured in EGM BulletKit medium (Cambrex) according to the manufacturer's recommendation. Human MCPIP SMART pools designed by Dharmacon (Lafayette, CO) were delivered into 70% confluent cells with the use of Lipofectamine™ and PLUS™ Reagents (Invitrogen) according to the manufacturer's protocol. Human MCPIP siRNA SMART pools targeting the sense sequence 5′-GUAAGAAGCCACUCACUUUUU-3′, 5′-GCAAGCGGGUGGUGUGCUAUU-3′, 5′-CCAACACGGUGCUGGGUGAUU-3′, 5′-AUACUAAGCUGUGUGGUGUUU-3′, and the antisense sequence 5′-AAAGUGAGUGGCUUCUUACUU-3′, 5′-UAGCACACCACCCGCUUGCUU-3′, 5′-UCACCCAGCACCGUGUUGGUU-3′, 5′-ACACCACACAGCUUAGUAUUU-3′ were selected. Human cdh12 siRNA SMART pools targeting the sense sequence 5′-GAGACAACGUCAUCCAUUAUU-3′, 5′-GGACAGCUACUUUACAAUAUU-3′, 5′-GGGCAACAAUUCUCCUUUAUU-3′, 5′-GCAGUAUAAUUUCUCCAUAUU-3′, and the antisense sequence 5′-UAAUGGAUGACGUUGUCUCUU-3′, 5′-UAUUGUAAAGUAGCUGUCCUU-3′, 5′-UAAAGGAGAAUUGUUGCCCUU-3′, 5′-UAUGGAGAAAUUAUACUGCUU-3′ were selected. Human cdh19 siRNA SMART pools targeting the sense sequence 5′-UAGGAACAAUCAUGGCAUAUU-3′, 5′-GAUAAUGGUACAAUCACUAUU-3′, 5′-GCUGAGGAGUAGUACCAUAUU-3′, 5′-CCAGCUAAGAUCUGAUUUAUU-3′ and the antisense sequence 5′-UAUGCCAUGAUUGUUCCUAUU-3′, 5′-UAGUGAUUGUACCAUUAUCUU-3′, 5′-UAUGGUACUACUCCUCAGCUU-3′, 5′-UAAAUCAGAUCUUAGCUGGUU-3′ were selected. HUVECs (5 × 104 cells/per well) were washed with Opti-MEM® I medium and incubated with Opti-MEM® I medium containing Lipofectamine/siRNA mixture (final concentration 100 nm of siRNA) for 6 h. Then, 2 ml of fresh EBM complete medium were added, and the cells were incubated for 24 h. To verify specificity of the knockdown effect, we used an oligonucleotide sequence 5′-UAGCGACUAAACACAUCAA-3′ (Dharmacon) with no known mammalian target as nonspecific siRNA.
Cell Migration Assays—The cell migration assay was performed as described previously (16). Briefly, HUVECs (5 × 104 cells/per well) were seeded into 6-well plates and grown to confluence. The cell monolayer was scratched with a plastic pipette tip to generate a cut of ∼1 mm in width, and the unattached cells were removed by washing twice. The remaining cells were transfected with the expression vector and incubated at 37 °C, 5% CO2 for 24 h. The number of cells that had migrated across the edge of the wound and into the denuded area was photographed and counted as migrating cells using the Metamorph Series 6.2 image program (Universal Imaging, West Chester, PA). Results were expressed as the average number of cells per field of view. The experiment was repeated three times.
BrdU Incorporation Assays—To determine the effect of MCPIP on cell proliferation, the rate of DNA synthesis was established by measuring BrdU incorporation in control and transfected HUVECs seeded in 8-well chamber glass slides. After incubation for 6 h with 10 μm BrdU, cells were fixed for 10 min with 3.7% paraformaldehyde and stained with an anti-BrdU antibody (Novus) for 60 min followed by staining with anti-rat IgG Cy2 antibody (1:500 dilution, Chemicon, Inc) for 30 min. The percentage of BrdU-positive nuclei (red) was determined by counting five randomly selected fields under ×20 magnification using the Metamorph Series 6.2 image program. The experiment was repeated three times.
In Vitro Capillary-like Tube Formation Assays—The ability of MCPIP to enhance HUVECs to form vascular network was tested in a standardized in vitro angiogenesis assay. Briefly, after transfection, HUVECs were harvested and seeded onto the surface of the polymerized fibril gels (1 × 104 cells/per well, Chemicon, Inc.) in 96-well plates, then incubated in EBM medium for 24 h. Tube formation was observed under a phase contrast microscope and photographed. Tube formation ability was quantified by counting the total number of cell clusters and branching in five randomly chosen microscopic fields per well under ×100 magnification. Results were expressed as the mean percentage of branching over total cell clusters and expressed as a ratio to the control. The experiment was repeated three times.
To examine whether MCP-1-induced angiogenic activity is mediated via MCPIP, HUVECs were incubated in EBM medium with the presence or absence of 100 nm of MCPIP siRNA. Then 100 ng/ml of the recombinant human MCP-1 were added to the medium for 24 h. Capillary-like tube formation was assayed as described above.
Detection of Apoptotic Cell Death in HUVECs—HUVECs (5 × 104/per well) were seeded onto 4-well chamber glass slides and were grown to confluence. After transfection with the MCPIP-GFP expression vector or GFP control for 24 h, cells were fixed with 3.7% paraformaldehyde and then pretreated with 0.1% Triton X-100 in 0.1% sodium citrate. The TUNEL assay was performed utilizing a TMR red in situ cell death detection kit (Roche Applied Science) per the manufacturer's instructions, and counterstaining of all nuclei was done with 4,6-diamidino-2-phenylindole (DAPI, Molecular Probes). The number of TUNEL-positive cells were counted and divided by the total number of cells in ten randomly selected fields of view under the fluorescence microscope. The experiment was repeated three times.
Gene Expression Profiling by Oligo GEArray Microarray— Angiogenesis-related gene expression profiling was performed using Oligo GEArray human angiogenesis microarray (Biosciences Corp.) according to the manufacturer's instructions. Briefly, total RNA was isolated from HUVECs using TRIzol reagent (Invitrogen). The biotin UTP-labeled cDNA probes were generated using 3 μg of total RNA. The array filters were hybridized with 6 μg of biotin-labeled probes at 60 °C overnight in the hybridization oven. GEArray membranes were washed and blocked with GEArray blocking solution, then incubated with alkaline phosphatase-conjugated streptavidin, and exposed to x-ray film (Kodak). Signal quantification of gene expression on the array was performed with the software supplied by the manufacturer.
