Abstract
Genetic and biochemical analyses show that IL-23p19 plays a central role in mediating bacteria-induced colitis in interleukin-10-deficient (IL-10–/–) mice. The molecular mechanisms responsible for the dysregulated innate host response leading to enhanced IL-23 gene expression in IL-10–/– mice are poorly understood. In this study, we investigated the role of Bcl3 in controlling LPS-induced IL-23p19 gene expression in bone marrow-derived dendritic cells (BMDC) isolated from IL-10–/– mice. We report higher IL-23p19 mRNA accumulation and protein secretion in LPS-stimulated BMDC isolated from IL-10–/– compared with WT mice. Lipopolysaccharide (LPS)-induced B cell leukemia 3 (Bcl3) expression was strongly impaired (90% decrease) in IL-10–/– BMDC compared with WT BMDC. Chromatin immunoprecipitation demonstrated enhanced RelA binding to the IL-23p19 promoter in IL-10–/– compared with WT BMDC. Bcl3 overexpression decreased LPS-induced IL-23p19 gene expression in IL-10–/– BMDC, which correlated with enhanced NF-κB p50 binding and decreased RelA binding to the gene promoter. Conversely, Bcl3 knockdown enhanced LPS-induced IL-23p19 gene expression in WT BMDC. Moreover, LPS-induced IL-23p19 gene expression was significantly enhanced in Bcl3–/– BMDC compared with WT BMDC. In conclusion, enhanced LPS-induced IL-23p19 gene expression in IL-10–/– mice is due to impaired Bcl3 expression leading to diminished p50 and enhanced RelA recruitment to the IL-23p19 promoter.
Inflammatory bowel diseases (IBDs)2 exemplified by Crohn disease and ulcerative colitis are chronic intestinal inflammatory disorders causing abdominal pain, diarrhea, and bleeding. Although the etiology of IBD still eludes the medical research field, experimental models of colitis have provided important clues about the pathogenesis of the disease. From this large body of work, it has become clear that a dysregulated host immune response to the endogenous intestinal microflora is a prerequisite to the development of the disease (1–4). The failure to implement a functional network of immunosuppressive cytokines dampening host responses to the abundant microbiota in conjunction with an overly active production of proinflammatory mediators likely contribute to the development of IBD (1, 5). The importance of a proper immunosuppressive response to microorganisms is clearly illustrated in interleukin-10-deficient (IL-10–/–) mice. These mice become intolerant to their intestinal microflora and spontaneously develop colitis through aberrant activation of effector T cells (6–8). Among the various inflammatory mediators released by the mucosal immune cells, IL-23p19 was identified as the key factor promoting T cell-mediated chronic intestinal inflammation (9). Indeed, IL-23p19 and IL-10 double knock-out mice fail to develop spontaneous colitis, whereas IL-12p35 and IL-10 double knock-out mice are as susceptible to colitis as IL-10–/– mice. Therefore, IL-23 but not IL-12 is the key mediator of the disease (9, 10). This new paradigm established IL-23 as an inducer of memory T lymphocytes producing IL-17 (Th17) and IL-6, both cytokines critical in mediating chronic inflammation. Interestingly, CD40- and Helicobacter hepaticus-induced colitis in Rag-1–/– mice is mediated by intestinal dendritic/myeloid cells producing IL-23, suggesting that this cytokine also plays a role in innate immune pathology (11, 12). Moreover, monoclonal anti-IL-23p19 neutralizing antibody prevented and treated established experimental colitis (13). Finally, genome-wide scan analysis indicates that genetic variants of IL-23R associate either positively or negatively with the development of the disease (14). Altogether, these findings established IL-23 as a key mediator of chronic intestinal inflammation and as a potential therapeutic target.
IL-10 is a potent multifunctional immunoregulatory cytokine that regulates the expression of numerous proinflammatory cytokines, chemokines, and costimulatory molecules expressed by various immune and non-immune cells (15). The molecular mechanism responsible for IL-10 inhibitory action is diverse and includes activation of the heme oxygenase/carbon monoxide pathway (16), inhibition of the NF-κB pathway (17–19), mitogen-activated protein kinase activity (20), STAT3 activation (21–23), and induction of SOCS-3 and B cell leukemia 3 (Bcl3) (24–26). Although the exact mechanism of IL-10-mediated immunosuppressive effects is still unclear, evidence points to a mechanism involving changes in the rate of transcriptional activity (27).
IL-23p19 is the critical mediator of experimental colitis in IL-10–/– mice, but the molecular mechanisms involved in the dysregulated production of this mediator remain unknown. In this study, we demonstrate that the increased IL-23p19 gene expression in IL-10–/– mice is due to a defective induction of Bcl3, thereby impairing recruitment of the transcriptionally inactive NF-κB p50 subunit while facilitating binding of the transcriptionally active RelA subunit to the gene promoter. These data indicate that Bcl3 plays a critical role in the resolution of LPS-induced IL-23p19 gene expression and may represent a potential target for the treatment of IBDs.
EXPERIMENTAL PROCEDURES
RNA Extraction and RT-PCR Analysis—RNA was isolated using TRIzol (Invitrogen), reverse transcribed (1 μg RNA), and amplified using specific primers for mouse IL-23p19, Bcl3, and GAPDH: IL-23p19 forward, 5′-GCCCCGTATCCAGTGTGA-3′; IL-23p19 reverse, 5′-GCTGCCACTGCTGACTAG-3′; Bcl3 forward, 5′-CCTTTGATGCCCATTTACTCTA-3′; Bcl3 reverse, 5′-AGCGGCTATGTTATTCTGGAC-3′; GAPDH forward, 5′-GGTGAAGGTCGGTGTGAACGGA-3′; and GAPDH reverse, 5′-GAGGGATCTCGCTCCTGGAAGA-3′. The PCR products (8 μl) were subjected to electrophoresis on 2% agarose gels containing GelStar fluorescent dye (Cambrex BioScience Rockland, Rockland, ME). Fluorescence staining was captured using an Alpha Imager 2000 (Alpha Innotech, San Leandro, CA). To precisely quantify the expression of IL-23p19, Bcl3, and GAPDH, real time PCR was conducted using an Applied Biosystems 7700 sequence detection system. For PCR 2-μl cDNA preparation, 150 nm final concentration of forward and reverse primers and 6 μl of QuantiTect SYBR Green PCR Master Mix (Qiagen) in a total of 12 μl were applied. The following PCR program was performed: 15 min at 95 °C (initial denaturation); 20 °C/s temperature transition rate up to 95 °C for 30 s, 45 s at 56 °C, 45 s at 72 °C, acquisition mode single, repeated 40 times (amplification). Specificity and linearity (efficacy) of amplification for each primer set (amplicon) was determined by melting curve analysis and calculation of the slope from serial diluted samples as suggested by the manufacturer (ABI, User Bulletin 2). Relative changes were determined using the ΔΔCT calculation method. The values were normalized to the internal control GAPDH.
