Abstract
Although the role of human IRF-5 in antiviral and inflammatory responses in vitro has been well characterized, much remains to be elucidated about murine IRF-5. Murine IRF-5, unlike the heavily spliced human gene, is primarily expressed as a full-length transcript, with only a single splice variant that was detected in very low levels in the bone marrow of C57BL/6J mice. This bone marrow variant contains a 288-nucleotide deletion from exons 4–6 and exhibits impaired transcriptional activity. The murine IRF-5 can be activated by both TBK1 and MyD88 to form homodimers and bind to and activate transcription of type I interferon and inflammatory cytokine genes. The importance of IRF-5 in the antiviral and inflammatory response in vivo is highlighted by marked reductions in serum levels of type I interferon and tumor necrosis factor α (TNFα) in Newcastle disease virus-infected Irf5–/– mice. IRF-5 is critical for TLR3-, TLR4-, and TLR9-dependent induction of TNFα in CD11c+ dendritic cells. In contrast, TLR9, but not TLR3/4-mediated induction of type I IFN transcription, is dependent on IRF-5 in these cells. In addition, IRF-5 regulates TNFα but not type I interferon gene transcription in Newcastle disease virus-infected peritoneal macrophages. Altogether, these data reveal the cell type-specific importance of IRF-5 in MyD88-mediated antiviral pathways and the widespread role of IRF-5 in the regulation of inflammatory cytokines.
Type I interferon (IFN)4 is rapidly produced in response to viral infection in almost all nucleated cells. Viral nucleic acids are recognized by two classes of cellular receptors as follows: toll-like receptors (TLR), which are present in the endosomal compartments of immune cells (1, 2), and the cytoplasmic RNA helicases RIG-I and MDA5 that detect 5′-triphosphate RNA (3) and dsRNA (4, 5), respectively. The TLR system senses viral dsRNA (TLR3) (6), viral single-stranded RNA (TLR7/8) (7), and unmethylated viral DNA with CpG motifs (TLR9) (8). In addition, recognition of cytosolic DNA by TLR-independent mechanisms has also been observed (9, 10). Binding of dsRNA to TLR3 triggers MyD88-independent signaling pathways, which activate two IKK-related kinases, Tank binding kinase 1 (TBK1) and IKKε (also called IKKi). The search for transcription factors activating the promoters of IFNA and IFNB genes led to the identification of IRF-3 and IRF-7, which have a critical role in the transcriptional activation of type I IFN genes (11–15). These cytoplasmic IRFs are specifically phosphorylated by TBK1 and IKKε, transported to the nucleus, and activate expression of type I IFN genes as well as certain IFN-induced genes (ISG) (16). Recognition of viral RNAs by cytoplasmic receptors RIG-I and MDA5 induces a distinct signaling cascade via the adapter IPS1 (also called MAVS, CARDif, and VISA), which also activates TBK1, IKKε, IRF-3, and IRF-7. In contrast, activation of the antiviral pathway by TLR7/8 or TLR9 is MyD88-dependent and requires IRAK1, IKKα, and TRAF6 but is independent of TBK1 and IKKε (17, 18). Generation of mice deficient in TLRs, RIG-I, IRF-3, or IRF-7 has revealed that although pathogen recognition may be mediated by distinct cellular receptors and signaling pathways, all pathways converge on IRF-3 and IRF-7 (1, 19). Activation of IRF-3 is sufficient for the stimulation of the IFNB gene transcription, whereas activation of the IFNA genes depends on IRF-7 (12, 20, 21).
Another member of the IRF family, IRF-5, has also been implicated in innate immunity. The HuIRF-5 variant 4 (GenBank™ accession number AY_504947.1) is expressed primarily in dendritic cells (DC) and B cells and shows some properties that are distinct from IRF-3 and IRF-7. Unlike IRF-3 and -7, the IRF-5 polypeptide contains two nuclear localization signals, and consequently low levels of nuclear IRF-5 can be detected in the nucleus of uninfected cells (22, 23). Human IRF-5 is expressed in multiple spliced variants, and some of these are transcriptionally inactive and may function as dominant negative mutants (24). The activation and nuclear transport of individual variants of HuIRF-5 also seems to be distinct (25, 26). In conjunction with the observed in vitro antiviral effects of HuIRF-5, further evidence for the role of IRF-5 in IFNα synthesis has come from genetic studies of systemic lupus erythromatosis (SLE), which is characterized by a constitutive expression of IFNα/β. Three different functional variants of IRF-5 have now been identified with defined risk haplotypes for SLE (27), as well as inflammatory bowel disease (28).
In vitro experiments have shown that in infected cells, HuIRF-5, like IRF-3 and IRF-7, is activated by phosphorylation, resulting in nuclear translocation and stimulation of IFNA gene expression (23). The transcriptional signatures of IRF-5 and IRF-7 in NDV-infected cells were shown to be both overlapping and distinct (22, 29). IRF-5 was found to specifically up-regulate early inflammatory cytokines and chemokines in addition to IFNα (29, 30). Distinctions also exist between the activation of IRF-3 and IRF-5. The TLR3-TRIF pathway, which activates both IRF-3 and IRF-7 by TBK-1 and IKKε, does not activate IRF-5. However, both IRF-5 and IRF-7 are activated by MyD88-dependent pathways through a TRAF6- and IRAK1-dependent mechanism (1, 31). Initial analysis of the role of IRF-5 in the innate antiviral response in Irf5–/– mice showed impairment in the TLR9-mediated induction of IL-6 and TNFα in splenic DC. However, unlike pDCs from Irf7–/– mice, which are severely compromised in CpG-DNA-induced IFN responses, pDCs from Irf5–/– mice did not show any defect in the induction of IFNα (19, 32). The impairment of TNFα, IL-6, and IL-12p40 was not limited to TLR9-mediated induction but was also observed in the TLR4 and TLR3 responses of splenic macrophages; however, the induction of type I IFN in Irf5–/– cells in response to these inducers was not examined (32). A subsequent paper by the same group reported that Irf5–/– mice are highly sensitive to viral infection and show lower levels of type I IFN in the serum. IFN production was also impaired in infected macrophages, whereas no decrease was seen in Irf5–/– MEFs (33). As demonstrated for human IRF-5 v4 (30), MuIRF-5 was also shown to play a role in apoptosis and tumorigenicity in nude mice (34). Recent observations have shown that IRF-5 and TLR7 play a role in IFNα induction by RNA-containing immune complexes present in the sera of SLE patients (35). Yasuda et al. (35) stimulated a mixture of cDC and pDC derived from the bone marrow of C57BL/6J and Irf5–/– mice with IgG from lupus sera, the TLR9 ligand CpG-A, or the TLR7 ligand R848 and show that production of IFNα and IL-6 was largely abolished in Irf5–/– cells, a result that is in contrast to the report by Takaoka et al. (32).
This study was initiated to clarify the apparent contradictions between the role of HuIRF-5 in the antiviral response (in particular the type I IFN response) in vitro and MuIRF-5 in vivo. To this effect, we have identified and characterized the IRF-5 transcripts expressed in C57BL/6J mice and determined the functional properties of MuIRF-5 in vitro and in vivo. Our results show that unlike HuIRF-5, which is expressed in multiple spliced variants, MuIRF-5 is expressed as a single major transcript. Low levels of a single spliced MuIRF-5 containing a 288-nucleotide deletion were detected only in the bone marrow of C57BL/6J mice. MuIRF-5 enhanced the NDV-induced transcriptional activation of the IFNA4 and IFNB promoters. TBK1- and MyD88-activated IRF-5 binds to the IFNA4 promoter and regulates transcription of both type I IFN genes and inflammatory cytokines. The role of IRF-5 in the antiviral response was also demonstrated in vivo; serum levels of type I IFN and TNFα were significantly lower in NDV-infected Irf5–/– mice than in the C57BL/6J mice, which is in agreement with the published data on VSV and HSV-1-infected Irf5–/– mice (33). IRF-5 plays an important role for NDV- and TLR7-induced TNFα production in peritoneal macrophages but not in bone marrow-derived macrophages and is not involved in type I IFN production in these cells. In contrast, IRF-5 is critical for TLR-mediated TNFα production in CD11c+ DC and for TLR9-mediated IFNα induction. Altogether, our data clearly reveal the importance of IRF-5 in antiviral and inflammatory responses to NDV and the TLR7 and TLR9 MyD88-mediated antiviral pathway.
EXPERIMENTAL PROCEDURES
Cell Culture, Virus, and Reagents—293T, RAW264.7 and L929 cells were obtained from the American Type Culture Collection (ATCC). Huh7 and Huh7.5 cells were obtained from Apath (St. Louis, MO) courtesy of C. Rice (Rockefeller University, New York). All cell lines were cultured in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma), 2 mm l-glutamine (Invitrogen), and gentamicin (Quality Biological) except RAW264.7 cells, which were grown in RPMI 1640 medium (Invitrogen). Newcastle disease virus (NDV) La Sota strain was acquired from ATCC and propagated in embryonated eggs. LPS was from Sigma; resiquimod (R848) was a generous gift from 3M (St. Paul, MN) and GL Synthesis Inc. (Worcester, MA), poly(I ·C) and poly(U) were from Roche Applied Science.
Mice and in Vivo Infection—Irf5–/– mice (generously provided by Dr. T. Mak, University of Toronto, Canada), were back-crossed to C57BL/6J mice for at least six generations. C57BL/6J mice were obtained from The Jackson Laboratory. Housing and all experiment were carried out in compliance with protocols approved by The Johns Hopkins University Animal Use and Care Committee. Mice were infected intraperitoneally with 1 × 107 plaque-forming units of NDV, and blood was collected at 6 and 24 h after infection.
Cytokine Analysis—Type I IFN levels in serum and cell culture supernatants were quantitated by bioassay on L929 cells using encephalomyocarditis virus as the challenging virus (36).
Samples from infected animals or cells were acidified to pH 4.5 for 3 days and neutralized prior to being assayed. Levels of TNFα and IL-6 were measured by ELISA according to manufacturer's instructions (Ebioscience or R&D Systems).
