Abstract
The opportunistic yeast Cryptococcus neoformans causes serious disease in humans and expresses a prominent polysaccharide capsule that is required for its virulence. Little is known about how this capsule is synthesized. We previously identified a β1,2-xylosyltransferase (Cxt1p) with in vitro enzymatic activity appropriate for involvement in capsule synthesis. Here, we investigate C. neoformans strains in which the corresponding gene has been deleted (cxt1Δ). Loss of CXT1 does not affect in vitro growth of the mutant cells or the general morphology of their capsules. However, NMR structural analysis of the two main capsule polysaccharides, glucuronoxylomannan (GXM) and galactoxylomannan (GalXM), showed that both were missing β1,2-xylose residues. There was an ∼30% reduction in the abundance of this residue in GXM in mutant compared with wild-type strains, and mutant GalXM was almost completely devoid of β1,2-linked xylose. The GalXM from the mutant strain was also missing a β1,3-linked xylose residue. Furthermore, deletion of CXT1 led to attenuation of cryptococcal growth in a mouse model of infection, suggesting that the affected xylose residues are important for normal host-pathogen interactions. Cxt1p is the first glycosyltransferase with a defined role in C. neoformans capsule biosynthesis, and cxt1Δ is the only strain identified to date with structural alterations of the capsule polysaccharide GalXM.
Cryptococcus neoformans is an opportunistic fungal pathogen of humans and other mammals that causes serious disease, including infections of both the lung and central nervous system (1). Neurological infections with C. neoformans typically occur in immunocompromised patients and are often debilitating or fatal. Anti-cryptococcal therapies that are currently available can resolve symptoms in many cases, but these drugs do not eradicate the organism from neural tissues (1). Recurrence is likely, and new therapies for this infection are needed.
The main virulence factor of C. neoformans is the carbohydrate capsule surrounding the cell. Strains of C. neoformans lacking this structure do not cause disease in animal models (2), suggesting that the mechanisms of capsule synthesis are potential drug targets (3). The capsule is composed of two polysaccharides, glucuronoxylomannan (GXM)2 and galactoxylomannan (GalXM), and a small amount of mannoproteins (3).
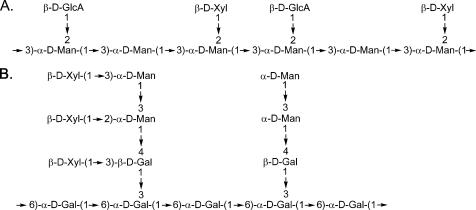
The structures of both GXM and GalXM were resolved by Cherniak and colleagues (4, 5) a decade ago. GXM is a large polysaccharide (1–7 megadaltons) (6) composed of α1,3-linked mannan with glucuronic acid and xylose side chains (4) (Fig. 1A). The mannose residues of GXM are also variably 6-O-acetylated (7). GalXM is a smaller polymer (∼100 kDa), consisting of an α1,6-galactan backbone with galactomannan side chains that are further substituted with variable numbers of xylose residues (5) (Fig. 1B).
FIGURE 1.
Repeating structures of GXM and GalXM from C. neoformans. A, the structure of serotype D GXM (4). Approximately 60% of the mannose residues are also 6-O-acetylated (7) (not shown). B, the structure of GalXM (5). Xyl, xylose; GlcA, glucuronic acid; Man, mannose; Gal, galactose.
GXM mediates diverse negative effects on the host immune response (8) and is well established as a critical virulence factor for C. neoformans. Our understanding of the importance of GalXM in virulence is not as complete, but recent studies suggest that it may play a role both in altering the immune response to C. neoformans (9) and in the virulence of this organism (10).
The synthetic pathways of the C. neoformans capsule polysaccharides, including the order in which the different sugar residues are added, have remained elusive. The structures of GXM and GalXM imply that a diverse set of glycosyltransferase enzymes will be needed to generate these polymers (3), but little is known about these fundamental catalysts of capsule synthesis. Recently, we reported the identification of a novel β1,2-xylosyltransferase (Cxt1p) from C. neoformans that transfers xylose to α1,3-linked dimannoside (11), an activity appropriate for the synthesis of both GXM and GalXM (see Fig. 1). Here, we report the generation of CXT1 deletion strains (cxt1Δ) and characterization of these strains, including evaluation of capsule polysaccharide structures and assessment of virulence in a murine model. Cxt1p plays a primary role in GalXM synthesis and is also involved in GXM production; its deletion leads to attenuation of growth in a mouse model of infection. This is the first cryptococcal glycosyltransferase shown to have a direct role in capsule biosynthesis, and the cxt1Δ mutant is the first C. neoformans strain with a demonstrated structural defect in GalXM.