Western Blots—The cells were harvested and lysed in 1× Laemmli buffer, and the protein concentration was measured using Bio-Rad quantification assay (Bio-Rad). Twenty micrograms of the protein extracts were subjected to electrophoresis on 10% SDS-polyacrylamide gels and transferred to Hybond-C Extra nitrocellulose membranes (Amersham Biosciences). Anti-Bcl3 (c-14; Santa Cruz Biotechnology) and GAPDH antibodies were used to detect Bcl3 and GAPDH, respectively. All of the antibodies were used at a 1:1,000 dilution in a solution containing 5% milk in TBS-T. Immunoreactive proteins were detected using an ECL detecting kit (Amersham Biosciences).
Cytokine Measurement—The cells were stimulated for 24 h with LPS (5 μg/ml), the supernatants were collected, and the cytokine levels were measured using commercially available kits specific for IL-23p19 (eBioscience, San Diego, CA) and IL-12p70 (PharMingen/BD Bioscience, San Diego, CA) according to the manufacturers' instructions. The cytokine levels were determined in triplicate culture supernatants in each separate experiment.
Cell Culture and Treatment of Bone Marrow-derived Dendritic Cells—Wild type (WT) IL-10–/– mice (129SvEv background) and WT Bcl3–/– mice (C57B6 background) between 6 and 10 weeks of age were used to generate bone marrow cells from femora and tibiae. Red blood cells were lysed using red blood cell lysis buffer (Sigma), and the cells were cultured in 24-well low adherence plates (Costar, Corning, NY) in complete medium containing RPMI 1640 plus 10% heat-inactivated fetal calf serum (HyClone, Logan, UT), 2 mm l-glutamine, 1 mm sodium pyruvate, 5 × 10–5 m 2-mercaptoethanol, and 50 μg/ml gentamycin in the presence of recombinant murine granulocyte-macrophage colony-stimulating factor (GM-CSF) and IL-4 (both 10 ng/ml; Peprotech, Rocky Hill, NJ). The floating cells were gently removed, the medium was refreshed at day 3, and the cells were collected at day 6. The cells were washed twice and incubated overnight in complete medium without IL-4/GM-CSF. Flow cytometry analysis demonstrated a homogenous cell population with greater than 85% CD11c+ cells and less than 2% T cells (data not shown). For cell stimulation, 2.5 × 106 cells (cytokine measurement) or 3 × 106 cells (proteins, RNA) were plated in 6- or 24-well plates, respectively. The cells were stimulated with LPS (5 μg/ml, from Escherichia coli 0111:B4; Sigma) in the presence or absence of recombinant murine IL-10 (12 h of preincubation, 10 ng/ml; Peprotech). For NF-κB inhibitory experiments, BMDC were pretreated with the IκB phosphorylation inhibitor BAY 11-7082 (25 μm) (Calbiochem, San Diego, CA) for 1 h followed by stimulation with LPS (5 μg/ml) at various time points.
Plasmids and siRNA Transfections—Transfection of BMDCs was performed using the Nucleofector (Amaxa Inc., Gaithersburg, MD) according to the manufacturer's instructions. The constitutive active cytomegalovirus Bcl3 plasmid (a generous gift of A. S. Baldwin, University of North Carolina at Chapel Hill), an empty control vector pcDNA, Bcl3 siRNA (knockdown experiments; sc-29790; Santa Cruz Biotechnology) and scrambled siRNA (negative control; sc-37007; Santa Cruz Biotechnology) were used for transfection of murine BMDC. Transfection efficacy of BMDC as measured using pmaxGFP (5 μg/transfection) was estimated at ∼50%. The BMDC were collected on day 5 and washed twice in phosphate-buffered saline. Subsequently, 3 × 106 cells were resuspended in 100 μlof human DC nucleofection solution (Amaxa), and the samples were transferred into certified cuvettes (Amaxa) and transfected by using the program X-001. The same programs were used for transfection with plasmids and siRNA. Following transfection, the BMDC were cultured in complete medium containing RPMI 1640 plus 1% heat-inactivated fetal calf serum (HyClone), 2 mm l-glutamine, 1 mm sodium pyruvate, 5 × 10–5 m 2-mercaptoethanol, and 50 μg/ml gentamycin without IL-4 or GM-CSF, and the cells were stimulated accordingly.
Chromatin Immunoprecipitation (ChIP) Analysis—Murine BMDCs were stimulated with LPS (5 μg/ml) at various time points, and ChIP assay was performed using a ChIP assay kit (Upstate-Cell Signaling Solutions, Temecula, CA) according to the manufacturer's specifications as described previously (23). Immunoprecipitation was carried out overnight with 2 μgof p65 (c-20; Santa Cruz Biotechnology) or RNA polymerase II (c-21; Santa Cruz Biotechnology), p50 (sc-114; Santa Cruz), and Bcl3 (c-14; Santa Cruz) antibodies. PCR was performed with total input DNA (5 μl) and immunoprecipitated DNA (5 μl) using IL-23p19 promoter-specific primers: IL-23p19 proximal promoter forward, 5′-CAGGCAGATTACACAGGAAGG-3′; IL-23p19 proximal promoter reverse, 5′-GCCCGCCCTTCACACTA-3′; IL-23p19 distal promoter forward, 5′-GGGTCATCATCGCTGTACTTT-3′; and IL-23p19 distal promoter reverse, 5′-CGGCCCTGGTTTTGAAG-3′. PCR was carried out in a volume of 50 μl containing final concentrations of 1× Taq buffer (Applied Biosystems, Foster City, CA), 50 nm primers, 0.5 mm dNTPs, and 1 unit of Thermo aquaticus polymerase (Applied Biosystems) using a 9700 Gene-Amp PCR system cycler (Applied Biosystems). The PCR temperatures used were 95 °C for 15 s, 56 °C for 45 s, and 72 °C for 45 s followed by an extension of 5 min at 72 °C. The PCR products were subjected to electrophoresis on 2% agarose gels containing GelStar fluorescent dye (Cambrex BioScience Rockland). Fluorescence staining was captured using an Alpha Imager 2000 (Alpha Innotech).