Chromatin Immunoprecipitation (ChIP) and Gel Retardation Assays—ChIP analysis was done essentially as described previously (17). HEK293 cells (3 × 107), transfected with pEGFP/N1 or pEGFP/MCPIP vector, were treated with 1% formaldehyde for 10 min and then lysed with lysis buffer (10 mmol/liter EDTA, 1% SDS, 1 mmol/liter phenylmethylsulfonyl fluoride, 1 μg/ml pepstatin, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 50 mm Tris/HCl, pH 8.1) followed by sonication for 15 s (Sonifire 450, Branson). The sheared preparations were incubated with a rabbit polyclonal antibody against MCPIP overnight at 4 °C. The immune complex was recovered with A/G beads, and cross-linking was reversed. After removal of the protein by treatment with proteinase K, DNA was recovered by phenol/chloroform and precipitated with sodium acetate. The recovered DNA was cloned into PCR-Blunt II-TOPO plasmid vector (Invitrogen) and sequenced. The sequences were located in the genome data base. Genes that were within 5 kb from the cloned sequence were identified. The expression of these candidate genes in HEK293 cells by transfection with MCPIP-GFP construct compared with GFP control was determined by RT-PCR with the following primers: cdh12, forward: 5′-AGGAGGTGGGGAGGAAGATA-3′, reverse: 5′-CATATGTGGCCAGTGAATCG-3′; cdh19, forward: 5′-ATCTGCACCCACTGGGACTT-3′, reverse: 5′-CTGCTCAGGAACATGATGG-3′. The cloned fragments from these candidate genes were tested for binding to the recombinant MCPIP by gel retardation assays as we described previously (18). Binding reactions were performed in a total volume of 25 μl containing 32P-labeled probe, 2 μg of poly(dI:dC), 0.3 mg/ml acetylated bovine serum albumin, 25 mm HEPES, pH 7.9, 80 mm KCl, 1 mm dithiothreitol, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride, 10% glycerol, 0.4 μg of purified MCPIP protein, ∼ 2 fmol of 32P-labeled probe with or without 100- or 300-fold molar excess of specific/nonspecific competitors. The mixtures were incubated for 25 min at room temperature before electrophoresis on a 4% nondenaturing polyacrylamide gel and autoradiography of the dried gel.
RT-PCR and Real-time PCR Assays—Total RNA was isolated from HUVECs using TRIzol reagent. cDNA was synthesized using the SuperScript First Strand Synthesis System (Invitrogen) and was then amplified by PCR with the following primers: MCPIP, forward: 5′-AGTCTGACGGGATCGTGGTT-3′, reverse: 5′-GGGAGACGTACGGGAGTGAG-3′; cdh12, forward: 5′-AGGAGGTGGGGAGGAAGATA-3′, reverse: 5′-CATATGTGGCCAGTGAATCG-3′; cdh19, forward: 5′-ATCTGCACCCACTGGGACTT-3′, reverse: 5′-CTGCTCAGGAACATGATGG-3′; HIF-α, forward: 5′-TCTGGATGCTGGTGATTTGG-3′, reverse: 5′-GTGAATGTGGCCTGTGCAGT-3′; VE Cadherin, forward: 5′-GTGTTCACGCATCGGTTGTT-3′, reverse: 5′-GGCTCATCTGGGTCCTCAAC-3′; VEGF, forward: 5′-CCCTGGCTTTACTGCTGTAC-3′, reverse: 5′-TCTGAACAAGGCTCACAGTG-3′. The PCR reaction consisted of 35 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 30 s, and 72 °C for 10 min. β-Actin (forward: 5′-AAATCGTGCGTGACATCAAAG-3′, reverse 5′-TGTAGTTTCATGGATGCCACAG-3′) was amplified as an internal control. PCR products were electrophoresed on a 1.5% agarose gel stained with ethidium bromide and analyzed using the imaging system (Alphaimager 2200). To confirm the results of RT-PCR, mRNA expression was also analyzed using iCycler real-time PCR system (Bio-Rad) with the use of the above primers for MCPIP, cdh12, cdh19, VE-cadherin, HIF-α, and VEGF. The expression level of each candidate gene was normalized by subtracting the corresponding β-actin threshold cycle (CT) values.
Western Blotting—HUVECs in culture under different experimental conditions were lysed, and the cell lysate was collected. An equal amount of protein (25 μg) from each condition was subjected to 12.5% SDS-PAGE. Western blotting was carried out using the indicated primary antibodies: rabbit polyclonal anti-MCPIP antibody (12), 1:500; rabbit polyclonal anti-VE-cadherin antibody, 1:1000; human cdh12 polyclonal antibody, 1:500 (R&D Systems); anti-cdh19 antibody, 1:2000 (Abnova), followed by incubation with a horseradish peroxidase-conjugated secondary IgG. Immunoreactive proteins were detected using an enhanced chemiluminescence (ECL) kit (Amersham Biosciences).
Statistical Analysis—Data are expressed as the mean ± S.D. of a given number of observations. Results were compared between groups by one-way analysis of variance analysis followed by Student's t tests using SPSS 10.0 software (SPSS Inc) under Windows XP. A p value of <0.05 was considered to be significant.