Plasmids—Full-length MuIRF-5 cDNA was generated from total RNA isolated from mouse splenocytes and RAW264.7 cells and cloned in pCDNA6.1. The bone marrow variant IRF-5 BMv cDNA was generated from RNA isolated from the bone marrow cells of C57BL/6J mice and cloned into pCDNA6.1. IRF-3, IRF-7 (52, 53), MyD88, and TBK1 expression plasmids were described previously (37, 38). pEFbos-RIG-I was a generous gift from Michael Gale (University of Washington, Seattle). Luciferase reporter plasmids containing promoters of Mu IFNA and -B genes, pGL-3-IFNB, -IFNA4, -IFNA5, -IFNA6, and -IFNA13 were a generous gift from Thomas Michiels (University of Louvain, Belgium). The RANTES luciferase reporter constructs were from John Hiscott (McGill University, Montreal, Canada). The ISRE (5× ISRE from ISG54 gene) was from Stratagene. The IL-6, TNF, IL-8, and IL-12 luciferase reporter genes were from K. Fitzgerald (Worcester, MA).
Peritoneal and Bone Marrow-derived Macrophages, Splenic DC Preparations—Peritoneal macrophages were isolated by adherence following peritoneal lavage 72 h after intraperitoneal injection of sterile thioglycolate solution (REMEL), seeded in RPMI 1640 medium (10% fetal calf serum, 2 mm glutamine, and gentamicin), and infected with 50 HAU of NDV for the indicated times. Medium was collected, and cytokines were assayed as described above. For bone marrow-derived macrophages (BMDM), bone marrow was flushed out of the femurs of WT and Irf5–/– mice with RPMI 1640 medium (10% fetal calf serum, 2 mm glutamine, 5% horse serum, 1 mm sodium pyruvate), and the macrophages were seeded into Petri dishes. For maintenance of the BMDM in culture RPMI 1640 medium was supplemented with 20% of supernatant taken from L929 cells (which produce macrophage colony-stimulating factor). Splenic CD11c+ DC were isolated by positive selection using anti-CD11c MACS beads (Miltenyi Biotec). For experiments cells were plated onto 6- or 96-well plates at a plating density of 2 × 106 or 2 × 104 per well, respectively.
RT, PCR, Quantitative PCR, and Sequencing—Total RNA was isolated using the RNeasy miniprep kit (Qiagen). One microgram of DNase-treated RNA was reverse-transcribed to cDNA with oligo(dT) primers and Superscript III (both Invitrogen). The cDNA was then amplified by PCR using primers specific for full-length IRF-5 (5′-TTAGATAAGCTTGCTATGAACCACTCAGCCCCAG containing a HindIII site and 3′-TAAATATCTAGATGCTTGCATGCCAACTGGGTG containing an XbaI site), exon 2–3 (5′-CTTCAGTGGGTCAACGGG and 3′-TGTACGGCTGAGGTGGCAT), exon 7–8 (5′-ATCCGTCTGTGCCAGTGTAAC and 3′-GCTTCTTCTCTCGGGGTTTG), and β-actin (5′-CATGTTTGAGACCTTCAACACC and 3′-GCACAGCTTCTCTTTGATGTCAC). Primers for exon 6–8 were described previously (32). Sequencing was conducted using T7 primers by Macrogen (Seoul, South Korea). Quantitative PCR analysis for IFNB gene expression was performed using SYBR green reagent (Invitrogen) on a DNA engine Opticon 2 cycler (Bio-Rad) using the following primers: IFNB (5′-CAGCAATTTTCAGTGTCAGAAGC and 3′-CATCCTGTCCTTGAGGCAGT) and β-actin (5′-CCTGGCACCCAGCACAAT and 3′-GCCGATCCACACGGAGTA). The specificity of amplification was assessed for each sample by melting curve analysis, and the size of the amplicon was checked by electrophoresis. PCR efficiency was calculated for each RNA with 10-fold dilutions of the cDNA with the formula E = 10–/slope–1 as described previously (39). Relative quantification was performed using standard curve analysis. All gene expression data were normalized with β-actin and are presented as a ratio of gene copy number per 100 copies of β-actin ± S.D.
Transient Transfection and Reporter Assays—Cells were transfected with indicated expression plasmids using Lipofectamine 2000 (Invitrogen). For luciferase assay, 293T cells (2 × 105 cells/well on a 96-well plate) were transfected with 10 ng/well Renilla luciferase plasmid, 40 or 50 ng of cytokine reporter constructs, and 0–100 ng of MyD88, TBK1, IRF-5, IRF-3, and IRF-7 plasmids. Empty vector DNA was used to keep the total amount of transfected DNA constant (250 ng/well of a 96-well plate). Where required, 16–18 h after transfection, cells were stimulated with NDV for 16 h. Luciferase activity was measured using the dual luciferase assay system (Promega) and normalized against Renilla luciferase activity.
Native and SDS-PAGE and Western Blotting—For the dimerization studies, 293T cells were transfected with the indicated amounts of expression plasmids, and where indicated, 18 h after transfection cells were stimulated with NDV for 6 h prior to lysis. Lysates (20 μg) were analyzed on native (nondenaturing) gels as described (40) and detected by immune blotting as described below. For SDS-PAGE, 10–20 μg of total proteins was resolved on 8 or 10% polyacrylamide gels and transferred to nitrocellulose or polyvinylidene difluoride membranes. Membranes were blocked for 1 h at room temperature with 5% dry skim milk powder in TBST before overnight incubation with anti-mouse IRF-5 (generated in rabbits against the VRFPSPEDIPSDKWR peptide by Affinity BioReagents), IRF-3, and β-actin antibodies (both from Santa Cruz Biotechnology). The membranes were subsequently incubated for 1 h at room temperature with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Amersham Biosciences), and immunodetection was visualized by ECL reagents (Amersham Biosciences) followed by autoradiography on HyBlot CL film (Denville Scientific).
Chromatin Immunoprecipitation Assay—This assay was performed using a chromatin immunoprecipitation assay kit (Upstate Biotechnology, Inc.) as described previously, (11); for full details see supplemental material. The immunoprecipitated DNA was amplified by PCR with promoter-specific primers for 30 cycles. IFNA4 (5′-TACGTTTGAGCCCAAGGTAGAC and 3′-GGCTGTGGGTTTGAGTCTTCTC), and IFNB (5′-AAAGGTACCTCACAGAGACCCTCTCCCAC and 3′-AAAAAGCTTGCTGGAAGCCAGGCTGGTGT).
Purification of Polyhistidine-tagged IRF-5 Using Probond Nickel-chelating Resin—293T cells (5 × 107) were transfected with a combination of expression plasmids for 24 h. Cells were then harvested, and His-tagged IRF-5 and associated proteins were purified using Ni2+ ProBind resin beads (Invitrogen) following the manufacturer's instructions; for full details see supplemental material. Eluted proteins were then washed with phosphate-buffered saline, resuspended in loading buffer, heated to 95 °C for 5 min, and resolved by SDS-PAGE. Proteins were detected by immunoblotting with specific antibodies.
RESULTS
Characterization of IRF-5 Transcripts in Inbred Strains of Mice—We have shown previously that HuIRF-5 is expressed as a number of spliced variants in established cell lines and peripheral blood mononuclear cells, with most of the deletions localized in exon 6 (24). In transient transfection assays only IRF-5 variant 4 (v4) was able to effectively stimulate transcriptional activity of IFNA and IFNB promoters. Two IRF-5 variants, v9, which encodes only the DNA binding peptide-(1–157) and v7-(110–389), encoding a truncated protein, could potentially serve as dominant negative mutants and interfere with endogenous IRF-5 activity (24–26). To determine whether similar variation in IRF-5 transcripts also exists in mice, we analyzed the IRF-5 transcripts found in C57BL/6J mice. Sequence analysis of MuIRF-5 clones generated from the splenic cells RNA of C57BL/6J mice, the genetic background of the Irf5–/– mice (32), has identified only a single transcript of MuIRF-5 identical to the MuIRF-5 sequence deposited in GenBank™ (NM_012057), which encodes a 498-amino acid protein.
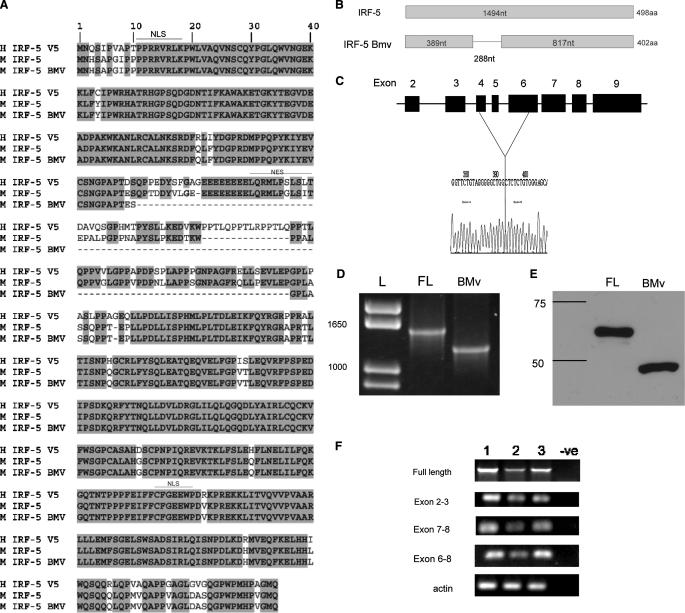
MuIRF-5 shares ∼87% amino acid sequence homology with HuIRF-5 v5 (GenBank™ accession number AA_U12877), which encodes the full-length IRF-5 protein with no internal deletions (Fig. 1A). The most striking difference between the human and mouse IRF-5 proteins is a 5-amino acid deletion in MuIRF-5. In addition to full-length MuIRF-5, we found a single splice variant expressed in the bone marrow at a very low frequency (supplemental Table 1), referred to hereafter as the bone marrow variant or BMv (GenBank™ accession number EU_401974). The BMv transcript contains a 288-nucleotide deletion, which excises part of exon 4, all of exon 5, and part of exon 6 (Fig. 1, A–C). When cloned into the pCDNA6.1 expression vector and expressed in 293T cells, full-length IRF-5 encodes a protein with an electrophoretic mobility of ∼55 kDa, whereas the BMv migrated as an ∼45-kDa protein (Fig. 1E).