Statistical Analysis—Statistical significance was evaluated by the two-tailed Student's t test for unpaired data. A p value of less than 0.05 was considered statistically significant.
RESULTS
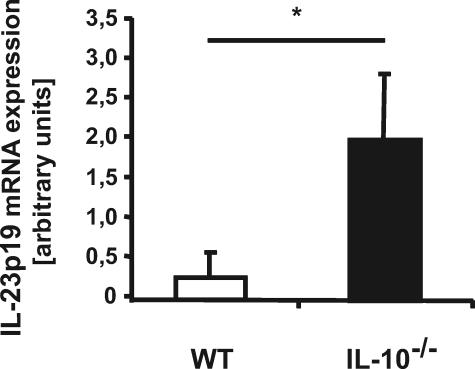
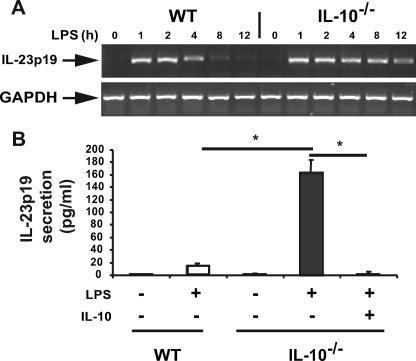
Experimental models of colitis as well as human genetic analysis have linked IL-23p19 to the development of IBD. Because IL-10–/– mice develop colitis when housed under specific pathogen-free conditions, we first investigated the expression of IL-23p19 in IL-10–/– versus WT mice. Germ-free IL-10–/– and WT mice were transferred to specific pathogen-free conditions, and 8 weeks later, mesenteric lymph nodes (MLN) were isolated, and IL-23p19 expression was analyzed by RT-PCR. Increased IL-23p19 mRNA expression was observed in MLN isolated from IL-10–/– mice compared with WT mice, suggesting a dysregulated pattern of expression in IL-10–/– mice (Fig. 1). Because IL-23p19 is mostly produced by antigen-presenting cells (28, 29), we sought to understand the mechanism of dysregulated IL-23p19 expression using BMDC cells generated from IL-10–/– and WT mice. Interestingly, LPS-induced IL-23p19 mRNA accumulation was prolonged in IL-10–/– BMDC compared with WT mice (Fig. 2A). Dysregulated IL-23p19 mRNA accumulation in IL-10–/– BMDC was followed by enhanced protein secretion (>10-fold) (Fig. 2B). Of note, preincubation with recombinant IL-10 (10 ng/ml) completely abrogated LPS-induced IL-23p19 secretion in IL-10–/– BMDC.
FIGURE 1.
IL-23p19 mRNA expression is elevated in MLN derived from IL-10–/– compared with WT mice. RT-PCR amplification of IL-23p19 from MLN of WT or IL-10–/– mice after being housed for 8 weeks under specific pathogen-free conditions. MLN were lysed in TRIzol, total RNA was extracted and reverse transcribed, and IL-23p19 mRNA expression was detected using real time PCR (Applied Biosystems 7700 sequence detection system). The results were normalized to the housekeeping gene GAPDH to ascertain similar loading. The results are the means ± S.D. of quadruplicate samples and are from one of three independent experiments (*, p < 0.05).
FIGURE 2.
Dysregulated LPS-induced IL-23p19 gene expression in IL-10–/– compared with WT mice. A, prolonged LPS-induced IL-23p19 mRNA expression in IL-10–/– compared with WT mice. Conventional reverse transcriptase-PCR results from WT or IL-10–/– BMDC after treatment with LPS (5 μg/ml) are shown for the indicated times. PCR products were separated on a 2% agarose gel and stained with GelStar. The results are representative of three independent experiments. B, IL-10 inhibits IL-23p19 protein secretion in BMDC derived from IL-10–/– mice. WT and IL-10–/– BMDC were prestimulated with IL-10 (10 ng/ml) overnight or left untreated. The cells were then stimulated with LPS (5 μg/ml) for 20 h, and the supernatants were collected. IL-23p19 protein secretion was detected using enzyme-linked immunosorbent assay (*, p < 0.05). The results show the means ± S.D. from triplicate cultures and are representative of three independent experiments.
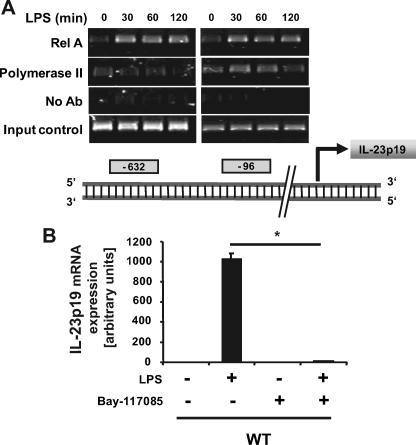
IL-10–/–;MyD88–/– mice failed to develop spontaneous colitis and displayed marked reduction of LPS-induced NF-κB signaling (30, 31). We therefore investigated whether dysregulated TLR/NF-κB signaling was responsible for enhanced IL-23p19 gene expression in IL-10–/– mice. LPS-induced IL-23p19 mRNA accumulation was markedly reduced in IL-10–/–;MyD88–/– BMDC compared with IL-10–/– cells (data not shown). Because TLR signals are mediated largely through the NF-κB family of transcription factors, we next investigated the role of NF-κB in LPS-induced IL-23p19 gene expression. Chromatin immunoprecipitation analysis showed that LPS induced RelA recruitment to the NF-κB sites located at position –96 and –632 of the IL-23p19 gene promoter (Fig. 3A). RNA polymerase II was recruited to the proximal NF-κB(–96) binding site following LPS stimulation. To determine the functional impact of LPS-induced NF-κB recruitment, BMDC were preincubated with the pharmacological inhibitor Bay 11–7085 and then stimulated with LPS. As shown in Fig. 3B, LPS-induced IL-23p19 mRNA accumulation was prevented in Bay 11–7085-treated WT BMDC cells, which was preceded by blockade of IκBα degradation (data not shown). Altogether, these findings demonstrate that TLR/NF-κB signaling is critical for LPS-induced IL-23p19 gene expression.