RESULTS
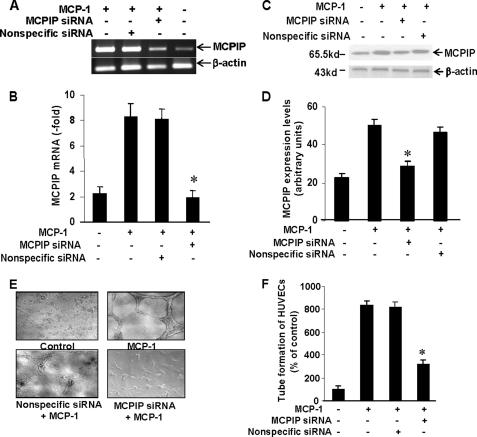
MCP-1 Induces Up-regulation of Angiogenesis-related Genes and in Vitro Angiogenesis via Transcription Factor MCPIP—To investigate whether transcription factor MCPIP might be involved in MCP-1-induced angiogenesis in endothelial cells, we tested whether MCP-1 could induce MCPIP in HUVECs. RT-PCR analysis revealed increased levels of MCPIP transcripts in HUVECs after treatment with MCP-1 (Fig. 1A). Real-time PCR and immunoblot analysis confirmed the up-regulated expression of MCPIP at both transcript and protein levels, respectively (Fig. 1, B–D). Both MCPIP mRNA and protein expression induced by MCP-1 in HUVECs were suppressed by treatment with siRNA specific for MCPIP, but not by nonspecific siRNA (Fig. 1, A–D). These results demonstrate that MCP-1 induced expression of MCPIP in HUVECs and effective knockdown of MCPIP by MCPIP-specific siRNA.
FIGURE 1.
MCP-1-induced angiogenesis is mediated via induction of MCPIP. HUVECs were treated with MCP-1 with or without transfection with MCPIP-specific or nonspecific siRNA for 24 h. Expression of MCPIP was detected by RT-PCR (A), real-time PCR (B), and immunoblot (C) analyses. β-Actin was amplified as an internal control for RT-PCR. D, histogram depicting the average MCPIP expression levels in the examined groups as assessed by immunoblot analysis. E, phase-contrast photomicrographs of HUVECs treated with MCP-1 with or without MCPIP-specific or nonspecific siRNA for 24 h (original magnification ×100), and the quantitative tube formation assay (F). *, p < 0.001 for treatment with MCP-1 and with nonspecific siRNA.
We examined the potential role of MCPIP in the control of MCP-1-induced angiogenesis using the in vitro angiogenesis assay. HUVECs treated with MCP-1 showed significantly increased numbers of capillary-like tube formation, and this tube formation was inhibited by the knockdown of MCPIP with siRNA for MCPIP, whereas nonspecific siRNA showed no effect (Fig. 1, E and F).
The effects of knockdown of MCPIP on the expression profile of MCP-1-induced angiogenesis-related genes were also examined using the Oligo GEArray human angiogenesis microarray, which contained a total of 113 genes that are involved in modulating angiogenesis (19–21). As summarized in Table 1, 32 of 113 genes were up-regulated in MCP-1-treated HUVECs compared with untreated-HUVECs (only genes whose expression was induced at least 2.0-fold are included). These up-regulated genes included angiopoietin-like 3, angiopoietin-like 4, VE-cadherin, the VEGF family, Tie-1, ephrin A2, MMP-1, TIMP-2, urokinase, and chemokine ligands. The addition of siRNA for MCPIP suppressed MCP-1-induced expression of most of these genes (Table 1).
TABLE 1.
Expression profile of angiogenesis-related genes in HUVECs treatment with MCP-1- and MCPIP-specific siRNA + MCP-1
|
Gene name
|
Fold inductiona
|
|
|---|---|---|
| MCP-1 | siRNA + MCP-1 | |
| Angiopoietin-like 3 | 10.0 | |
| Angiopoietin-like 4 | 5.0 | |
| Cadherin 5 | 5.0 | |
| CD13/Gp156 | 5.0 | |
| Chemokine (CXC motif) ligand 11 | 5.0 | 2.4 |
| Shingolipid G-protein-coupled receptor 1 | 5.0 | 2.4 |
| Endoglin | 5.0 | |
| Laminin α5 | 5.1 | 2.1 |
| TIMP-1 | 4.7 | |
| Endostatin | 4.6 | |
| Prostaglandin-endoperoxide synthase 1 | 4.6 | 5.0 |
| Akt-1 | 4.3 | 2.4 |
| PECAM-1 | 4.2 | |
| Tie-1 | 4.2 | |
| VEGF-C | 4.1 | |
| Thrombospondin-1 | 4.1 | 2.5 |
| MMP-2 | 3.9 | 2.0 |
| VEGF-B | 3.5 | |
| ECGF-1 | 3.5 | 3.5 |
| Chemokine (CXC motif) ligand 10 | 3.4 | 4.7 |
| TEK tyrosine kinase | 3.2 | 2.1 |
| Angiopoietin 1 | 3.2 | |
| Platelet-derived growth factor α | 3.1 | 2.0 |
| Collagen type IV-α3 | 3.0 | |
| Interleukin 8 | 2.7 | |
| Ephrin A2 | 2.5 | |
| Jagged 1 | 2.3 | 2.2 |
| Chemokine (CXC motif) ligand 2 | 2.2 | 2.4 |
| Kinase insert domain receptor | 2.1 | |
| Urokinase | 2.1 | |
| Chemokine (CXC motif) ligand 1 | 2.0 | |
| Epidermal growth factor | 2.0 | |
Only genes whose expression was induced at least 2.0-fold are included. Most of angiogenesis-related genes-induced by MCP-1 were suppressed by MCPIP-specific siRNA
MCP-1-induced angiogenesis has been reported to be mediated through up-regulation of hypoxia-inducible factor-1α (HIF-1α) and subsequent induction of VEGF (11). The findings that siRNA for MCPIP suppressed MCP-1-induced expression of VEGF (Table 1) strongly suggest MCP-1-induced VEGF expression is mediated via MCPIP. Real-time PCR analysis confirmed that MCP-1 induced HIF-1α and VEGF production in HUVECs and siRNA for MCPIP, but not nonspecific siRNA, suppressed the MCP-1-induced HIF-1α and VEGF expression (Fig. 2, A and B). Furthermore, transfection of HUVECs with MCPIP-GFP expression vector, but not GFP control, induced HIF-1α and VEGF production (Fig. 2, A and B).
FIGURE 2.
Real-time PCR analysis of HIF-α (A) and VEGF (B) expression in HUVECs treated with MCP-1 with or without MCPIP-specific or nonspecific siRNA. *, p < 0.05 for treatment with MCP-1 and with nonspecific siRNA. #, p < 0.05 versus GFP vector-transfected HUVECs.