FIGURE 1.
Characterization of mouse IRF-5 and its splice variants. A, protein sequence alignment of human IRF-5 variant 5 with mouse IRF-5 and the mouse IRF-5 BMv. The deletion in IRF-5 BMv is shown schematically both for the protein (B) and the genomic DNA sequences (C). L, molecular weight markers (ladder). Expression of ectopic MuIRF-5 (full-length (FL)) and IRF-5 BMv proteins in transfected 293T cells was identified by immune blotting (D), and mRNA expression was identified by RT-PCR (E). F, to detect the possible presence of IRF-5 splice variants in C57BL/6J (lane 1), BALB/c (lane 2), and NZB mice (lane 3), regions representing full-length IRF-5 and distinct exons were amplified by two-step RT-PCR using total splenic RNA.
Like IRF-3 (41), IRF-5 contains two domains, an N-terminal DNA binding domain and a C-terminal transactivation domain, which is responsible for IRF-3 dimerization and C-terminal-specific serine phosphorylation. In both IRF-3 and IRF-5, these two regions are connected by a 70-amino acid proline-rich linker that contains nuclear export signals. The role of the linker in IRF-3 is unknown, but all the major changes in the different HuIRF-5 variants occur in this region. Similarly, the 96-amino acid deletion in MuIRF-5 BMv encompasses the majority of this linker region. To determine whether the presence of the IRF-5 BMv is specific to the high IFN producing C57BL/6J strain, we have also examined the profile of IRF-5 transcripts in the spleens of two other inbred strains of mice as follows: the low type I IFN producing BALB/c strain and the NZB strain, one of the animal models for SLE. Elevated expression of multiple splice variants of IRF-5 has been found to be a genetic risk factor for autoimmune diseases such as SLE in humans (27, 42). An RT-PCR analysis of full-length IRF-5 transcripts and the regions between exons 2–3, 6–8, and 7–8 shows that there were no additional splice variants present in the spleens of these two strains of mice (Fig. 1F). Although we cannot completely rule out the possibility that there is very low level expression of splice variants in these mice, these data indicate that the full-length transcript is the major form of IRF-5 present in these strains. Similarly, the analyses of IRF-5 transcripts in various mouse cell lines show the presence of only full-length transcripts (data not shown), which is in marked contrast to HuIRF-5.
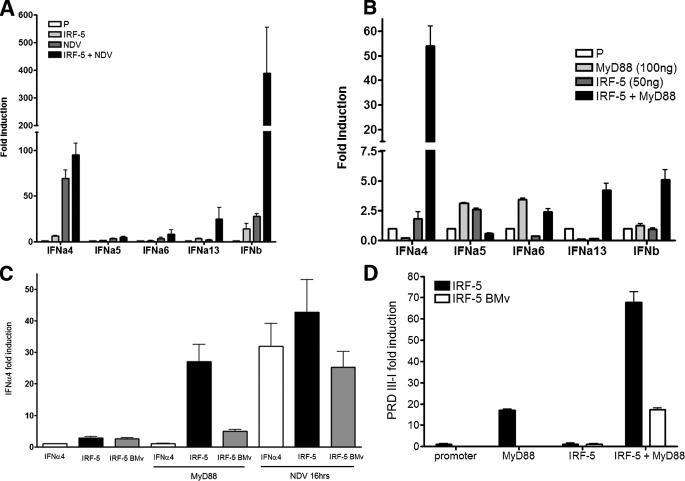
Activated MuIRF-5 Stimulates Transcription of IFNA and -B Promoters—As we have shown previously, HuIRF-5 v4, stimulates transcriptional activity of IFNA gene promoters in reporter assays and induces several IFNA genes in NDV-infected 2fTGH cells (which lack IRF-7) (23, 41). We therefore examined whether MuIRF-5 is also capable of stimulating type I IFN promoters in infected cells by employing the luciferase reporter assay. Promoter activation was measured in 293T cells cotransfected with IFNA and IFNB reporter plasmids in the presence or absence of IRF-5 in both uninfected and NDV-infected cells. NDV infection resulted in a large enhancement of the IRF-5-mediated stimulation of the IRFB (12-fold) possibly because of interaction with endogenous IRF-3 and only a minor effect on the induction of the IFNA4 promoter, but it had no effect on any of the other IFNA promoters (Fig. 2A).
FIGURE 2.
MuIRF-5 preferentially stimulates the murine IFNA4 gene promoter. A, 293T cells were transfected with the respective type I IFN promoter-luciferase reporter plasmids (50 ng), MuIRF-5 expressing plasmids (50 ng), or empty pcDNA6 vector and Renilla luciferase plasmid (10 ng). 20 h post-transfection cells were infected with NDV for 16 h and assayed for luciferase activity that was then normalized against Renilla luciferase. B, 293T cells were transfected with IRF-5 (50 ng) in the presence or absence of MyD88 (100 ng), with Renilla luciferase plasmid (10 ng) and type I IFN promoter-luciferase reporters (50 ng). C, 293T cells were transfected with IRF-5 full-length or IRF-5 BMv (100 ng) and MyD88 (50 ng) plasmids, IFNA4 reporter plasmid, and Renilla luciferase plasmid. Where indicated, cells were also infected with NDV for 16 h. D, 293T cells were transfected with IRF-5 full-length or IRF-5 BMv (10 ng), MyD88 (40 ng), PRDIII-I reporter plasmid, and Renilla luciferase plasmid. B and D, samples were assayed for luciferase activity 24 h post-transfection and normalized against Renilla luciferase. Data shown are combined from 2 or 3 independent experiments in A and C assays and triplicate repeats for B and D experiments.
It has been shown that both Hu- and MuIRF-5 are activated by the TLR7 and TLR9 signaling pathways and that this activation is MyD88-dependent (1, 31). We therefore used the overexpression of MyD88 to mimic TLR activation. Overexpression of MyD88 or IRF-5 alone did not enhance significantly stimulation of the type I IFN promoters (Fig. 2B). MyD88-activated IRF-5 induced an increase in transcriptional activity of the IFNB promoter (5-fold) as well as the IFNA13 promoter, with no effect on IFNA5 or IFNA6. The major effect was seen on the IFNA4 promoter, where a significant enhancement (20-fold) in transcriptional activity was observed with MyD88-activated IRF-5 compared with either IRF-5 or MyD88 alone. Furthermore, the stimulation of IFNA4 and IFNB promoters was proportional to the amount of transfected IRF-5 both in infected and uninfected cells (supplemental Fig. 1, A–C), and the effect was cell line-independent as IRF-5 was able to stimulate the IFNA4 promoter in C57BL/6J, MEFs, NIH/3T3, L929, and RAW264.7 cells (data not shown). As the most significant stimulation by MyD88-activated MuIRF-5 in this assay was observed for the IFNA4 promoter, subsequent work was focused mainly on this promoter.
As the IRF-5 BMv splice variant, described in Fig. 1, contains an internal deletion of most of exons 4–6, we next examined whether this variant retains transcriptional activity using the reporter assays described above. Comparisons of the ability of MyD88-activated IRF-5 and IRF-5 BMv to stimulate the IFNA4 promoter show that IRF-5 BMv is a much weaker activator of this reporter, indicating that the proline-rich linker of the IRF-5 peptide is functionally important (Fig. 2C). Because our data indicated that IRF-5 also binds to the PRDIII-I region of the IFNB promoter,5 we have examined whether MyD88-activated MuIRF-5 can stimulate a reporter plasmid in which luciferase expression is activated by PRDIII-I repeats. As shown in Fig. 2D, MyD88-activated IRF-5 effectively stimulated the transcriptional activity of the PRDIII-I reporter, whereas activation of this reporter by the BMv was negligible, further confirming the importance of this linker region for the function of MuIRF-5.
The activation of IRF-7 by the MyD88 pathway is well established (1). We therefore compared the activation of IFNA4 by MyD88-activated IRF-5 and IRF-7. Under the transfection conditions used, stimulation of the IFNA4 promoter by ectopic expression of IRF-5 was low (2–3-fold), whereas the overexpression of IRF-7 resulted in significant promoter stimulation (supplemental Fig. 1B) indicating that the activity of the ectopic IRF-7 contributes significantly to the high transcriptional activity of MyD88-activated IRF-7 in transfected cells.
It has been shown that the promoters of the IFNA4 and IFNB genes can be stimulated by both activated IRF-3 and IRF-7 (15, 43). These IRFs are activated by specific serine phosphorylation mediated by TBK1 and IKKε kinases (44, 45), which are stimulated by TLR3, TLR4, and RIG-I/MDA-5 signaling pathways (46). To determine whether TBK1 also activates MuIRF-5, the IFNA4 and IFNB reporter plasmids were cotransfected with IRF-5 in the presence and absence of a TBK1 expression vector. However, ectopic expression of TBK1 alone resulted in strong promoter stimulation, and the presence of IRF-5 resulted only in a small (2–3-fold) enhancement (supplemental Fig. 2A). Ectopic TBK1 also effectively stimulated the IFNB promoter, whereas the presence of IRF-5 resulted in an insignificant enhancement (supplemental Fig. 2B). Taken together these results demonstrate that in cells overexpressing TBK1, IRF-5 expression does not contribute significantly to the transcriptional activation.