FIGURE 3.
LPS-induced IL-23p19 mRNA expression is TLR/NF-κB dependent. A, LPS-induced RelA and RNA polymerase II recruitment to the IL-23p19 NF-κB-binding sites. Bioinformatic analysis using transcription element search software revealed two potential NF-κB-binding sites in the murine IL-23p19 promoter region at position –96 and –632 relative to the transcription start site. WT BMDC were stimulated with LPS (5 μg/ml) at the indicated time points, and ChIP assays were performed using anti-RelA or anti-RNA polymerase II antibodies as described under “Experimental Procedures.” The results are representative of three independent experiments. PCR primers to the –632 (left panel) or the –96 (right panel) were used. B, LPS-induced IL-23p19 mRNA expression is NF-κB-dependent. BMDC from WT mice were preincubated (1 h) with the NF-κB inhibitor Bay 11-7082 and then stimulated in the absence or presence of LPS (5 μg/ml) for 2 h. Total RNA was extracted and analyzed for the levels of IL-23p19 and GAPDH mRNA using real time PCR (*, p < 0.05). The results are the means ± S.D. of triplicate samples from one of three independent experiments. Ab, antibody.
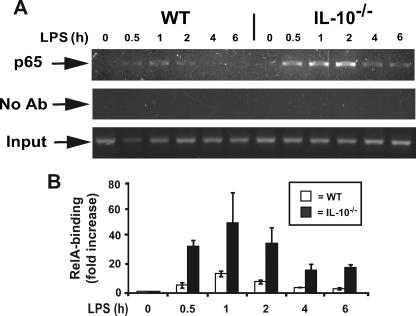
We next sought to determine the mechanism responsible for dysregulated LPS-induced IL-23p19 gene expression in IL-10–/– mice. NF-κB DNA binding activity was compared between LPS-stimulated WT and IL-10–/– BMDC using an enzyme-linked immunosorbent assay-based assay. We observed a 50% increase in RelA binding activity at 30 and 60 min in LPS-stimulated IL-10–/– BMDC compared with WT cells (data not shown). To gain more insight into the relationship between NF-κB DNA binding activity and endogenous IL-23p19 gene expression, we compared RelA recruitment to the IL-23p19 gene promoter between WT and IL-10–/– BMDC using ChIP assay. Increased RelA recruitment to the IL-23p19 promoter was observed in LPS-stimulated IL-10–/– BMDC compared with WT cells (Fig. 4A). Semi-quantitative analysis of the ChIP assay using an ABI Prism 7700 sequence detection system showed an increase of up to 6-fold in LPS-stimulated IL-10–/– BMDC compared with WT cells (Fig. 4B). Because the kinetic and extent of IκB degradation and NF-κB nuclear translocation is similar between WT and IL-10–/– BMDC (23) and because DNA binding activity is minimally impaired, we investigated whether an IL-10-inducible nuclear factor was responsible for controlling RelA recruitment to IL-23p19 gene promoter. We focused our attention on B cell leukemia 3 (Bcl3), an IL-10-inducible gene interacting with the p50 NF-κB subunit (26, 32–35).
FIGURE 4.
IL-10–/– mice exhibit prolonged LPS-induced RelA (p65) binding to the IL-23p19 promoter after LPS stimulation. A, prolonged LPS-induced RelA (p65) binding in IL-10–/– compared with WT BMDC. The cells were isolated from WT and IL-10–/– mice and stimulated with LPS (5 μg/ml) for the indicated times. ChIP assays were performed using anti-RelA antibodies as described under “Experimental Procedures.” PCR products were separated on a 2% agarose gel and stained with GelStar. The results are representative of three independent experiments. B, Semi-quantitative analysis of RelA binding to the IL-23p19 promoter (–632). WT (white bars) and IL-10–/– (black bars) BMDC were stimulated with LPS (5 μg/ml) at different time points, and ChIP assays were performed as described above. Relative fold changes were determined by semi-quantitative assay using the ABI 7700 sequence detection system as described under “Experimental Procedures.” The results are representative of three independent experiments.
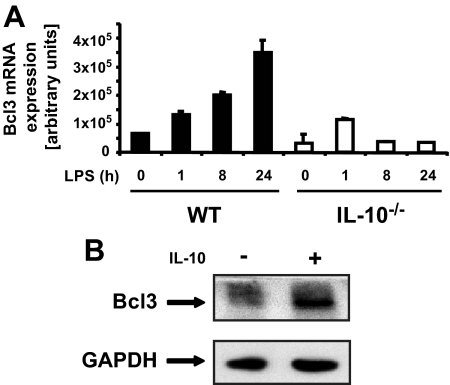
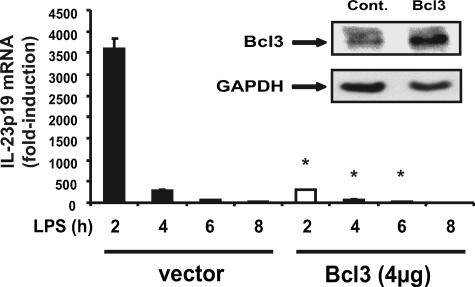
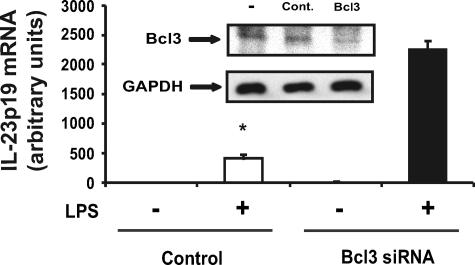
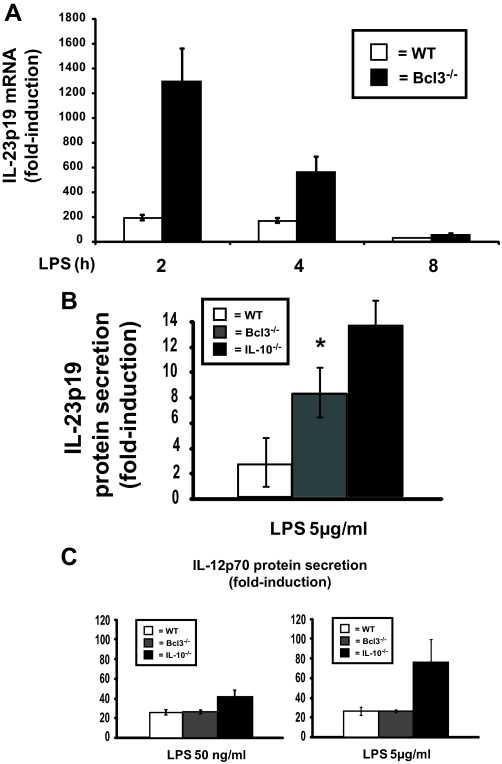
As shown in Fig. 5A, Bcl3 mRNA accumulation progressively increased following LPS stimulation in WT BMDC, whereas levels remained relatively low in IL-10–/– BMDC. The addition of exogenous IL-10 strongly increased Bcl3 protein accumulation in IL-10–/– BMDC (Fig. 5B). These findings indicate that decreased Bcl3 expression in IL-10–/– BMDC correlates with increased LPS-induced IL-23p19 expression. To test the functional impact of Bcl3 on IL-23p19 expression, IL-10–/– BMDC were transfected with a Bcl3 expression vector and then stimulated with LPS for various time points. Using an enhanced green fluorescent protein plasmid control, transfection efficiency was found to be up to 50% in BMDC with no significant cell death (data not