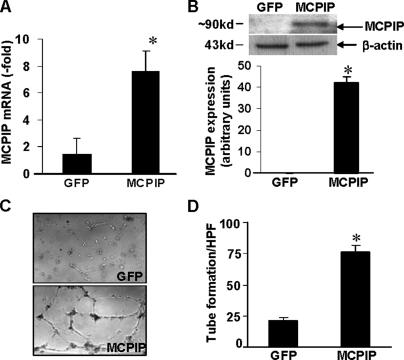
Expression of MCPIP Up-regulates Angiogenesis-related Genes and Promotes Capillary-like Tube Formation by HUVECs—We next examined whether expression of MCPIP in HUVECs directly up-regulates angiogenesis-related genes and induces angiogenesis. When HUVECs were transfected with MCPIP-GFP vector, the increased expression of MCPIP was found after 24 h at both mRNA and protein levels as measured by real-time PCR and immunoblot analyses, respectively (Fig. 3, A and B). When RNA harvested from GFP- or MCPIP-GFP-expressing HUVECs was subjected to angiogenesis gene array analysis, we observed that MCPIP induced up-regulation of 31 of 113 genes that are known to contribute to the increased angiogenic properties of endothelial cells (Table 2). These up-regulated genes included ephrin A1, ephrin B2, ephrin A3, IL-1β, notch homolog 4, angiopoietin-2, neuropilin-1, urokinase, PDGF-α, TIMP-2, MMP-9, and chemokine ligands. When HUVECs transfected with MCPIP were planted onto the surface of the polymerized fibrin gels for 24 h, we observed the typical capillary-like tube formation, whereas no significant angiogenic responses were observed in HUVECs transfected with the GFP expression control plasmid (Fig. 3C). Quantitative analysis of tube formation showed that tube formation in HUVECs transfected with MCPIP-GFP was much higher than that observed with GFP control (Fig. 3D). These results suggest that expression of MCPIP can directly induce endothelial cell capillary-like tube formation.
FIGURE 3.
Expression of MCPIP induces capillary-like tube formation in HUVECs. HUVECs were transfected with the MCPIP-GFP expression vector or GFP control for 24 h, and expression of MCPIP was detected by real-time PCR (A) and immunoblot (B) analyses. *, p < 0.001 versus GFP vector-transfected HUVECs. C, phase-contrast photomicrographs (original magnification ×100) of HUVECs seeded on the surface of the polymerized fibrin gels for 24 h after transfection with MCPIP-GFP expression vector or GFP control. D, mean number of tube branch points in randomly selected 5 high power fields (×40) of views was quantified. *, p < 0.05 versus GFP vector-transfected HUVECs.
TABLE 2.
Expression profile of angiogenesis-related genes in GFP/hMCPIP-over GFP-infected HUVECs
| Gene name | Fold induction |
|---|---|
| Ephrin-A1 | 12 |
| Interleukin 1β | 11.7 |
| Notch Homolog 4 | 11 |
| Ephrin B2 | 8.6 |
| Platelet-derived growth factor α | 7.6 |
| Tissue inhibitor of metalloproteinase 2 | 6.8 |
| Ephrin A3 | 5.8 |
| Midkine (neurite growth promoting factor 2) | 5.1 |
| Thrombospondin 1 | 5 |
| Colony-stimulating factor 3 | 5 |
| Angioprotein 2 | 4.4 |
| Chemokine (CXC motifs) ligand 9 | 4.3 |
| Angiogenic factor with path and FHA domains | 4 |
| Matrix metalloproteinase 9 | 3.8 |
| Hypoxia-inducible factor 1 | 3.6 |
| Chemokine (CXC motifs) ligand 2 | 3.5 |
| Chemokine (CXC motifs) ligand 3 | 3.4 |
| Chemokine (C-C motif) ligand 11 | 3.2 |
| Epidermal growth factor | 3.2 |
| Neuropilin 1 | 3.1 |
| Collagen type IV α3 | 2.6 |
| Angioprotein 1 | 2.5 |
| Tumor necrosis factor superfamily 12A | 2.5 |
| Chemokine (CXC motifs) ligand 5 | 2.5 |
| Chemokine (C-C motif) ligand 2 | 2.5 |
| Chemokine (CXC motifs) ligand 1 | 2.4 |
| Angioprotein-like 4 | 2.4 |
| Urokinase | 2.2 |
| VEGF | 2.0 |
| Interleukin 8 | 2.0 |
| Jagged 1 | 2.0 |
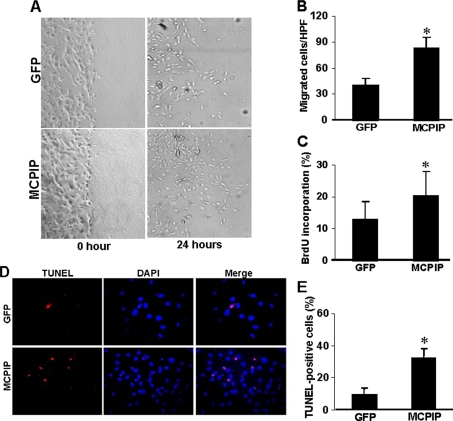
Influence of MCPIP on Endothelial Cell Behavior—Capillary-like tube formation in fibrin gels depends on the migratory and proliferative potential of endothelial cells. This process begins with the formation of endothelial cell sprouts initiated by apoptosis, followed by the proliferation and migration of neighboring endothelial cells along preformed extensions (15, 22). As MCPIP can induce capillary-like tube formation, we tested whether MCPIP might enhance angiogenesis-related properties of HUVECs. After 24 h of incubation, HUVECs transfected with MCPIP-GFP displayed significantly increased cell migration compared with cells transfected with GFP control (Fig. 4, A and B). DNA synthesis, as determined by BrdU incorporation, was also compared in HUVECs transfected with MCPIP-GFP or the GFP control. Results showed that MCPIP caused increased DNA synthesis (Fig. 4C). TUNEL assay and DAPI counterstaining were performed to detect apoptotic cell death in HUVECs after transfection with MCPIP-GFP expression vector or GFP control. After 24 h, HUVECs transfected with MCPIP-GFP showed a higher number of TUNEL-positive cells compared with cells transfected with the GFP control (Fig. 4, D and E). These results indicate that expression of MCPIP causes the induction of angiogenesis-related properties of HUVECs.