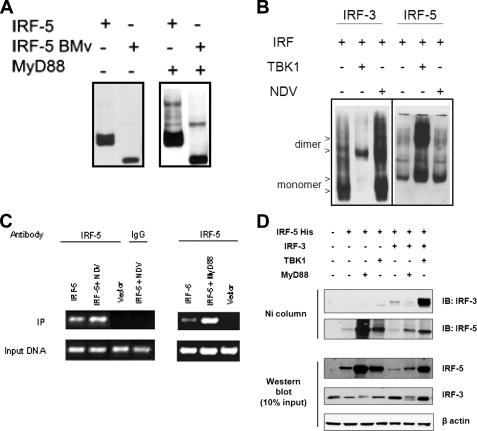
Activated IRF-5 Forms Homodimers and Binds to the IFNA4 Promoter—Activation of the IRF-3 protein by specific serine phosphorylation results in a conformational change, which facilitates homodimer formation (40). We have previously shown that HuIRF-5 v4 undergoes specific serine phosphorylation in NDV-infected cells and forms homodimers as well as heterodimers with IRF-3 and IRF-7 (29). We have therefore examined whether ectopic expression of MyD88 or TBK1 and NDV infection can also induce formation of MuIRF-5 homodimers by using nondenaturing gels to detect the change in protein mobility following dimerization (25, 40). Formation of IRF-5 homodimers was detected in cells cotransfected with MyD88 and MuIRF-5 (Fig. 3A). In addition, in cells transfected with the IRF-5 BMv, MyD88 also induced formation of IRF-5 BMv homodimers. These data indicate that the proline-rich region of the IRF-5 peptide is not critical for dimer formation.
FIGURE 3.
MyD88-activated IRF-5 dimerizes and binds to the IFNA4 promoter. A, 293T cells were transfected with MyD88, full-length IRF-5, or IRF-5 BMv (all 0.5 μg) plasmids as indicated. Cells were lysed 24 h after transfection, proteins separated on a 7.5% native gel, and dimers detected by immunoblotting with anti-IRF-5 antibody. B, 293T cells were transfected as indicated with IRF-5 or IRF-3 and TBK1 plasmids (all 0.8 μg). 18 h after transfection cells were infected with NDV for 6 h. Proteins were analyzed on native and SDS-polyacrylamide gels and visualized by immunodetection. β-Actin levels were determined as a control for equal protein loading. C, in vivo binding of IRF-5 to the murine IFNA4 promoter was analyzed using the chromatin immunoprecipitation assay. The 293T cells were transfected with IFNA4 reporter (150 ng) and IRF-5 (110 ng) plasmids and either cotransfected with MyD88 plasmid (1.2 μg) or 18 h after transfection infected with NDV for 6 h. The chromatin immunoprecipitation assay was performed 24 h post-transfection as described under “Experimental Procedures.” Transfection with empty vector and immunoprecipitation with a nonspecific IgG antibody were used as controls. D, 293T cells (5 × 107) were transfected with a combination of IRF-3, IRF-5, MyD88, and TBK-1 plasmids, and 24 h after the transfection, cells were lysed under nondenaturing conditions, and IRF-5 was purified on Ni+-charged resin as described under supplemental Experimental Procedures. IB, immunoblot.
It was shown that NDV-induced type I IFN synthesis proceeds through the activation of the RIG-I pathway in most cells (47), where both the TBK1 and IKKε kinases have a critical role for the expression of type I IFN genes (38, 45). Ectopic expression of TBK1 very effectively induced formation of IRF-5 homodimers, whereas the formation of IRF-5 dimers was nearly undetectable in cells infected with NDV (Fig. 3B). As a positive control we show that both TBK-1 and NDV infection induced formation of IRF-3 homodimers. These data indicate that the levels of activated TBK1 induced by NDV infection are sufficient to induce dimerization of IRF-3 but not of IRF-5.
To further evaluate the role of IRF-5 in the induction of the IFNA4 gene, we have analyzed the ability of MyD88-activated IRF-5 to bind to the IFNA4 promoter using chromatin immunoprecipitation. The ability of NDV infection to stimulate IRF-5 binding to the IFNA4 VRE was also examined, as we had observed that IRF-5 enhanced NDV-mediated activation of the IFNA4 promoter in the luciferase reporter assay. Cells were either cotransfected with IRF-5 and MyD88 or IRF-5-transfected cells were infected with NDV. The DNA-protein complexes were precipitated with anti-IRF-5 antibody, and the precipitated DNA was amplified by PCR using primers specific for the IFNA4 VRE. The PCR conditions used gave rise to linear amplification (data not shown). As shown in Fig. 3C, fragments containing the IFNA4 VRE were effectively amplified from DNA-protein complexes immunoprecipitated from MyD88-transfected cells, but low levels of amplification were seen also in the absence of MyD88. These data indicate that in the uninduced cells overexpressing MuIRF-5, low levels of nuclear MuIRF-5 bind the IFNA4 promoter. When the chromatin precipitation was done in NDV-infected cells, we also detected binding of IRF-5 to the IFNA4 VRE, but the binding was increased only by about 2-fold compared with uninfected cells (Fig. 3C). No binding of IRF-5 to the IFNA4 VRE could be detected when the cells were transfected with empty vector, or following precipitation with nonspecific IgG antibodies, although input DNA amplification shows equal loading of the samples. These data indicate that in cells expressing ectopic IRF-5, the low levels of IRF-5 constitutively present in the nucleus are able to bind to the IFNA4 promoter. In contrast to NDV infection, MyD88 activation of IRF-5 leads to a significant increase of IRF-5 binding to the IFNA4 promoter. Furthermore, IRF-5 was found to bind both the IFNA4 and IFNB promoters in RAW264.7 cells stimulated with the TLR7 ligand poly(U) but not in cells stimulated with poly(I ·C) or LPS, demonstrating that IRF-5 binds to type I IFN promoters in response to TLR7 stimulation but not TLR3 or TLR4 stimulation (supplemental Fig. 3C). Taken together, these data correlate with the results of the transient transfection assays, and they indicate that under the same experimental conditions the activation of IRF-5 by MyD88 (TLR) is more effective in inducing both homodimer formation and binding of activated IRF-5 to the IFNA4 promoter than activation by NDV infection.
We have shown that HuIRF-5 v4 forms heterodimers with IRF-3; however, binding of IRF-5 v5 and IRF-3 was not detected (25, 48). Analysis of the lysates from IRF-5-transfected cells on nondenaturing gels shows that IRF-5 forms dimers in both TBK1- and MyD88-transfected cells; however, the relative levels of IRF-5 dimers induced by MyD88 activation were significantly lower than levels of TBK1-induced dimers (Fig. 3, A and B). Analysis of the lysates from cells cotransfected with IRF-3, IRF-5, and TBK1 shows the formation of IRF-3 homodimers as well as the presence of a broad IRF-5 double band that could potentially represent the distinct mobility of an IRF-5 homodimer and an IRF-5/IRF-3 heterodimer (data not shown).
To determine whether MuIRF-5 can bind IRF-3, we have transfected IRF-5 either alone, with MyD88 or TBK1 plasmids, and in the presence or absence of IRF-3, lysed the cells in nondenaturing conditions, and used Ni2+-charged resin to pull down the His-tagged MuIRF-5 as well as any proteins bound to IRF-5, which can then be detected by Western blotting. It can be seen in Fig. 3D that the relative levels of ectopic IRF-5 in the cell lysates were greatly enhanced when IRF-5 was cotransfected with MyD88 or TBK1 and that MyD88- or TBK1-activated IRF-5 bound more effectively to the Ni2+ resin than inactive IRF-5. As shown in Fig. 3D, no association between the endogenous IRF-3 and ectopic IRF-5 or MyD88-activated IRF-5 was detected, whereas in TBK1-transfected cells low levels of endogenous IRF-3 were pulled down together with IRF-5. In cells cotransfected with both IRF-3 and IRF-5, high levels of ectopic IRF-3 copurified with IRF-5, in the presence of ectopic TBK1, whereas very little IRF-3 copurified with IRF-5 in MyD88-transfected cells (Fig. 3D). These data indicate that TBK1-activated IRF-3 and IRF-5 can heterodimerize; however, the relative levels of heterodimers formed between MyD88-activated IRF-5 and inactive IRF-3 were about the same as detected between ectopic IRF-5 and endogenous IRF-3. The data also indicate that inactive IRF-5 binds poorly to the Ni2+ resin. Western blot analysis of the input lysates shows that the relative levels of ectopic IRF-5 were substantially increased in the presence of MyD88 and TBK1. Surprisingly, the levels of endogenous and transfected IRF-3 were lower in cells cotransfected with MyD88, and levels of ectopic IRF-5 were lower in cells cotransfected with IRF-5, IRF-3, and MyD88. These differences were not because of variability in the amount of analyzed protein as the levels of β-actin were comparable in all samples, or transfection efficiency because the amount of transfected DNA was kept constant. Whether the overexpression of ectopic MyD88 induces IRF-3 or IRF-5 degradation is under investigation.