shown). Interestingly, LPS-induced IL-23p19 mRNA accumulation was strongly reduced (>90%) in Bcl3-transfected IL-10–/– BMDC (Fig. 6). Western blot analysis confirmed Bcl3 expression in transfected IL-10–/– BMDC (Fig. 6). To validate these findings, we knocked down Bcl3 expression in WT BMDC using siRNA. LPS-induced IL-23p19 mRNA accumulation was significantly increased in WT BMDC transfected with Bcl3 siRNA (Fig. 7). Western blot analysis confirmed that Bcl3 siRNA effectively decreased Bcl3 expression. Control siRNA had minimal effect in BMDC (Fig. 7). To directly link Bcl3 to the regulation of IL-23p19 gene expression, we generated BMDC from Bcl3–/– mice. Fig. 8A shows increased LPS-induced IL-23p19 mRNA expression in Bcl3–/– compared with WT BMDC. Concomitant to mRNA levels, IL-23p19 secretion was higher in LPS-stimulated Bcl3–/– BMDC compared with WT cells (Fig. 8B; 9-fold versus 3-fold; p < 0.05) although to a lesser extent than stimulated IL-10–/– BMDC (14-fold versus 9-fold; p < 0.05). In contrast secretion of IL-12p70, a Bcl3-independent gene (34), is similar between Bcl3–/– BMDC and WT cells, regardless of LPS concentration (Fig. 8C). These findings identify Bcl3 as an important regulator of IL-23p19 gene expression.
FIGURE 5.
LPS-induced Bcl3 expression is impaired in IL-10–/– compared with WT mice. A, impaired LPS-induced Bcl3 expression in IL-10–/– mice. BMDC were isolated from WT or IL-10–/– mice and stimulated with LPS (5 μg/ml) for the indicated times. Total RNA was then extracted, and Bcl-3 mRNA levels were analyzed using real time PCR. The data were normalized using the housekeeping gene GAPDH. The results are the means ± S.D. of triplicate samples from one of three independent experiments. B, Bcl3 expression in IL-10–/– mice can be restored by stimulation with IL-10. BMDC were isolated from IL-10–/– mice and stimulated with IL-10 (10 ng/ml) for 24 h. Total protein was extracted, and 20 μg was subjected to SDS-PAGE followed by immunoblotting with Bcl3 and GAPDH specific antibodies. The results are representative of three independent experiments.
FIGURE 6.
IL-10–/–-derived BMDC overexpressing Bcl3 displayed abrogated IL-23p19 mRNA expression. Bcl3 overexpression abrogated LPS-induced IL-23p19 mRNA expression in IL-10–/– mice. BMDC were isolated from IL-10–/– mice and transfected with a constitutively expressing Bcl3 plasmid or empty vector control (Cont.). BMDC were stimulated with LPS (5 μg/ml) for the indicated times. Total RNA was extracted and analyzed for levels of IL-23p19 and GAPDH mRNA using real time PCR. Statistical analysis was performed comparing 2-, 4-, 6-, and 8-h values between vector only and Bcl3-transfected mice (*, p < 0.05). The inset shows Bcl3 protein expression by immunoblot in Bcl3-transfected IL-10–/– derived BMDC. The results are representative of three independent experiments.
FIGURE 7.
Bcl3 knockdown leads to enhanced LPS-induced IL-23p19 expression in WT BMDC. BMDC were isolated from WT mice and transfected with Bcl3 siRNA or scrambled siRNA (control, Cont.) using the Amaxa nucleofector system. The cells were then stimulated with LPS (5 μg/ml) for 4 h, total RNA was extracted, and IL-23p19 mRNA expression was detected using real time PCR. The data were normalized using the housekeeping gene GAPDH. Statistical analysis was performed comparing LPS-stimulated BMDC (*, p < 0.05). The inset shows decreased Bcl3 protein expression in Bcl3 siRNA-transfected WT-derived BMDC. The results are the means ± S.D. of triplicate samples obtained from one of three independent experiments.
FIGURE 8.
Enhanced LPS-induced IL-23p19 expression in Bcl3–/– BMDC. A, IL-23p19 mRNA expression in BMDC derived from WT and Bcl3–/– mice. BMDC were isolated from WT and Bcl3–/– mice and stimulated with LPS (5 μg/ml) for the indicated times. Total RNA was extracted and analyzed by RT-PCR for Bcl3 mRNA expression. The data were normalized using the housekeeping gene GAPDH. The results are means ± S.D. of triplicate samples from one of three independent experiments. B, IL-23p19 and C, IL-12p70 protein secretions in BMDC derived from WT, Bcl3–/– and IL-10–/– mice. BMDC were generated from WT (white bars), Bcl3–/– (gray bars), and IL-10–/– (black bars) mice and stimulated LPS (50 ng/ml or 5 μg/ml) for 20 h, and the supernatants were collected. IL-23p19 and IL-12p70 protein secretion was detected using enzyme-linked immunosorbent assay. (*, p < 0.05). The results are the means ± S.D. of triplicate samples from one of three independent experiments.