EXPERIMENTAL PROCEDURES
Materials—Media components were from Difco, nourseothricin (ClonNat) was from Werner BioAgents (Germany), and oligonucleotides were from Integrated DNA Technologies. Reagents for DNA isolation and biolistic transformation were from Bio-Rad. DH5α cells, TOPO TA® cloning kits, and some restriction enzymes were from Invitrogen; other restriction enzymes were from New England Biolabs. Kits for purification of DNA fragments and PCR products were from Qiagen. Unless noted, all other reagents were from Sigma.
Strains and Cell Growth—C. neoformans wild-type strains JEC20 and JEC21 (serotype D MAT a and α, respectively) were provided by Joseph Heitman (Duke University) and the CAP67 mutant strain (MATα serotype D cap59) was provided by June Kwon-Chung (National Institutes of Health). Strains were grown in YPD medium (1% (w/v) yeast extract, 2% (w/v) peptone, 2% (w/v) dextrose) at 30 °C with continuous shaking (200 rpm) or on YPD agar plates. Growth at 30 and 37 °C was also assessed on YPD plates with and without 0.05% SDS.
Amplification and Cloning of CXT1 into TOPO pCR2.1—The sequence of the CXT1 gene (GenBank™ accession number 3254907) and its flanking regions was obtained from the sequence of chromosome 12 (accession number AE017352) within the C. neoformans serotype D genomic sequence in the NCBI data base. Genomic DNA from JEC21 was isolated as previously described (12). This was used as the template for PCR amplification of CXT1 and its flanking sequences using a sense PCR primer (CXT1S, TGGGGTCCATAAAATGTCGTT) corresponding to a region 1.3-kb upstream of the CXT1 start codon and an antisense primer (CXT1A, CCGCAGAGTAGAGAAGGAGG) corresponding to 1 kb downstream of the gene. The 5-kb PCR product was cloned into the TOPO pCR2.1 vector according to the manufacturer's directions, and the resulting plasmid (TOPO-CXT1) was maintained in the DH5α Escherichia coli strain.
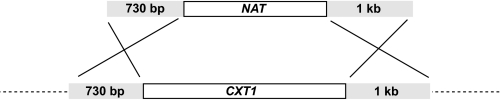
CXT1 Gene Disruption—The genomic copy of CXT1 was replaced with a nourseothricin acetyltransferase (NAT) (13) resistance gene using a replacement cassette consisting of the NAT sequence flanked by the 5′- and 3′-untranslated regions of CXT1 (Fig. 2). To generate this construct, the TOPO-CXT1 vector was digested with BsmI and SspI to remove the CXT1 open reading frame, and the ends of the cut plasmid were blunted using T4 DNA polymerase. The NAT gene was released from the GMC-200 plasmid (provided by Gary Cox, Duke University Medical Center) using HpaI and EcoRV and the resulting fragment was gel purified and bluntend cloned between the BsmI and SspI sites of TOPO-CXT1. Primers CXT1S and CXT1A (above) were then used to PCR amplify this region to generate linear DNA, which was used to biolistically transform the C. neoformans CAP67 strain as previously described (14). A separate replacement cassette was generated in the same manner, except the CXT1 gene was excised from TOPO-CXT1 with AvrII and SspI, removing only the 3′ two-thirds of the gene. This was used to replace CXT1 in a JEC21 background.
FIGURE 2.
Generation of CXT1 deletion strains. Shown are the CXT1 deletion construct (above) and corresponding genomic region (below) with lengths of homologous targeting sequences (gray) indicated.
Screening of CXT1 Gene Disruption Candidates by PCR and Confirmation by Southern Blot—Genomic DNA was extracted from drug-resistant transformants as previously described (15) and screened by PCR in the presence of 5% dimethyl sulfoxide (13) to confirm the absence of the CXT1 gene and the presence of the NAT marker in its place (supplemental Fig. 1). Genomic DNA from WT, cxt1Δ, and isogenic wild-type strains derived from JEC21 was also digested with NcoI (which cuts within the CXT1 coding sequence but not that of NAT) and blotted using standard techniques (16) with probes corresponding to CXT1 and NAT. DNA blotting confirmed gene replacement and that the marker cassette was present only once in the genome, with no additional ectopic insertions (data not shown).