FIGURE 4.
Expression of MCPIP induces angiogenesis-related properties in HUVECs. A, confluent HUVECs monolayers were wounded by scraping and transfected with the expression vector for MCPIP-GFP or GFP control in serum-free medium. Cell migration to the wound surface was monitored from 0 to 24 h and quantitated at 24 h (B). Proliferation was detected by BrdU incorporation (C), and apoptosis was determined by TUNEL staining (D and E) in HUVECs transfected with expression vector for MCPIP-GFP or GFP control for 24 h. *, p < 0.05 versus GFP-vector transfected HUVECs.
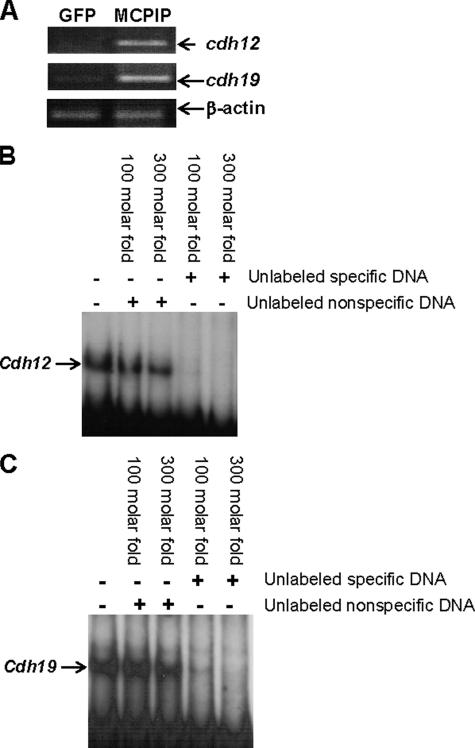
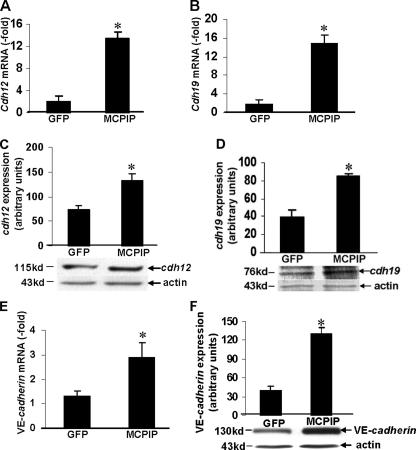
cdh12 and cdh19 Are in Vivo Targets of MCPIP—To identify the potential in vivo target genes for MCPIP, ChIP assays were performed in HEK293 cells transfected with the MCPIP-GFP expression vector. Sequencing of the MCPIP-bound genomic fragments revealed binding to cdh12 and cdh19 genes, indicating that these are in vivo targets of MCPIP. Because in vivo binding of a protein to DNA segments in the genome without affecting transcription is possible, we tested whether MCPIP expression up-regulates the expression of cdh12 and cdh19 genes. The transcript levels of cdh12 and cdh19 were elevated in HEK293 cells transfected with the MCPIP-GFP expression vector compared with cells transfected with the GFP control (Fig. 5A). Furthermore, gel retardation assays showed that the cloned fragments from cdh12 and cdh19 genes, where in vivo binding of MCPIP in the genome was indicated by the ChIP analysis, bound to recombinant MCPIP protein (Fig. 5, B and C). This binding was competed out by an excess of unlabeled specific gene fragments, but not by nonspecific DNA, demonstrating that this MCPIP binding was DNA sequence-specific. These results strongly suggest that cdh12 and cdh19 are in vivo targets of MCPIP.
FIGURE 5.
Induction of cdh12 and cdh19 by MCPIP and specific binding of MCPIP to cdh12 and cdh19. A, HEK293 cells were transfected with the MCPIP-GFP expression vector or GFP control for 12 h. RNA was isolated and subjected to RT-PCR using primers for cdh12 and cdh19 sequences. β-Actin was amplified as an internal control. B and C, gel retardation assays were performed as described under “Experimental Procedures” with a 32P-labeled 415-bp cdh12 gene fragment and a 207-bp cdh19 fragment with or without the indicated excess of unlabeled specific gene fragment or nonspecific DNA.
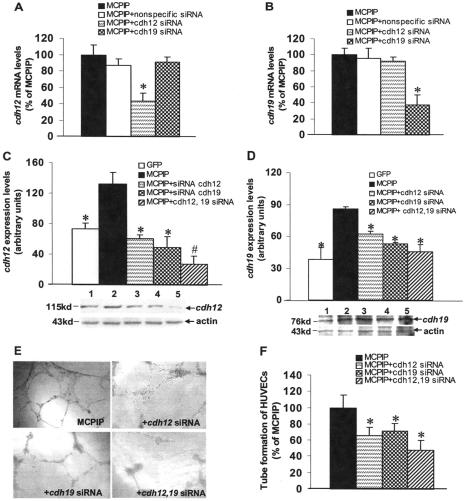
Contribution of cdh12 and cdh19 to MCPIP-induced Angiogenesis in Vitro—As cadherins have been shown to play a central role in the initiation of cellular response and the assembly of the vascular network (23), we tested whether MCPIP-mediated induction of cdh12 and cdh19 might be involved in MCPIP-induced angiogenesis by HUVECs. Real-time PCR analysis showed that expression of cdh12 and cdh19 were induced in HUVECs after transfection with MCPIP-GFP expression vector compared with cells transfected with the GFP control (Fig. 6, A and B), and the induction of cdh12 and cdh19 by protein expression was further confirmed by immunoblot analysis (Fig. 6, C and D), suggesting that MCPIP indeed up-regulated expression of cdh12 and cdh19 in HUVECs. Vascular endothelial (VE)-cadherin, an endothelial cell-specific cadherin required for angiogenesis, was also found to be induced by MCPIP at both transcript and protein levels (Fig. 6, E and F). siRNA specific for cdh12 and cdh19 markedly suppressed MCPIP-induced expression of cdh12 and cdh19 at both transcript and protein levels (Fig. 7, A–D). Specificity of knockdown was indicated by the real-time PCR analysis showing that siRNA for cdh12 did not affect the transcription of cdh19, and siRNA for cdh19 did not affect the transcription of cdh12 (Fig. 7, A and B). However, the levels of both cdh12 and cdh19 proteins decreased in HUVECs by siRNA specific for either cdh12 or cdh19 (Fig. 7, C and D). Knockdown of both cdh12 and cdh19 drastically diminished the levels of both cdh12 and cdh19 proteins induced by enhanced expression of MCPIP (Fig. 7, C and D). Thus, it is possible that siRNA for cdh12 and cdh19 not only affect transcription of their respective genes, but also translation of mRNA for both cdh12 and cdh19. siRNA specific for either cdh12 or cdh19 significantly inhibited MCPIP-induced capillary-like tube formation in vitro, and knockdown of both cdh12 and cdh19 by siRNA showed enhanced inhibition of MCPIP-induced cdh12 and cdh19 expression and capillary-like tube formation (Fig. 7, E and F).