Bcl3 has been shown to enhance p50 inhibitory activity and consequently down-modulate NF-κB transcriptional activity (26, 32–35). To gain more insight into the relationship between Bcl3, p50, and IL-23p19 gene expression, we performed ChIP assays in Bcl3-transfected IL-10–/– BMDC. As seen in Fig. 9, p50 recruitment to the IL-23p19 gene promoter was progressively decreased over time in LPS-stimulated IL-10–/– BMDC, whereas p50 levels remained elevated in Bcl3-transfected cells. Decreased p50 binding in IL-10–/– BMDC correlated with enhanced RelA binding to the gene promoter and concomitant increased in IL-23p19 mRNA accumulation (Fig. 9). Of note, increased p50 binding in Bcl3-transfected IL-10–/– BMDC leads to a decreased p65 and sustained Bcl3 binding to the IL-23p19 gene promoter.
FIGURE 9.
Bcl3 overexpression reduced LPS-induced RelA binding and enhanced p50 recruitment to the IL-23p19 promoter. BMDC were isolated from IL-10–/– mice and transfected with a constitutively expressing Bcl3 plasmid or an empty vector control (Cont.) for 16 h. BMDC were then stimulated with LPS (5 μg/ml) for the indicated times, and ChIP assays were performed using anti-p50, anti-p65, and anti-Bcl3 antibodies as described under “Experimental Procedures.” PCR was performed using primers specific for the NF-κB consensus site within the IL-23p19 gene promoter (–632). PCR products were loaded on a 2% agarose-gel and visualized with GelStar. The results are shown from one of three representative experiments.
In summary our data suggest that decreased Bcl3 expression in IL-10–/– BMDC impairs p50 recruitment/binding to the IL-23p19 gene promoter and favors RelA loading, leading to an increased gene expression.
DISCUSSION
In this study, we investigated the molecular mechanism responsible for the control of IL-23p19 gene expression. We report dysregulated levels of IL-23p19 secretion in BMDC derived from IL-10–/– compared with WT mice following LPS stimulation. Similarly, IL-23p19 mRNA expression is strongly enhanced in mesenteric lymph nodes isolated from colitic IL-10–/– mice compared with healthy WT mice. This indicates that in the absence of endogenous IL-10, innate response to luminal bacterial products is dysregulated and leads to higher IL-23p19 gene expression. Importantly, administration of neutralizing IL-23p19 antibody prevents the early onset of colitis in a colitic transfer model (13). In addition, an increased TLR-dependent IL-23p19 mRNA expression was observed in IL-10–/–-derived macrophages (36, 37). Consequently, our finding that LPS induced higher IL-23p19 gene expression in BMDC isolated from IL-10–/– mice correlates with the key role of this cytokine in this model of spontaneous colitis.
In an effort to understand the mechanism responsible for dysregulated IL-23p19 gene expression in IL-10–/– BMDC, we investigated the impact of LPS-induced NF-κB signaling in BMDC. Using the NF-κB pharmacological inhibitor Bay 11-7082, we demonstrated that LPS utilizes the NF-κB signaling cascade to induce IL-23p19 gene expression in BMDC. In addition, we report that LPS enhanced RelA recruitment to two NF-κB consensus elements (–632 and –96) located in the IL-23p19 gene promoter. This is in line with a recent report showing that the IL-23p19 promoter region contains two NF-κB-binding sites essential for LPS-induced transcriptional activity (38). Interestingly, enhanced RelA recruitment to the IL-23p19 promoter was observed in IL-10–/– BMDC mice compared with WT mice. This enhanced RelA recruitment correlated with prolonged LPS-induced IL-23p19 mRNA expression and elevated IL-23p19 secretion. LPS-induced IL-23p19 secretion is abrogated when IL-10–/– BMDC receive exogenous IL-10, suggesting that this immunosuppressive molecule triggers the expression of a factor controlling IL-23p19 gene expression. Because LPS-induced Bcl3 mRNA and protein expression was impaired in IL-10–/– BMDC compared with WT cells, we focused our attention on this IκB-like protein. This protein interacts exclusively with the NF-κB transcriptionally inactive p50 and p52 subunits (39–41). Here, we demonstrated a functional relationship between Bcl3 expression levels and LPS-induced IL-23p19 gene expression using various molecular strategies. First, LPS-induced IL-23p19 gene expression is strongly inhibited in Bcl3-transfected IL-10–/– BMDC. This inhibitory effect was associated with enhanced Bcl3/p50 binding to the IL-23p19 gene promoter and concomitant decreased p65 binding. This is in line with a recent observation showing that Bcl3 overexpression enhanced p50 homodimer binding to NF-κB-binding sites in the tumor necrosis factor promoter (32).
Second, knock-down experiments using Bcl3 RNA interference showed enhanced LPS-induced IL23p19 gene expression in WT BMDC. Finally, Bcl3–/– BMDC displayed enhanced LPS-induced IL23p19 gene expression compared with WT cells, although to a lesser extent than in IL-10–/– cells. This suggests that Bcl3-independent factors are also involved in the regulation of LPS-induced IL-23p19 gene expression. Bcl3-deficient mice exhibit various immunological defects including decreased T cell-dependent immunoglobulin class switching, defective generation of influenzaspecific antibodies, and increased susceptibility to a variety of other pathogenic infections, including Toxoplasma gondii, Listeria monocytogenes, and Streptococcus pneumonia (34, 42, 43). Interestingly, as opposed to IL-10–/– mice, Bcl3–/– mice fail to spontaneously develop colitis, suggesting a broader immunoregulatory impact of IL-10 on intestinal homeostasis. Although LPS-induced IL-23p19 gene expression is higher in Bcl3–/– mice BMDC compared with WT cells, the former also secrete higher amounts of IL-10 (data not shown). Therefore, higher IL-10 secretion in Bcl3 mice may dampen any spontaneous intestinal phenotype in these mice. It would be of high interest to investigate the impact of Bcl3 on chemically (dextran sodium sulfate or 2,4,6-trinitrobenzenesulfonic acid) induced experimental colitis. A recent report by Zhang et al. (44) demonstrated that p52–/–;Bcl3–/– mice developed a profound breakdown in central tolerance that resulted in fatal multiorgan inflammation, whereas the loss of only p52 or Bcl3 had only minor effects. Although the impact of p52 and Bcl3 on intestinal homeostasis was not investigated, their data implicate Bcl3 and NF-κB in the control of immunological self-tolerance, which also plays a major role in IBD.