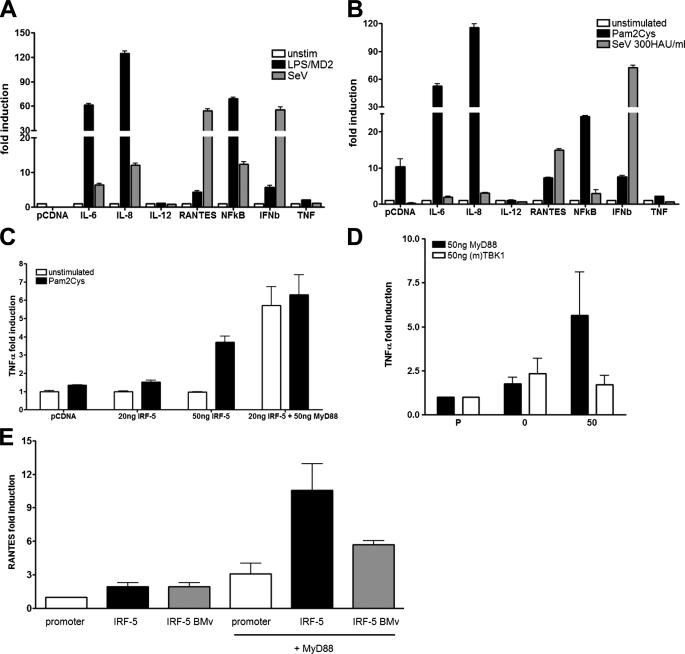
Role of IRF-5 in TLR-dependent Induction of Inflammatory Cytokines—Our previous results indicated that overexpression of HuIRF-5 in virus-infected fibroblasts or B cell lines induced a number of inflammatory cytokine and chemokine genes in addition to type I IFNs (22, 29). Furthermore, it was reported that Irf5–/– mice showed a major defect in TLR4- and TLR9-dependent induction of inflammatory cytokines (32). We were therefore interested in determining whether we could establish a role for MuIRF-5 in the TLR-mediated induction of inflammatory cytokines in vitro. To this effect we first analyzed the activation of different cytokine promoters in 293T cells stably expressing TLR2 or TLR4. Both TLR2 (Fig. 4A) and TLR4 ligands (Fig. 4B) were effective inducers of the IL-6, IL-8, and NFκB promoters, with only small inductions observed for the RANTES and IFNB promoters. In contrast, Sendai virus, which does not signal through either TLR, efficiently induced the IFNB and RANTES promoters and low levels of the IL-6, IL-8, and NFκB promoters. As the syntheses of TNFα and IL-6 were down-regulated in macrophages from Irf5–/– mice upon LPS and CG DNA stimulation (32), we have also examined whether ectopic expression of IRF-5 would facilitate the activation of these promoters. Stimulation of TLR2 by Pam2Cys or overexpression of IRF-5 alone did not activate the TNFA promoter; however, ectopic expression of IRF-5 in conjunction with TLR2 stimulation resulted in a 3-fold increase in the promoter activity (Fig. 4C). MyD88-activated IRF-5 significantly increased the activity of TNFA promoter (5–6-fold) in unstimulated cells, and no further enhancement was seen upon TLR2 stimulation. Similarly IRF-5 enhanced the transcriptional activity of the IL-6 promoter (supplemental Fig. 3A) and the promoter containing multiple ISREs (data not shown) in TLR2-stimulated cells. The fact that TLR2 stimulation and MyD88 activation of IRF-5 result in the same degree of activation of TNFA and IL-6 promoters indicates that the TLR2-mediated activation of IRF-5 is through MyD88 and that IRF-5 is the mediator of the inflammatory cytokines induced by TLR2 signaling. Indeed, cotransfection of IRF-5 with either MyD88 or TBK1 as activators indicates that it is only through MyD88 that IRF-5 activates the TNFA promoter effectively (Fig. 4D).
FIGURE 4.
Role of IRF-5 in TLR-dependent induction of inflammatory cytokines. 293T cells stably expressing TLR4 (A) or TLR2 (B) were transfected with luciferase reporters containing promoters of different inflammatory cytokines (40 ng), IRF-5 (100 ng), and Renilla luciferase (40 ng) plasmids. 8 h after transfection, cells were infected with Sendai virus (300 HAU/ml) or stimulated with Pam2Cys (10 nm) or LPS (10 ng/ml) in the presence of supernatants from MD2-expressing cells for 16 h. C, cells were transfected with the indicated amounts of IRF-5 and MyD88, Renilla luciferase (40 ng) plasmids, and TNFA reporter plasmid (40 ng) as described above. Transfected cells were left unstimulated or stimulated with 10 nm Pam2Cys for 16 h. D, 293T cells were transfected with IRF-5 (100 ng) and MyD88 or TBK1 (50 ng) plasmids, TNFA reporter, and Renilla luciferase plasmids. E, 293T cells were transfected as indicated with full-length IRF-5 or IRF-5 BMv (100 ng) and MyD88 (50 ng) plasmids, RANTES reporter plasmid, and Renilla luciferase plasmid. D and E samples were assayed for luciferase activity 24 h post-transfection and normalized against Renilla luciferase. Data shown are combined triplicates from two independent experiments.
It has been previously demonstrated that activated IRF-3 is an effective inducer of the RANTES promoter that, like the IFNB promoter, contains IRF- and NFκB-binding sites (49). NDV-activated HuIRF-5 v4 was shown to be an effective inducer of inflammatory cytokines including RANTES (22). We have therefore examined whether MyD88-activated MuIRF-5 and IRF-5 BMv can also stimulate the promoter of the RANTES gene. Although IRF-5, IRF-5 BMv, or MyD88 alone did not significantly induce the RANTES promoter, MyD88-activated IRF-5 was an effective inducer (4-fold stimulation), and as seen for the type I IFN promoters, IRF-5 BMv showed a reduced transcriptional activation of this promoter (Fig. 4E). Altogether these data indicate that the MyD88-activated antiviral and inflammatory pathways employ IRF-5.
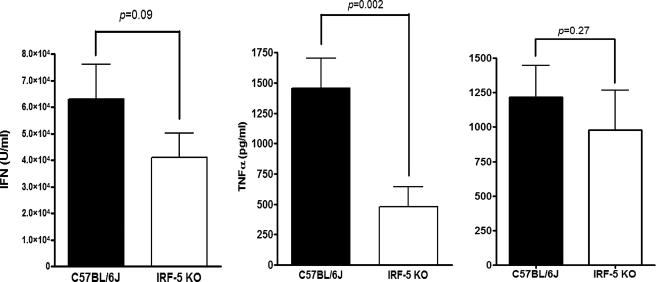
Effect of Homozygous MuIRF-5 Deletion on the Antiviral and Inflammatory Responses—To confirm the role of IRF-5 in the antiviral and anti-inflammatory response to viral infection we have used mice, which have a homozygous deletion of the IRF-5 gene. Previous reports suggested that whereas IRF-5 deletion in cDC, macrophages, and pDC impaired the TLR-mediated induction of several inflammatory cytokines, induction of IFNα in pDCs was unaffected, at least for CpG-A DNA stimulation (32). The same laboratory recently reported that Irf5–/– mice showed enhanced sensitivity to VSV infection and decreased serum levels of type I IFN (33). Given our in vitro data, which clearly established IRF-5 as a key mediator of IFNA gene induction, we wished to clarify the role of IRF-5 in both the antiviral and inflammatory responses. We therefore compared these responses in NDV-infected Irf5–/– mice and wild type (WT) C57BL/6J mice. High serum IFN levels could be detected in WT mice at 6 h postinfection with 107 plaque-forming units of NDV, whereas the levels of biologically active type I IFN were significantly lower in Irf5–/– mice (Fig. 5). Similarly there was a highly significant decrease in the serum levels of TNFα; however, no differences in serum levels of IL-6 were observed, indicating that although IRF-5 plays a role in the antiviral IFN and TNFα response to NDV infection in vivo it does not play a significant role in IL-6 induction in this particular model.
FIGURE 5.
Role of IRF-5 in the antiviral and inflammatory response to NDV infection in vivo. IRF-5-deficient mice (Irf5–/–) and C57BL6/J (wt) mice were infected with NDV (50 HAU) for 6 h and then the collected serum was analyzed for type I IFN by bioassay (left), TNFα (middle), and IL-6 by ELISA (right). Data shown are combined from four independent experiments, wild type n = 12 and Irf5–/– n = 15 mice.
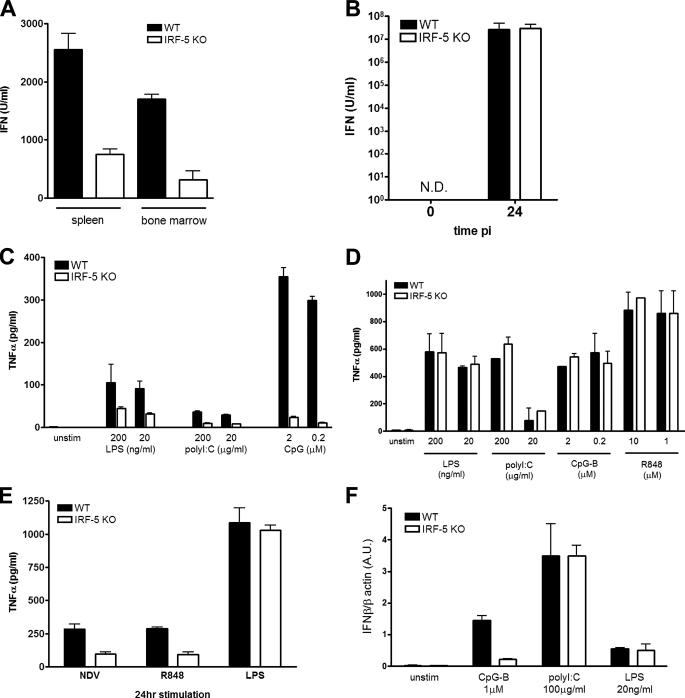
Viral infection can stimulate type I IFN production in most infected cells; however, pDCs were shown to be the major source of circulating IFNα (50, 51). In vitro experiments have shown that although the synthesis of type I IFN in pDC is primarily mediated by activation of TLR7 or TLR9, in other cells types viral infection induces IFN synthesis primarily by the RIG-I/MDA5 pathway (1). To determine which immune cells require IRF-5 for the antiviral and inflammatory response, we have analyzed its role in ex vivo experiments. Comparison of IFN synthesis in NDV-infected bone marrow cells and splenocytes from WT and Irf5–/– mice shows that IRF-5 contributes to IFN synthesis in the mixed cell populations from both compartments (Fig. 6A). The analysis of an inducible cytokine profile was repeated in peritoneal macrophages infected with NDV for 24 h, with no observable difference in type I IFN (Fig. 6B) and IL-6 (data not shown) synthesis between WT and Irf5–/– macrophages and decreased TNFα production in Ifr5–/– macrophages (Fig. 6C). A decrease in TNFα synthesis induced by TLR7 stimulation, but not after TLR4 stimulation, was also observed (Fig. 6C).
FIGURE 6.