FIGURE 6.
MCPIP induces cdh12 and cdh19 expression in HUVECs. A and B, HUVECs were transfected with MCPIP-GFP expression vector or GFP control for 24 h, and expression of cdh12 and cdh19 was detected by real-time PCR. The expression of both cdh12 and cdh19 at protein levels was demonstrated by immunoblot analysis (C and D). VE-cadherin, an endothelial cell-specific cadherin required for angiogenesis, was also found to be induced by MCPIP at both transcript and protein levels, assayed by real-time PCR (E) and Western blot (F). *, p < 0.05 versus GFP vector-transfected HUVECs.
FIGURE 7.
Contribution of cdh12 and cdh19 expression to MCPIP-mediated angiogenesis. A and B, real-time PCR analysis of cdh12 and cdh19 mRNA in HUVECs transfected with the expression vector for MCPIP-GFP with or without cdh12- and cdh19-specific or nonspecific siRNA showed that only siRNA specific for the particular cdh gene showed knockdown. *, p < 0.05 versus MCPIP vector-transfected HUVECs. C and D, histograms depicting the average cdh12 and cdh19 expression levels in the examined groups shown in the Western blot. Lane 1, GFP; lane 2, MCPIP; lane 3, MCPIP+cdh12 siRNA; lane 4, MCPIP+cdh19 siRNA; lane 5, MCPIP+cdh12,19 siRNA. *, p < 0.05; #, p < 0.01 versus MCPIP vector-transfected HUVECs. E, phase-contrast photomicrographs (original magnification ×100) of HUVECs seeded on the surface of the polymerized fibrin gels for 24 h after transfection with MCPIP expression vector, MCPIP+cdh12 siRNA, MCPIP+cdh19 siRNA, or MCPIP+cdh12, 19 siRNA. F, mean number of tube branch points in randomly selected 5 high-power field (×100) of views were quantified and expressed as a percentage of MCPIP-treated group; *, p < 0.05 versus MCPIP vector-transfected HUVECs.
We next examined whether MCP-1 induces expression of cdh12 and cdh19 in HUVECs during the development of tube formation. Real-time PCR and immunoblot analysis revealed a significant increase in cdh12 and cdh19 at transcripts and protein levels in HUVECs after treatment with MCP-1 for 24 h, and these increases were markedly suppressed by treatment with siRNA specific for MCPIP but not nonspecific siRNA (Fig. 8, A–D), suggesting that MCP-1-induced angiogenesis is associated with MCPIP-mediated induction of cdh12 and cdh19.
FIGURE 8.
MCP-1 treatment induces expression of cdh12 and cdh19 in HUVECs and siRNA specific for MCPIP inhibits their expression. A and B, HUVECs were treated with MCP-1 (100 ng/ml), in the presence or absence of MCPIP-specific or nonspecific siRNA for 24 h, and mRNA expression of cdh12 and cdh19 was assessed by real time-PCR demonstrating that knockdown of MCPIP inhibited MCP-1 induced expression of cdh12 and cdh19.*, p < 0.05 versus MCP-1- or nonspecific siRNA-treated HUVECs. β-Actin was amplified as an internal control. C and D, histograms depicting the average cdh12 and cdh19 expression levels in the examined groups shown in the Western blot. Lane 1, control; lane 2, MCP-1; lane 3, MCP-1+nonspecific siRNA; lane 4, MCP-1+MCPIP siRNA. *, p < 0.05 versus MCP-1-treated HUVECs or MCP-1 with nonspecific siRNA-treated HUVECs.
DISCUSSION
MCP-1 is known to facilitate angiogenesis (6, 7, 9–11). However, the mechanism by which MCP-1 mediates angiogenesis is unknown. Here we report that MCPIP, a novel transcription factor induced by MCP-1 in human peripheral blood monocytes (12), is also induced by MCP-1 in HUVECs and that this transcription factor mediates angiogenesis induced by MCP-1. Our study identified cdh12 and cdh19 as in vivo targets of MCPIP. Knockdown of MCPIP significantly suppressed MCP-1-induced expression of cdh12 and cdh19 in HUVECs. Moreover, knockdown of either cdh12 or cdh19 with specific siRNA inhibited MCPIP-induced angiogenesis. These findings strongly suggest MCP-1-induced angiogenesis is mediated via MCPIP, at least in part through transcriptional activation of cdh12 and cdh19.
Angiogenesis is a complex process that involves the activation of quiescent endothelial cells to the migratory and proliferative phenotype, and differentiation to the angiogenic phenotype (22). Recently, apoptosis of endothelial cells has been implicated in the initiation of angiogenesis and in the regression of neo-vessels (13–15). Several reports suggest that MCP-1-induced angiogenesis is mediated by monocyte chemotaxis (6–9) or through pathways involving VEGF and activation of RhoA small G protein (10, 11). MCPIP was initially identified as a transcription factor induced by MCP-1 in monocytes and it was shown to have a proapoptotic activity (12, 24). In the present study we demonstrate that MCPIP expression in HUVECs promotes cell proliferation and migration. The MCPIP-induced angiogenesis was found to be accompanied by induction of apoptotic cell death and the disappearance of the endothelial cell monolayer. When MCPIP-specific siRNA was added to HUVECs treated with MCP-1, they significantly inhibited the MCP-1-mediated angiogenesis, demonstrating the angiogenic activity of MCPIP.