Strain Backcrosses—To obtain a cxt1Δ strain in an encapsulated strain background, the CAP67 cxt1Δ strain was crossed to JEC20 on V8 agar as described in Ref. 17, and the resulting spores were plated on solid medium containing nourseothricin. An encapsulated, nourseothricin-resistant, MATa strain from this cross was then backcrossed to JEC21 eight times with spore selection on nourseothricin plates. After the ninth cross, spores were plated onto YPD plates and an isogenic MATα pair was obtained, differing only at the CXT1 locus. To obtain an isogenic pair in the CAP67 background, an acapsular, nourseothricin-resistant, MATa colony from the first cross above was crossed three times to the CAP67 strain.
Crude Membrane Preparation and Xylosyltransferase Activity Analysis—Wild-type and cxt1Δ strains were grown in 50 ml of YPD medium and crude membranes were prepared as previously described (11). Xylosyltransferase activity of the membranes was assayed based on the transfer of [14C]xylose from UDP-[14C]Xyl to a Man-α1,3-Man (Man2) acceptor. The product was separated from unconsumed UDP-Xyl and detected by scintillation counting and thin-layer chromatography, also as previously described (11).
Immunofluorescence—Wild-type and cxt1Δ strains were stained as in Ref. 18 with anti-GXM monoclonal antibodies 1255, F12D2, and 3C2 (from Tom Kozel, University of Nevada at Reno (19)) and an Alexa Fluor 546-tagged anti-mouse secondary antibody (Invitrogen). Samples were visualized on a Zeiss Axioskop2 MOT Plus microscope (Carl Zeiss) under phase-contrast and fluorescence-filtered conditions.
Purification of GXM—GXM was purified essentially as previously described (20). 500 ml of capsule induction medium (35 mm MOPS, pH 7.1, 12 mm NaHCO3, 2% (w/v) dextrose, 1.5 g/liter asparagine, 1.7 g/liter yeast nitrogen base without amino acids and ammonium sulfate) was inoculated with the appropriate strains and incubated for 5 days at 30 °C. The cultures were next autoclaved and centrifuged at 18,000 × g for 1 h at room temperature to pellet the cells. The supernatant was filtered with a 0.22-μm membrane, adjusted to 0.2 m NaCl, and 15 g of hexadecyltrimethylammonium bromide were added to the solution with stirring. After dissolution, 1 liter of 0.05% hexadecyltrimethylammonium bromide was added slowly with stirring and the solution left at room temperature overnight to allow precipitation of GXM. The resulting mixture was subjected to centrifugation (18,000 × g, 1 h, 4 °C), the supernatant fraction was discarded, and the pellet was suspended in 10% ethanol and re-centrifuged. The pellet was then dissolved in 200 ml of 2 m NaCl by stirring at room temperature overnight, and the GXM was precipitated by slowly adding ∼600 ml of cold 95% ethanol and incubating for 6 h at 4°C. The precipitate was recovered by centrifugation and the pellet was dissolved overnight in 150 ml of 2 m NaCl. The resulting solution was dialyzed (3,500 MWCO) against 4 liters of diH2O for 5 days with daily water changes, frozen, and lyophilized to dryness.