Cell type-specific role of IRF-5 in the antiviral and inflammatory response. A, total white blood cells from spleen and bone marrow cells (2 × 106 cells). B, peritoneal macrophages (2 × 106 cells) from WT and Irf5–/– mice were infected with NDV (50 HAU) for 24 h. The levels of type I IFN in the culture medium were measured by bioassay. C, peritoneal macrophages from WT and Irf5–/– mice were infected with NDV (50 HAU) and treated with R848 (10 μm) or LPS (20 ng/ml) for 24 h, and TNFα levels in the supernatant were measured by ELISA. D, BMDC. E, purified splenic CD11c+ DC (1 × 105 cells) from WT and Irf5–/– mice were stimulated for 16 h with the respective TLR ligands, and levels of TNFα in the medium were measured by ELISA. F, purified splenic CD11c+ DC (1 × 106 cells) from WT and Irf5–/– mice were stimulated for 2 h with respective TLR ligands. Cells were harvested; total RNA was isolated, and relative levels of IFNβ were measured by real time PCR. Transcript levels are shown in arbitrary units (A.U.) compared with β-actin.
To further examine the role of IRF-5 in inflammatory and IFN responses to TLR ligands, we generated BMDM by incubation of bone marrow cells with granulocyte-macrophage colony-stimulating factor. BMDM were stimulated with ligands for TLR3, TLR4, TLR7, and TLR9, and TNFα levels were measured by ELISA. No differences in TLR3-, TLR4-, TLR7-, or TLR9-activated TNFα or IFN synthesis were observed between BMDM from WT and Irf5–/– mice (Fig. 6D and data not shown). We next examined these responses in splenic CD11c+ DC, a population of cells that contains both cDC and pDC (52). In contrast to the BMDM, where no effect of IRF-5 was observed, the splenic CD11c+ DC population from Irf5–/– mice showed impaired TNFα production in response to TLR3, TLR4, and in particular CpG-DNA B class oligodeoxynucleotides that signal via TLR9 (Fig. 6E). When the same population of cells and inducers were analyzed for IFN synthesis, the absence of IRF-5 led to a marked defect in CpG-B-mediated induction of IFNB (Fig. 6F) but not that induced by TLR3 or TLR4 pathway. A similar defect in IFNB induction was seen when the CD11c+ DC from Irf5–/– mice was stimulated with the TLR7 ligand R848 (data not shown).
From these data, we conclude that the antiviral response to NDV infection in vivo is not entirely mediated by the RIG-I pathway, which does not effectively activate IRF-5. The function of IRF-5 in the inflammatory response is also broader and less cell type- and inducer-restricted than its role in the antiviral response. IRF-5 plays a role in TNFα synthesis in peritoneal macrophages and CD11c+ DC in response to both viral infection and TLR3 and TLR4 stimulation, although its antiviral role is limited to cDC and TLR7 and TLR9 activation.
DISCUSSION
Remarkable progress has been made in recent years in identifying cellular receptors detecting invading pathogens as well as in understanding the signaling pathways leading to the transcription of type I IFN genes. Two of the IRFs, IRF-3 and IRF-7, were shown to have a major role in the transcriptional activation of type I IFN genes (43, 53–55), and although recognition of viral nucleic acids is mediated by distinct cellular receptors and signaling pathways, they all converge on IRF-3 and IRF-7 (1, 19). The implication of a role of IRF-5 in the inflammatory response gave new insights into the regulation of the antiviral response (56, 57). In vitro studies showing that HuIRF-5 stimulates the transcription of chemokines, inflammatory cytokines and IFNA genes suggested that IRF-5 is required for an optimal inflammatory and antiviral response (22). The fact that HuIRF-5 is expressed as a large number of spliced variants, some of which are not activated by viral infection, suggested its potentially complex role in vivo (24–26). However, the initial studies using Irf5–/– mice showed a significant decrease in inducible expression of inflammatory cytokines but no defect in TLR-mediated type I IFN induction (32). Thus the role of IRF-5 in the induction of the antiviral response remained unclear.
Previous studies had identified several distinctly spliced HuIRF-5 isoforms (designated variants 1–9) that showed variation in insertions or deletions within exon 6, cell type-specific expression, and distinct cellular localization (24–26). In contrast, the characterization of IRF-5 transcripts in the spleen cells of C57BL/6J, BALB/c, and NZB mice and several murine cell lines has shown the presence of a single dominant transcript with close homology to the full-length human IRF-5 variant 5. However the existence of low levels of alternatively spliced variants expressed in distinct cell types cannot be completely eliminated, as a single variant (IRF-5 BMv) containing a deletion from exon 4 to 6 was expressed at very low levels in the bone marrow of C57BL/6J mice (Fig. 1 and supplemental Table 1). These data indicate that MuIRF-5 unlike the HuIRF-5 is not heavily spliced but is expressed as a single dominant mRNA and minor alternately spliced mRNAs. Minor alternatively spliced variants of IRF-1, IRF-3, and IRF-7 were also detected previously (58–60).
HuIRF-5 v4 is one of the most transcriptionally active and best studied variants of HuIRF-5 and is able to efficiently stimulate the expression of IFNA1, -A4, -A8, and -A14 genes in NDV-infected cells in the absence of IRF-7 (23). The transcriptional profiles of inflammatory and particularly type I IFN genes induced by IRF-5 and IRF-7 were found to be both overlapping and distinct suggesting that the two factors have both common and nonredundant roles (23, 25, 29). The activation of HuIRF-5 was shown to be inducer- and TLR-specific, with activation occurring after NDV or VSV infection, but not after poly(I ·C) stimulation (23). Furthermore, although signaling through TLR7 and TLR9 resulted in the activation and nuclear translocation of HuIRF-5, signaling induced by poly(I ·C) binding to TLR3 did not activate HuIRF-5 (31, 32). These data suggested that activation of IRF-5 occurs in a more restricted manner than that seen with IRF3 and IRF7.
Transcriptional activation by MuIRF-5 shows a slightly different pattern; in NDV-infected cells MuIRF-5 activated the murine IFNA4 and IFNB promoters, but no significant activation of the other murine IFNA promoters was detected (Fig. 2A). Furthermore, the activation of MuIRF-5 by NDV infection was inefficient, with no significant formation of MuIRF-5 homodimers and only a small increase in the binding of IRF-5 to the IFNA4 VRE in cells (Fig. 3). Thus, in this respect, MuIRF-5 resembles HuIRF-5 v5, which was not phosphorylated and did not form dimers in NDV-infected cells, rather than HuIRF-5 v4 (25). Although the formation of HuIRF-5v4 and IRF-3 heterodimers in infected cells was clearly demonstrated (48) binding of IRF-5 v5 and IRF-3 was not observed (25). We show that the TBK1-activated MuIRF-5 forms both homodimers and heterodimers with activated IRF-3. Recruitment of activated IRF-5 to the IFNA4 promoters was also increased in cells overexpressing IRF-3 (data not shown).
The NDV-induced activation of IRF-3 was shown to proceed through the RIG-I pathway, leading to the activation of TBK1 and phosphorylation of IRF-3 on a specific cluster of serines in the C terminus of the protein (44, 45). The role of the RIG-I/MDA5 pathway in the activation of IRF-5 by NDV infection needs further evaluation; in vivo infection and ex vivo stimulation of macrophages suggested that IRF-5 was important in the innate inflammatory response following infection, yet in vitro transfection assays showed no significant cooperation between RIG-I and IRF-5. As expected, the NDV-mediated induction of IFNB promoter was nearly negligible in the RIG-I-defective Huh7.5 cell line, and the activation of IRF-5 by ectopic RIG-I after NDV infection was lower than the activation of IRF-3 (supplemental Fig. 4). There is also no evidence that TBK1 plays any role in the TLR7/9-mediated activation of IRF-7 as, in mice, IKKα rather than TBK1 was shown to be critical for the TLR7- and TLR9-mediated activation of IRF-7 and IFNA genes (17, 18, 61). Therefore, although TBK1 can induce formation of IRF-5 homodimers, the serine phosphorylation required for the activation of IRF-5 in the MyD88-dependent pathway is dependent on IRAK1 (31). Both IRF-7 and IRF-5 associate with MyD88 as well as with TRAF6, and this association is critical for the activation of both IRFs (17, 32, 61). We have shown that MyD88-induced homodimerization of MuIRF-5 is less efficient than TBK1-induced homodimerization, yet MyD88 is a more effective activator of IRF-5 than TBK1. These data indicate that formation of IRF-5 homodimers may not be a true indicator of IRF-5 activation. The MyD88-activated MuIRF-5 binds efficiently to the IFNA4 VRE in cells (Fig. 3) and activates both IFNA and IFNB promoters in a dose-dependent manner (Fig. 2B and supplemental Fig. 1). Furthermore, the binding of activated endogenous MuIRF-5 to the IFNA4 VRE was also seen upon activation of TLR7 by the poly(U) ligand in RAW264.7 cells (supplemental Fig. 3C). Additional experiments are required to determine whether IRF-5, like IRF-7, is ubiquitinated and interacts with IRAK1 and IKKα and whether IRF-5 phosphorylated by either of these kinases is transcriptionally active.
The in vivo data presented in this study also confirms the role of IRF-5 in the antiviral and inflammatory response to NDV infection. It was shown that in vivo NDV stimulates the type I IFN response via the RIG-I pathway (47); however, NDV infection of IPS-1–/– mice, in which the RIG-I pathway is abrogated, demonstrated that IFN production, although significantly decreased, was not completely eliminated (62). Similarly, the serum levels of type I IFN and TNFα were significantly lower in Irf5–/– mice infected with NDV than in the WT mice, but IFN synthesis was not completely blocked (Fig. 6). These two observations indicate that the NDV-induced IFN response in vivo is mediated by two independent signaling pathways, a result that is in agreement with previous reports (62). Because pDC are the major source of the circulating type I IFN, our data indicate that IRF-5 may play some role in the NDV-stimulated production of IFN in pDC. It was recently shown that systemic NDV infection stimulates expression of several IFNA genes in pDC and to lesser extend in cDC and macrophages (62). Our data also indicate that IFN production is decreased in NDV-infected Irf5–/– splenocytes as well as in splenic CD11c+DC (which contain both cDC and pDC) in response to TLR9 but not to TLR4 or TLR3 activation. In contrast there was no effect on IFN production in response to NDV infection in Irf5–/– peritoneal macrophages or to TLR stimulation of BMDM. Macrophages were shown to utilize the RIG-I pathway in response to a systemic NDV infection (62). Thus our data expand these observations and show that in the antiviral response to a systemic NDV infection, IRF-5 plays a critical role in the TLR pathway in cDC and pDC and not in the RIG-I pathway in macrophages. The inhibition of IFNα synthesis in IPS-1–/– mice was most significant in the early times post-infection, and therefore it was suggested that the antiviral response to NDV in cDC and macrophages precedes the pDC response (62). Thus the activation of IRF-5 may contribute to the overall duration of the antiviral response. Experiments are under way to examine this possibility.