Endothelial cells express a variety of genes during vascular development or angiogenesis (19–21). Thus, our discovery of MCPIP as a novel angiogenic factor prompted us to examine whether MCPIP plays a key role in MCP-1-induced expression of genes potentially involved in angiogenesis. Consistent with a role in endothelial sprouting and tube formation, many of the genes identified to be up-regulated by MCP-1 included molecules associated with cell communication and morphogenesis. These genes include the growth factors and receptors (angiopoietin-1, angiopoietin-like 3, angiopoietin-like 4, PDGF-α, VEGF, ECGF, EphA2), cytokines and chemokines (CXCL-1, CXCL-2, CXCL-10, and IL-8), adhesion molecules and matrix proteins (VE-cadherin, collagen type IV-α3, and laminin α5) as well as proteases and their inhibitors (MMP-2, TIMP-1, and urokinase). Such genes are now recognized to modulate the biochemical processes involved in angiogenesis (19–21). For example, angiopoietin-1 plays an essential role in regulating angiogenesis (25). Angiopoietin-like 3 and -4 are both agonists of the Tie2 receptor whose signaling is critical to regulating vascular stabilization and remodeling (25). EphA2 was reported to be important in regulating endothelial cell assembly and migration through phosphoinositide 3-kinase-mediated activation of Rac1 GTPase (26). Similarly, inflammatory chemokine IL-8 has been shown to have proangiogenic activity (27). To test whether these up-regulated genes induced by MCP-1 are mediated via MCPIP, RNA interference experiments were performed. Microarray analysis revealed that expression of most of these up-regulated genes was significantly suppressed by siRNA specific for MCPIP. Furthermore, we demonstrated that transfection of MCPIP in HUVECs with an MCPIP expression vector resulted in up-regulation of angiogenesis-related genes. Up-regulation of EphA3, EphB2, IL-1β, and notch homolog 4 by MCPIP that we observed, fit well with recent findings demonstrating coordinated participation of these gene products in angiogenesis (28–31).
Cadherins are commonly activated by vascular remodeling-related molecules and play a central role in the initiation of cellular response and the assemblies of the vascular network (23). ECs express two major cadherins, VE- and N-cadherins. The importance of VE-cadherin in vascular development has been well established (32), whereas N-cadherin is thought to function in adherence junctions between endothelial cells and mural cells (pericytes and vascular smooth muscle cells) (33). Although N-cadherin has been known to be abundantly expressed in endothelial cells (34), its role in endothelial cell function, including angiogenesis, has remained largely elusive. Recently, N-cadherin has been found to play a fundamental role in angiogenesis by modulating adherence junction components and EC behavior (35). The endothelial-specific knockout of N-cadherin in mice led to an aberrant vasculature both in the embryo and in the yolk sac, resulting in embryonic lethality at mid-gestation (35). Prior to the present study, there had been no previous documentation of involvement of cdh12 and cdh19 (both belong to N-cadherin family) in endothelial sprouting or angiogenesis. The ChIP assay, the widely used approach to identify the in vivo targets of transcription factors, revealed that cdh12 and cdh19 are in vivo targets of MCPIP in HEK293 cells. That cdh12 and cdh19 are the targets of MCPIP in HUVECs was shown by the induction of these genes through expression of MCPIP that also caused capillary-like tube formation. The inductions of cdh12 and cdh19 and tube formation were suppressed by knockdown of MCPIP with specific siRNA. Furthermore, down-regulation of cdh12 and cdh19 by specific siRNA significantly attenuated the capillary-like tube formation induced by the expression of MCPIP. These observations strongly suggest that MCPIP promotes angiogenesis at least in part via enhanced expression of cdh12 and cdh19. Further studies will be required to fully understand the newly discovered role of cdh12 and cdh19 in the regulation of angiogenesis.
MCP-1-induced angiogenesis has been reported to be mediated through up-regulation of HIF-1α and subsequent induction of VEGF (11). In the present study, the Oligo GEArray microarray showed up-regulation of HIF-1α (3.6-fold) in HUVECs transfected with the MCPIP-GFP expression vector over GFP control, and knockdown of MCPIP prevented this HIF-1α induction. A significant induction of VEGF was also observed in HUVECs treated with MCP-1 or transfected with the MCPIP-GFP expression vector. Knockdown of MCPIP with specific siRNA suppressed MCP-1-induced VEGF expression, suggesting that MCP-1-induced up-regulation of HIF-1α and induction of VEGF are mediated through the transcription factor MCPIP.
In conclusion, we have demonstrated that transcription factor MCPIP regulates the expression of cdh12 and cdh19 and is a modulator of angiogenesis. The present studies provide new insights into the mechanism by which MCP-1 induces angiogenesis. In future studies, however, it will be important to ascertain whether MCPIP actually accelerates angiogenesis in vivo.
This work was supported, in whole or in part, by National Institutes of Health Grant HL-69458. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: MCP, monocyte chemotactic protein; HUVEC, human umbilical vein endothelial cells; MCPIP, MCP-1-induced protein; siRNA, small interfering RNA; GFP, green fluorescent protein; TUNEL, terminal deoxynucleotidyltransferase-mediated dUTP nick end-labeling; ChIP, chromatin immunoprecipitation assay; VEGF, vascular endothelial growth factor; BrdU, bromodeoxyuridine; IL, interleukin.