In summary, our study identifies the IL-10-inducible molecule Bcl3 as a key regulator of LPS-induced IL-23p19 production in BMDC. Bcl3 appears to control IL-23p19 gene expression through the regulation of p50 binding to the gene promoter. Understanding the regulation of IL-23p19, a major cytokine involved in chronic inflammatory diseases, will be key to the development of future clinical treatments for diseases like inflammatory bowel diseases.
Acknowledgments
We thank Brigitte Allard for excellent technical assistance and Dr. Ulrich Siebenlist (NIAID, National Institutes of Health, Bethesda, MD) for the Bcl3–/– mice.
This work was supported, in whole or in part, by National Institutes of Health Grants ROI DK 47700 and RO1 DK 73338 (to C. J.). This work was also supported by Deutsche Forschungsgemeinschaft Grant MU2301/2-1 (to M. M.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IBD, inflammatory bowel disease; BMDC, bone marrow derived dendritic cells; IL, interleukin; LPS, lipopolysaccharide; NF-κB, nuclear factor-κB; TLR, Toll-like receptor; WT, wild type; STAT, signal transducers and activators of transcription; RT, reverse transcription; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GM-CSF, granulocyte-macrophage colony-stimulating factor; siRNA, small interfering RNA; ChIP, chromatin immunoprecipitation; MLN, mesenteric lymph nodes; Bcl3, B cell leukemia 3.
References
- 1.Haller, D., and Jobin, C. (2004) J. Pediatr. Gastroenterol. Nutr. 38 123–136 [DOI] [PubMed] [Google Scholar]
- 2.Kraehenbuhl, J. P., and Corbett, M. (2004) Science 303 1624–1625 [DOI] [PubMed] [Google Scholar]
- 3.Sartor, R. B. (2006) Nat. Clin. Pract. Gastroenterol. Hepatol. 3 390–407 [DOI] [PubMed] [Google Scholar]
- 4.Xavier, R. J., and Podolsky, D. K. (2007) Nature 448 427–434 [DOI] [PubMed] [Google Scholar]
- 5.Strober, W., Murray, P. J., Kitani, A., and Watanabe, T. (2006) Nat. Rev. Immunol. 6 9–20 [DOI] [PubMed] [Google Scholar]
- 6.Kuhn, R., Lohler, J., Rennick, D., Rajewsky, K., and Muller, W. (1993) Cell 75 263–274 [DOI] [PubMed] [Google Scholar]
- 7.Sellon, R. K., Tonkonogy, S., Schultz, M., Dieleman, L. A., Grenther, W., Balish, E., Rennick, D. M., and Sartor, R. B. (1998) Infect. Immun. 66 5224–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berg, D. J., Davidson, N., Kuhn, R., Muller, W., Menon, S., Holland, G., Thompson-Snipes, L., Leach, M. W., and Rennick, D. (1996) J. Clin. Investig. 98 1010–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yen, D., Cheung, J., Scheerens, H., Poulet, F., McClanahan, T., McKenzie, B., Kleinschek, M. A., Owyang, A., Mattson, J., Blumenschein, W., Murphy, E., Sathe, M., Cua, D. J., Kastelein, R. A., and Rennick, D. (2006) J. Clin. Investig. 116 1310–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker, C., Dornhoff, H., Neufert, C., Fantini, M. C., Wirtz, S., Huebner, S., Nikolaev, A., Lehr, H. A., Murphy, A. J., Valenzuela, D. M., Yancopoulos, G. D., Galle, P. R., Karow, M., and Neurath, M. F. (2006) J. Immunol. 177 2760–2764 [DOI] [PubMed] [Google Scholar]
- 11.Hue, S., Ahern, P., Buonocore, S., Kullberg, M. C., Cua, D. J., McKenzie, B. S., Powrie, F., and Maloy, K. J. (2006) J. Exp. Med. 203 2473–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uhlig, H. H., McKenzie, B. S., Hue, S., Thompson, C., Joyce-Shaikh, B., Stepankova, R., Robinson, N., Buonocore, S., Tlaskalova-Hogenova, H., Cua, D. J., and Powrie, F. (2006) Immunity 25 309–318 [DOI] [PubMed] [Google Scholar]
- 13.Elson, C. O., Cong, Y., Weaver, C. T., Schoeb, T. R., McClanahan, T. K., Fick, R. B., and Kastelein, R. A. (2007) Gastroenterology 132 2359–2370 [DOI] [PubMed] [Google Scholar]
- 14.Duerr, R. H., Taylor, K. D., Brant, S. R., Rioux, J. D., Silverberg, M. S., Daly, M. J., Steinhart, A. H., Abraham, C., Regueiro, M., Griffiths, A., Dassopoulos, T., Bitton, A., Yang, H., Targan, S., Datta, L. W., Kistner, E. O., Schumm, L. P., Lee, A. T., Gregersen, P. K., Barmada, M. M., Rotter, J. I., Nicolae, D. L., and Cho, J. H. (2006) Science 314 1461–1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore, K. W., de Waal Malefyt, R., Coffman, R. L., and O'Garra, A. (2001) Annu. Rev. Immunol. 19 683–765 [DOI] [PubMed] [Google Scholar]
- 16.Lee, T. S., and Chau, L. Y. (2002) Nat. Med. 8 240–246 [DOI] [PubMed] [Google Scholar]
- 17.Wang, P., Wu, P., Siegel, M. I., Egan, R. W., and Billah, M. M. (1995) J. Biol. Chem. 270 9558–9563 [DOI] [PubMed] [Google Scholar]
- 18.Schottelius, A. J., Mayo, M. W., Sartor, R. B., and Baldwin, A. S., Jr. (1999) J. Biol. Chem. 274 31868–31874 [DOI] [PubMed] [Google Scholar]