Taniguchi and co-workers (32) had previously examined IFNα production in pDCs from Irf5–/– mice stimulated with CpG-A oligonucleotides and have observed no difference. Based on these observations, they concluded that there was no role for IRF-5 in IFN production. However, our findings that IFNα is reduced in CD11c+ DC following CpG-B stimulation (Fig. 6F) highlight the importance of IRF-5 in TLR9 signaling and IFN production in other cell types. Although our CD11c+ population contains both mDC and pDCs, pDCs do not induce IFN in response to CpG-B oligonucleotides (63). MDCs have been shown to induce IFNβ in response to CpG B-class oligonucleotides (64). Therefore, in our culture conditions, only the mDCs induced IFNβ in response to the B-class CpG. Interesting and somewhat unexpected is the finding of the role of IRF-5 in TLR3- and TLR4-mediated activation of TNFA and IL-6 expression, because neither TLR3 nor TLR4 pathway activates IRF-5. Further studies need to clarify the mechanism by which inactive IRF-5 contributes to the expression of the inflammatory cytokines.
While this study was being completed it was reported that the Irf5–/– mice are highly susceptible to viral infection (33). Yanai et al. (33) demonstrate that Irf5–/– mice infected with VSV or HSV-1 exhibited higher mortality and produced lower levels of IFNα/β than wild type controls. These results concur with our observations and clearly indicate that IRF-5 has a central role in the antiviral response in vivo.
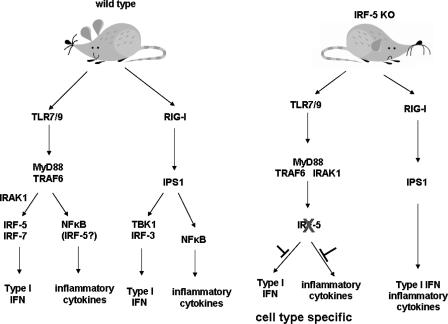
In summary, this study clarifies the role of IRF-5 in the type I IFN response with regard to virus and MyD88 activation through analysis of the functional properties of MuIRF-5 in vitro and in vivo. Surprisingly, despite the sequence similarities between the human and mouse IRF-5 gene, MuIRF-5 is expressed as a single dominant transcript, which is in contrast to the multiple splice variants described for HuIRF-5. We show the ability of both MyD88 and TBK1 to activate MuIRF-5 to form homodimers, bind to the IFNA4 and IFNB promoters in cells, and stimulate their transcriptional activity. We also show that NDV infection and the RIG-I pathway are weaker activators of IRF-5 than TLR7/9-activated MyD88 pathway. We demonstrate that the antiviral and inflammatory response to NDV infection in vivo is impaired in Irf5–/– mice and show in ex vivo experiments that down-regulation of IFNα and TNFα synthesis is highly cell type- and inducer-specific. These data indicate that IRF-5 has a key role in the TLR but not in the RIG-I mediated antiviral and inflammatory response, a proposed model that we present in Fig. 7. Clearly the role of IRF-5 in the innate immune response warrants further investigation, as its deregulation may play a broad role in modulation of the immune response and autoimmunity (28, 42). Thus regulation of IRF-5 levels and its function may provide a potential therapeutic target for inflammatory disease as well as viral infection.
FIGURE 7.
The role of IRF-5 in the innate antiviral response. Following infection, viral nuclei acids are recognized by membrane-bound TLR and cytoplasmic RIG-I/MDA5 receptors. Our data, in conjunction with data of others (32), have shown that IRF-5 is activated by TLR7- or TLR9-, MyD88-dependent pathway. Activated IRF-5 stimulates expression of IFNA and IFNB genes as well as the inflammatory cytokines. However, although the role of IRF-5 in the activation of type I IFN genes is restricted to the MyD88 pathway and to cells that express the TLR7 or TLR9 receptors, the activation of the inflammatory genes by TLR4 and TLR3 seems also to be dependent on IRF-5, and consequently the role of IRF-5 in the induction of these inflammatory cytokines is broader and less cell type-restricted. There is no evidence that IRF-5 plays a substantial, if any, role in the RigI, IPS1 (VISA/MAVS/CARDif) pathway, and whether it can be activated by the MAD5 pathways is yet to be determined.
Supplementary Material
Acknowledgments
We thank Dr. Tak Mak for generosity in providing the Irf5–/– mice, Michael Gale and Thomas Michiel for their generous donation of plasmids, and Charles Rice for the Huh7 and Huh7.5 cells. The technical work of the late Merrill Kellum has been greatly appreciated.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI067632 (to P. M. P.) and AI067497 (to K. A. F.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement”in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Experimental Procedures, Table 1, and Figs. 1–4.
The nucleotide sequence(s) reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) EU_401974.
Footnotes
The abbreviations used are: IFN, interferon; IRF, interferon regulatory factor; PRD, positive regulatory domain; RIG-I, retinoic acid-inducible gene I; MyD88, myeloid differentiation factor 88; TBK1, tank-binding kinase 1; TRAF6, tumor necrosis factor receptor-associated factor 6; IRAK1, interleukin 1 receptor-associated kinase 1; IKKα, inhibitor of NFκB kinase α; DC, dendritic cell; pDC, plasmacytoid dendritic cell; NDV, Newcastle disease virus; ELISA, enzyme-linked immunosorbent assay; RT, reverse transcription; BMv, bone marrow variant; BMDM, bone marrow-derived macrophage; WT, wild type; TNFα, tumor necrosis factor α; dsRNA, double-stranded RNA; SLE, systemic lupus erythromatosis; VSV, vesicular stomatitis virus; RANTES, regulated on activation normal T cell expressed and secreted; TLR, toll-like receptor; LPS, lipopolysaccharide; ISRE, interferon-stimulated response element; HAU, hemagglutination units.
P. M. Pitha and K. A. Fitzgerald, unpublished data.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783–801 [DOI] [PubMed] [Google Scholar]
- 2.O'Neill, L. A. (2006) Curr. Opin. Immunol. 18 3–9 [DOI] [PubMed] [Google Scholar]
- 3.Hornung, V., Ellegast, J., Kim, S., Brzozka, K., Jung, A., Kato, H., Poeck, H., Akira, S., Conzelmann, K. K., Schlee, M., Endres, S., and Hartmann, G. (2006) Science 314 994–997 [DOI] [PubMed] [Google Scholar]
- 4.Yoneyama, M., Kikuchi, M., Matsumoto, K., Imaizumi, T., Miyagishi, M., Taira, K., Foy, E., Loo, Y. M., Gale, M., Jr., Akira, S., Yonehara, S., Kato, A., and Fujita, T. (2005) J. Immunol. 175 2851–2858 [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730–737 [DOI] [PubMed] [Google Scholar]
- 6.Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001) Nature 413 732–738 [DOI] [PubMed] [Google Scholar]
- 7.Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis e Sousa, C. (2004) Science 303 1529–1531 [DOI] [PubMed] [Google Scholar]
- 8.Lund, J., Sato, A., Akira, S., Medzhitov, R., and Iwasaki, A. (2003) J. Exp. Med. 198 513–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stetson, D. B., and Medzhitov, R. (2006) Immunity 24 93–103 [DOI] [PubMed] [Google Scholar]
- 10.Ishii, K. J., Coban, C., Kato, H., Takahashi, K., Torii, Y., Takeshita, F., Ludwig, H., Sutter, G., Suzuki, K., Hemmi, H., Sato, S., Yamamoto, M., Uematsu, S., Kawai, T., Takeuchi, O., and Akira, S. (2006) Nat. Immunol. 7 40–48 [DOI] [PubMed] [Google Scholar]
- 11.Au, W. C., and Pitha, P. M. (2001) J. Biol. Chem. 276 41629–41637 [DOI] [PubMed] [Google Scholar]
- 12.Juang, Y. T., Lowther, W., Kellum, M., Au, W. C., Lin, R., Hiscott, J., and Pitha, P. M. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 9837–9842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, R., Mamane, Y., and Hiscott, J. (2000) J. Biol. Chem. 275 34320–34327 [DOI] [PubMed] [Google Scholar]
- 14.Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., Nakaya, T., Katsuki, M., Noguchi, S., Tanaka, N., and Taniguchi, T. (2000) Immunity 13 539–548 [DOI] [PubMed] [Google Scholar]
- 15.Schafer, S. L., Lin, R., Moore, P. A., Hiscott, J., and Pitha, P. M. (1998) J. Biol. Chem. 273 2714–2720 [DOI] [PubMed] [Google Scholar]
- 16.Paun, A., and Pitha, P. M. (2007) Adv. Virus Res. 69 1–66 [DOI] [PubMed] [Google Scholar]
- 17.Uematsu, S., Sato, S., Yamamoto, M., Hirotani, T., Kato, H., Takeshita, F., Matsuda, M., Coban, C., Ishii, K. J., Kawai, T., Takeuchi, O., and Akira, S. (2005) J. Exp. Med. 201 915–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoshino, K., Sugiyama, T., Matsumoto, M., Tanaka, T., Saito, M., Hemmi, H., Ohara, O., Akira, S., and Kaisho, T. (2006) Nature 440 949–953 [DOI] [PubMed] [Google Scholar]