References
- 1.Griffioen, A. W., and Molema, G. (2000) Pharmacol. Rev. 52 237-268 [PubMed] [Google Scholar]
- 2.Pandya, N. M., Dhalla, N. S., and Santani, D. D. (2006) Vasc. Pharmacol. 44 265-274 [DOI] [PubMed] [Google Scholar]
- 3.Arras, M., Ito, W. D., Scholz, D., Winkler, B., Schaper, J., and Schaper, W. (1998) J. Clin. Investig. 101 40-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herold, J., Pipp, F., Fernandez, B., Xing, Z., Heil, M., Tillmanns, H., and Braun-Dullaeus, R. C. (2004) Hum. Gene Ther. 15 1-12 [DOI] [PubMed] [Google Scholar]
- 5.Charo, I. F., and Taubman, M. B. (2004) Circ. Res. 95 858-866 [DOI] [PubMed] [Google Scholar]
- 6.Ito, W. D., Arras, M., Winkler, B., Scholz, D., Schaper, J., and Schaper, W. (1997) Circ. Res. 80 829-837 [DOI] [PubMed] [Google Scholar]
- 7.Voskuil, M., von Royen, N., Hoefer, I. E., Seidler, R., Guth, B. D., Bode, C., Schaper, W., Piek, J. J., and Buschmann, I. R. (2003) Am. J. Physiol. Heart Circ Physiol. 284 H1422-H1428 [DOI] [PubMed] [Google Scholar]
- 8.Deindl, E., and Schaper, W. (2005) Cell Biochem. Biophys. 43 1-15 [DOI] [PubMed] [Google Scholar]
- 9.Moldovan, N. I., Goldschmidt-Clermont, P. J., Parker-Thornburg, J., Shapiro, S. D., and Kolattukudy, P. E. (2000) Circ. Res. 87 378-384 [DOI] [PubMed] [Google Scholar]
- 10.Salcedo, R., Ponce, M. L., Young, H. A., Wasserman, K., Ward, J. M., Kleinman, H. K., Oppenhein, J. J., and Murphy, W. J. (2000) Blood 96 34-40 [PubMed] [Google Scholar]
- 11.Hong, K. H., Ryu, J., and Han, K. H. (2005) Blood 105 1405-1407 [DOI] [PubMed] [Google Scholar]
- 12.Zhou, L., Azfer, A., Niu, J., Graham, S., Choudhury, M., Adamski, F. M., Younce, C., Binkley, P. F., and Kolattukudy, P. E. (2006) Circ. Res. 98 1177-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peters, K., Troyer, D., Kummer, S., Kirkpatrick, C. J., and Rauterberg, J. (2002) Microvasc. Res. 64 334-338 [DOI] [PubMed] [Google Scholar]
- 14.Tertemiz, F., Kayisl, U. A., Aric, A., and Demir, R. (2005) Biol. Reprod. 72 727-735 [DOI] [PubMed] [Google Scholar]
- 15.Duval, H., Harris, M., Li, J., Johnson, N., and Print, C. (2003) Angiogenesis 6 171-183 [DOI] [PubMed] [Google Scholar]
- 16.Myers, C., Charboneau, A., and Boudreau, N. (2000) J. Cell Biol. 148 343-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nelson, E. A., Walker, S. R., Alvarez, J. V., and Frank, D. A. (2004) J. Biol. Chem. 279 54724-54730 [DOI] [PubMed] [Google Scholar]
- 18.Kamper, J. T., Kamper, U., Rogers, L. M., and Kolattukudy, P. E. (1994) J. Biol. Chem. 269 9195-9204 [PubMed] [Google Scholar]
- 19.Oettgen, P. (2001) Circ. Res. 89 380-388 [DOI] [PubMed] [Google Scholar]
- 20.Hamik, A., Wang, B., and Jain, M. K. (2006) Arterioscler. Thromb. Vasc. Biol. 26 1936-1947 [DOI] [PubMed] [Google Scholar]
- 21.Kontos, C. D., and Annex, B. H. (2007) Curr. Opin. Mol. Ther. 9 145-152 [PubMed] [Google Scholar]
- 22.Carmeliet, P. (2005) Nature 438 932-936 [DOI] [PubMed] [Google Scholar]
- 23.Blaschuk, O. W., and Rowlands, T. M. (2000) Cancer Metastasis Rev. 19 1-5 [DOI] [PubMed] [Google Scholar]
- 24.Bidzhekov, K., Zernecke, A., and Weber, C. (2006) Circ. Res. 98 1107-1109 [DOI] [PubMed] [Google Scholar]
- 25.Morisada, T., Kubota, Y., Urano, T., Suda, T., and Oike, Y. (2006) Endothelium 13 71-79 [DOI] [PubMed] [Google Scholar]
- 26.Zhang, J., and Hughes, S. E. (2006) J. Pathol. 208 453-461 [DOI] [PubMed] [Google Scholar]
- 27.Heidemann, J., Ogawa, H., Dwinell, M. B., Rafiee, P., Maaser, C., Gockel, H. R., Otterson, M. F., Ota, D. M., Lugering, N., Domschke, W., and Binion, D. G. (2003) J. Biol. Chem. 278 8508-8515 [DOI] [PubMed] [Google Scholar]
- 28.Cheng, N., Brantley, D. M., and Chen, J. (2002) Cytokine Growth Factor Rev. 13 75-85 [DOI] [PubMed] [Google Scholar]
- 29.Cheng, N., Brantley, D. M., Liu, H., Lin, Q., Enriquez, M., Gale, N., Yancopoulos, G., Gerretti, D. P., Daniel, T. O., and Chen, J. (2002) Mol. Cancer Res. 1 2-11 [PubMed] [Google Scholar]
- 30.Voronov, E., Shouval, D. S., Krelin, Y., Cagnano, E., Benharroch, D., Iwakura, Y., Dinarello, C. A., and Apte, R. N. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 2645-2650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iso, T., Hamamori, Y., and Kedes, L. (2003) Arteriosc. Thromb. Vasc. Biol. 23 380-387 [DOI] [PubMed] [Google Scholar]
- 32.Carmeliet, P., Lampugnani, M. G., Moons, L., Breviario, F., Compernolle, V., Bono, F., Balconi, G., Spagnuolo, R., Oostuyse, B., Dewerchin, M., Zanetti, A., Angellilo, A., Mattot, V., Nuyens, D., Lutgens, E., Clotman, F., de Ruiter, M. C., Gittenberger-de Groot, A., Poelmann, R., Lupu, F., Herbert, J. M., Collen, D., and Dejana, E. (1999) Cell 98 147-157 [DOI] [PubMed] [Google Scholar]
- 33.Gerhardt, H., Wolburg, H., and Redies, C. (2000) Dev. Dyn. 218 472-479 [DOI] [PubMed] [Google Scholar]
- 34.Salomon, D., Ayalon, O., Patel-King, R., Hynes, R. O., and Geiger, B. (1992) J. Cell Sci. 102 7-17 [DOI] [PubMed] [Google Scholar]
- 35.Luo, Y., and Radice, G. L. (2005) J. Cell Biol. 169 29-34 [DOI] [PMC free article] [PubMed] [Google Scholar]