- 19.Bhattacharyya, S., Sen, P., Wallet, M., Long, B., Baldwin, A. S., Jr., and Tisch, R. (2004) Blood 104 1100–1109 [DOI] [PubMed] [Google Scholar]
- 20.Kontoyiannis, D., Kotlyarov, A., Carballo, E., Alexopoulou, L., Blackshear, P. J., Gaestel, M., Davis, R., Flavell, R., and Kollias, G. (2001) EMBO J. 20 3760–3770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams, L., Bradley, L., Smith, A., and Foxwell, B. (2004) J. Immunol. 172 567–576 [DOI] [PubMed] [Google Scholar]
- 22.El Kasmi, K. C., Holst, J., Coffre, M., Mielke, L., de Pauw, A., Lhocine, N., Smith, A. M., Rutschman, R., Kaushal, D., Shen, Y., Suda, T., Donnelly, R. P., Myers, M. G., Jr., Alexander, W., Vignali, D. A., Watowich, S. S., Ernst, M., Hilton, D. J., and Murray, P. J. (2006) J. Immunol. 177 7880–7888 [DOI] [PubMed] [Google Scholar]
- 23.Hoentjen, F., Sartor, R. B., Ozaki, M., and Jobin, C. (2005) Blood 105 689–696 [DOI] [PubMed] [Google Scholar]
- 24.Qin, H., Wilson, C. A., Roberts, K. L., Baker, B. J., Zhao, X., and Benveniste, E. N. (2006) J. Immunol. 177 7761–7771 [DOI] [PubMed] [Google Scholar]
- 25.Qasimi, P., Ming-Lum, A., Ghanipour, A., Ong, C. J., Cox, M. E., Ihle, J., Cacalano, N., Yoshimura, A., and Mui, A. L. (2006) J. Biol. Chem. 281 6316–6324 [DOI] [PubMed] [Google Scholar]
- 26.Kuwata, H., Watanabe, Y., Miyoshi, H., Yamamoto, M., Kaisho, T., Takeda, K., and Akira, S. (2003) Blood 102 4123–4129 [DOI] [PubMed] [Google Scholar]
- 27.Murray, P. J. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 8686–8691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langrish, C. L., McKenzie, B. S., Wilson, N. J., de Waal Malefyt, R., Kastelein, R. A., and Cua, D. J. (2004) Immunol. Rev. 202 96–105 [DOI] [PubMed] [Google Scholar]
- 29.Parham, C., Chirica, M., Timans, J., Vaisberg, E., Travis, M., Cheung, J., Pflanz, S., Zhang, R., Singh, K. P., Vega, F., To, W., Wagner, J., O'Farrell, A. M., McClanahan, T., Zurawski, S., Hannum, C., Gorman, D., Rennick, D. M., Kastelein, R. A., de Waal Malefyt, R., and Moore, K. W. (2002) J. Immunol. 168 5699–5708 [DOI] [PubMed] [Google Scholar]
- 30.Karrasch, T., Kim, J. S., Muhlbauer, M., Magness, S. T., and Jobin, C. (2007) J. Immunol. 178 6522–6532 [DOI] [PubMed] [Google Scholar]
- 31.Rakoff-Nahoum, S., Hao, L., and Medzhitov, R. (2006) Immunity 25 319–329 [DOI] [PubMed] [Google Scholar]
- 32.Carmody, R. J., Ruan, Q., Palmer, S., Hilliard, B., and Chen, Y. H. (2007) Science 317 675–678 [DOI] [PubMed] [Google Scholar]
- 33.Dechend, R., Hirano, F., Lehmann, K., Heissmeyer, V., Ansieau, S., Wulczyn, F. G., Scheidereit, C., and Leutz, A. (1999) Oncogene 18 3316–3323 [DOI] [PubMed] [Google Scholar]
- 34.Riemann, M., Endres, R., Liptay, S., Pfeffer, K., and Schmid, R. M. (2005) J. Immunol. 175 3560–3568 [DOI] [PubMed] [Google Scholar]
- 35.Wessells, J., Baer, M., Young, H. A., Claudio, E., Brown, K., Siebenlist, U., and Johnson, P. F. (2004) J. Biol. Chem. 279 49995–50003 [DOI] [PubMed] [Google Scholar]
- 36.Kamada, N., Hisamatsu, T., Okamoto, S., Sato, T., Matsuoka, K., Arai, K., Nakai, T., Hasegawa, A., Inoue, N., Watanabe, N., Akagawa, K. S., and Hibi, T. (2005) J. Immunol. 175 6900–6908 [DOI] [PubMed] [Google Scholar]
- 37.Schuetze, N., Schoeneberger, S., Mueller, U., Freudenberg, M. A., Alber, G., and Straubinger, R. K. (2005) Int. Immunol. 17 649–659 [DOI] [PubMed] [Google Scholar]
- 38.Carmody, R. J., Ruan, Q., Liou, H. C., and Chen, Y. H. (2007) J. Immunol. 178 186–191 [DOI] [PubMed] [Google Scholar]
- 39.Kerr, L. D., Duckett, C. S., Wamsley, P., Zhang, Q., Chiao, P., Nabel, G., McKeithan, T. W., Baeuerle, P. A., and Verma, I. M. (1992) Genes Dev. 6 2352–2363 [DOI] [PubMed] [Google Scholar]
- 40.Nolan, G. P., Fujita, T., Bhatia, K., Huppi, C., Liou, H. C., Scott, M. L., and Baltimore, D. (1993) Mol. Cell. Biol. 13 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Inoue, J., Takahara, T., Akizawa, T., and Hino, O. (1993) Oncogene 8 2067–2073 [PubMed] [Google Scholar]
- 42.Franzoso, G., Carlson, L., Poljak, L., Shores, E. W., Epstein, S., Leonardi, A., Grinberg, A., Tran, T., Scharton-Kersten, T., Anver, M., Love, P., Brown, K., and Siebenlist, U. (1998) J. Exp. Med. 187 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schwarz, E. M., Krimpenfort, P., Berns, A., and Verma, I. M. (1997) Genes Dev. 11 187–197 [DOI] [PubMed] [Google Scholar]
- 44.Zhang, X., Wang, H., Claudio, E., Brown, K., and Siebenlist, U. (2007) Immunity 27 438–452 [DOI] [PMC free article] [PubMed] [Google Scholar]