- 19.Honda, K., and Taniguchi, T. (2006) Nat. Rev. Immunol. 6 644–658 [DOI] [PubMed] [Google Scholar]
- 20.Yeow, W. S., Au, W. C., Juang, Y. T., Fields, C. D., Dent, C. L., Gewert, D. R., and Pitha, P. M. (2000) J. Biol. Chem. 275 6313–6320 [DOI] [PubMed] [Google Scholar]
- 21.Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., Shimada, N., Ohba, Y., Takaoka, A., Yoshida, N., and Taniguchi, T. (2005) Nature 434 772–777 [DOI] [PubMed] [Google Scholar]
- 22.Barnes, B. J., Kellum, M. J., Field, A. E., and Pitha, P. M. (2002) Mol. Cell. Biol. 22 5721–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barnes, B. J., Moore, P. A., and Pitha, P. M. (2001) J. Biol. Chem. 276 23382–23390 [DOI] [PubMed] [Google Scholar]
- 24.Mancl, M. E., Hu, G., Sangster-Guity, N., Olshalsky, S. L., Hoops, K., Fitzgerald-Bocarsly, P., Pitha, P. M., Pinder, K., and Barnes, B. J. (2005) J. Biol. Chem. 280 21078–21090 [DOI] [PubMed] [Google Scholar]
- 25.Cheng, T. F., Brzostek, S., Ando, O., Van Scoy, S., Kumar, K. P., and Reich, N. C. (2006) J. Immunol. 176 7462–7470 [DOI] [PubMed] [Google Scholar]
- 26.Lin, R., Yang, L., Arguello, M., Penafuerte, C., and Hiscott, J. (2005) J. Biol. Chem. 280 3088–3095 [DOI] [PubMed] [Google Scholar]
- 27.Graham, R. R., Kyogoku, C., Sigurdsson, S., Vlasova, I. A., Davies, L. R., Baechler, E. C., Plenge, R. M., Koeuth, T., Ortmann, W. A., Hom, G., Bauer, J. W., Gillett, C., Burtt, N., Cunninghame Graham, D. S., Onofrio, R., Petri, M., Gunnarsson, I., Svenungsson, E., Ronnblom, L., Nordmark, G., Gregersen, P. K., Moser, K., Gaffney, P. M., Criswell, L. A., Vyse, T. J., Syvanen, A. C., Bohjanen, P. R., Daly, M. J., Behrens, T. W., and Altshuler, D. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 6758–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dideberg, V., Kristjansdottir, G., Milani, L., Libioulle, C., Sigurdsson, S., Louis, E., Wiman, A. C., Vermeire, S., Rutgeerts, P., Belaiche, J., Franchimont, D., Van Gossum, A., Bours, V., and Syvanen, A. C. (2007) Hum. Mol. Genet. 16 3008–3016 [DOI] [PubMed] [Google Scholar]
- 29.Barnes, B. J., Richards, J., Mancl, M., Hanash, S., Beretta, L., and Pitha, P. M. (2004) J. Biol. Chem. 279 45194–45207 [DOI] [PubMed] [Google Scholar]
- 30.Barnes, B. J., Kellum, M. J., Pinder, K. E., Frisancho, J. A., and Pitha, P. M. (2003) Cancer Res. 63 6424–6431 [PubMed] [Google Scholar]
- 31.Schoenemeyer, A., Barnes, B. J., Mancl, M. E., Latz, E., Goutagny, N., Pitha, P. M., Fitzgerald, K. A., and Golenbock, D. T. (2005) J. Biol. Chem. 280 17005–17012 [DOI] [PubMed] [Google Scholar]
- 32.Takaoka, A., Yanai, H., Kondo, S., Duncan, G., Negishi, H., Mizutani, T., Kano, S., Honda, K., Ohba, Y., Mak, T. W., and Taniguchi, T. (2005) Nature 434 243–249 [DOI] [PubMed] [Google Scholar]
- 33.Yanai, H., Chen, H. M., Inuzuka, T., Kondo, S., Mak, T. W., Takaoka, A., Honda, K., and Taniguchi, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 3402–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Couzinet, A., Tamura, K., Chen, H. M., Nishimura, K., Wang, Z., Morishita, Y., Takeda, K., Yagita, H., Yanai, H., Taniguchi, T., and Tamura, T. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 2556–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yasuda, K., Richez, C., Maciaszek, J. W., Agrawal, N., Akira, S., Marshak-Rothstein, A., and Rifkin, I. R. (2007) J. Immunol. 178 6876–6885 [DOI] [PubMed] [Google Scholar]
- 36.Cheung, S. C., Chattopadhyay, S. K., Hartley, J. W., Morse, H. C., III, and Pitha, P. M. (1991) J. Immunol. 146 121–127 [PubMed] [Google Scholar]
- 37.Fitzgerald, K. A., Palsson-McDermott, E. M., Bowie, A. G., Jefferies, C. A., Mansell, A. S., Brady, G., Brint, E., Dunne, A., Gray, P., Harte, M. T., McMurray, D., Smith, D. E., Sims, J. E., Bird, T. A., and O'Neill, L. A. (2001) Nature 413 78–83 [DOI] [PubMed] [Google Scholar]
- 38.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Latz, E., Golenbock, D. T., Coyle, A. J., Liao, S. M., and Maniatis, T. (2003) Nat. Immunol. 4 491–496 [DOI] [PubMed] [Google Scholar]
- 39.Rutledge, R. G., and Cote, C. (2003) Nucleic Acids Res. 31 e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwamura, T., Yoneyama, M., Yamaguchi, K., Suhara, W., Mori, W., Shiota, K., Okabe, Y., Namiki, H., and Fujita, T. (2001) Genes Cells 6 375–388 [DOI] [PubMed] [Google Scholar]
- 41.Dragan, A. I., Hargreaves, V. V., Makeyeva, E. N., and Privalov, P. L. (2007) Nucleic Acids Res. 35 3525–3534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Graham, R. R., Kozyrev, S. V., Baechler, E. C., Reddy, M. V., Plenge, R. M., Bauer, J. W., Ortmann, W. A., Koeuth, T., Gonzalez Escribano, M. F., Pons-Estel, B., Petri, M., Daly, M., Gregersen, P. K., Martin, J., Altshuler, D., Behrens, T. W., and Alarcon-Riquelme, M. E. (2006) Nat. Genet. 38 550–555 [DOI] [PubMed] [Google Scholar]
- 43.Marie, I., Durbin, J. E., and Levy, D. E. S. (1998) EMBO J. 17 6660–6669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fitzgerald, K. A., Rowe, D. C., Barnes, B. J., Caffrey, D. R., Visintin, A., Latz, E., Monks, B., Pitha, P. M., and Golenbock, D. T. (2003) J. Exp. Med. 198 1043–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma, S., ten Oever, B. R., Grandvaux, N., Zhou, G. P., Lin, R., and Hiscott, J. (2003) Science 300 1148–1151 [DOI] [PubMed] [Google Scholar]
- 46.Kawai, T., and Akira, S. (2006) Nat. Immunol. 7 131–137 [DOI] [PubMed] [Google Scholar]
- 47.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101–105 [DOI] [PubMed] [Google Scholar]
- 48.Barnes, B. J., Field, A. E., and Pitha-Rowe, P. M. (2003) J. Biol. Chem. 278 16630–16641 [DOI] [PubMed] [Google Scholar]
- 49.Lin, R., Heylbroeck, C., Genin, P., Pitha, P. M., and Hiscott, J. (1999) Mol. Cell. Biol. 19 959–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siegal, F. P., Kadowaki, N., Shodell, M., Fitzgerald-Bocarsly, P. A., Shah, K., Ho, S., Antonenko, S., and Liu, Y. J. (1999) Science 284 1835–1837 [DOI] [PubMed] [Google Scholar]
- 51.Asselin-Paturel, C., Boonstra, A., Dalod, M., Durand, I., Yessaad, N., Dezutter-Dambuyant, C., Vicari, A., O'Garra, A., Biron, C., Briere, F., and Trinchieri, G. (2001) Nat. Immunol. 2 1144–1150 [DOI] [PubMed] [Google Scholar]
- 52.Edwards, A. D., Diebold, S. S., Slack, E. M., Tomizawa, H., Hemmi, H., Kaisho, T., Akira, S., and Reis e Sousa, C. (2003) Eur. J. Immunol. 33 827–833 [DOI] [PubMed] [Google Scholar]
- 53.Au, W. C., Moore, P. A., LaFleur, D. W., Tombal, B., and Pitha, P. M. (1998) J. Biol. Chem. 273 29210–29217 [DOI] [PubMed] [Google Scholar]
- 54.Au, W. C., Moore, P. A., Lowther, W., Juang, Y. T., and Pitha, P. M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 11657–11661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ronco, L. V., Karpova, A. Y., Vidal, M., and Howley, P. M. (1998) Genes Dev. 12 2061–2072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barnes, B., Lubyova, B., and Pitha, P. M. (2002) J. Interferon Cytokine Res. 22 59–71 [DOI] [PubMed] [Google Scholar]
- 57.Paun, A., and Pitha, P. M. (2007) Biochimie (Paris) 89 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Harada, H., Kondo, T., Ogawa, S., Tamura, T., Kitagawa, M., Tanaka, N., Lamphier, M. S., Hirai, H., and Taniguchi, T. (1994) Oncogene 9 3313–3320 [PubMed] [Google Scholar]
- 59.Karpova, A. Y., Ronco, L. V., and Howley, P. M. (2001) Mol. Cell. Biol. 21 4169–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, L., and Pagano, J. S. (1997) Mol. Cell. Biol. 17 5748–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J., Uematsu, S., Takeuchi, O., and Akira, S. (2004) Nat. Immunol. 5 1061–1068 [DOI] [PubMed] [Google Scholar]
- 62.Kumagai, Y., Takeuchi, O., Kato, H., Kumar, H., Matsui, K., Morii, E., Aozasa, K., Kawai, T., and Akira, S. (2007) Immunity 27 240–252 [DOI] [PubMed] [Google Scholar]
- 63.Honda, K., Ohba, Y., Yanai, H., Negishi, H., Mizutani, T., Takaoka, A., Taya, C., and Taniguchi, T. (2005) Nature 434 1035–1040 [DOI] [PubMed] [Google Scholar]
- 64.Negishi, H., Fujita, Y., Yanai, H., Sakaguchi, S., Ouyang, X., Shinohara, M., Takayanagi, H., Ohba, Y., Taniguchi, T., and Honda, K. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 15136–15141 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.