Purification of GalXM—GalXM was purified based on the methods of Vaishnav et al. (5). 200 ml of YPD medium was inoculated with either CAP67 or CAP67 cxt1Δ and incubated at 30 °C with shaking for 5 days. Cells were sedimented (9000 × g, 10 min) and the supernatant fraction was concentrated to ∼10 ml with a Pall Jumbosep spin concentrator containing a 10-kDa MWCO membrane. After addition of 30 ml of buffer CA (10 mm Tris-HCl, pH 7.2, 0.5 m NaCl, 1 mm CaCl2, 1 mm MnCl2), the sample was re-concentrated to 10 ml, mixed with 20 ml of buffer CA, and filtered through a 0.22-μm filter to remove any remaining cells or debris. The filtered sample was applied to a column containing 40 ml of concanavalin A resin (Type IV) that had been pre-equilibrated with buffer CA. The sample was re-circulated (∼20 ml/h) through the column for 18 h at 4 °C using a multichannel peristaltic pump, and the column was then eluted and washed with 200 ml of buffer CA, collecting all volume. The wash/eluate was dialyzed overnight against 4 liters of diH2O, lyophilized to dryness, re-suspended in ∼15 ml of buffer DW (10 mm Tris-HCl, pH 7.6), and concentrated to 1 ml. After four rounds of re-suspension and re-concentration to remove salt, the sample (∼15 ml) was applied to a column containing 230 ml of DE-52 anion exchange resin (Whatman) that had been equilibrated with buffer DW. The column was washed with 250 ml of the same buffer, discarding the flow-through, and eluted with a 1-liter linear gradient from buffer DW to buffer DE (10 mm Tris-HCl, pH 7.6, 1 m NaCl), collecting 10-ml fractions. 500 μl of each fraction was assayed for carbohydrate using the phenol-sulfuric acid method (21). The first peak of carbohydrate eluting from the column was pooled, lyophilized, and re-suspended in buffer GF (10 mm Tris-HCl, pH 7.2, 100 mm NaCl). The sample was filtered and applied to a 316-ml HiPrep 26/60 Sephacryl S-300 gel filtration column (GE Healthcare) that was preequilibrated with Buffer GF. The column was eluted with a 253-ml (0.8 column volume) isocratic gradient of Buffer GF, with the first 95 ml (void volume) going to waste. The remaining 158 ml were collected into 4-ml fractions and assayed for carbohydrate as above. The single carbohydrate peak was pooled, dialyzed for 5 days against diH2O, and lyophilized.
NMR Spectroscopy of GXM—GXM samples were dissolved in water (5 mg/ml) and sonicated to reduce the molecular weight. The samples were then deuterium exchanged by lyophilization from D2O (99.9% D, Aldrich) and dissolved in 0.7 ml of D2O (99.96% D, Cambridge Isotope Laboratories). Proton, gradient enhanced COSY (gCOSY), and HSQC NMR spectra were acquired on a Varian Inova-500 MHz spectrometer at 70 °C using standard Varian pulse sequences. Proton chemical shifts were measured relative to acetone (δH = 2.225, δC = 31.07 ppm), and xylose H-1 peak volumes were measured by integration of the HSQC spectra. Samples were also deacetylated by adjusting to pH 11 with concentrated NH4OH, followed by incubation at 25 °C for 24 h, dialysis, and lyophilization. The acetylated samples were again deuterium exchanged and dissolved in 0.7 ml of D2O. TOCSY, gCOSY, HSQC, and NOESY spectra were recorded at 70 °C. TOCSY and NOESY mixing times were 60 and 250 ms, respectively. 1H and 13C NMR assignments for GXM are described in supplementary data Table S1.
NMR Spectroscopy of GalXM—GalXM samples were deuterium-exchanged by lyophilization from D2O and dissolved in 0.7 ml of D2O. Proton, gCOSY, TOCSY, NOESY, and HSQC NMR spectra were acquired on Varian Inova-500 MHz and 600 MHz spectrometers at 70 °C. TOCSY and NOESY mixing times were 100 and 300 ms, respectively. 1H and 13C NMR assignments for GalXM are indicated in supplementary data Tables S2 and S3.
Glycosyl Linkage Analysis of GalXM—GalXM samples were permethylated, depolymerized, reduced, and acetylated; the resulting partially methylated alditol acetates were then analyzed by gas chromatography-mass spectrometry as described by York and colleagues (22). Briefly, an aliquot of each sample was suspended in 200 μl of dimethyl sulfoxide and stirred for 2 days before permethylation by the method of Ciucanu and Kerek (23). Following permethylation, this material was hydrolyzed with 2 m trifluoroacetic acid (2 h in a sealed tube at 121 °C), reduced with NaBD4, and acetylated using acetic anhydride/trifluoroacetic acid. The resulting partially methylated alditol acetates were analyzed on a Hewlett Packard 5890 GC interfaced to a 5970 MSD (mass selective detector, electron impact ionization mode); separation was performed on a 30M Supelco 2330 bonded phase fused silica capillary column.
In Vivo Growth in Mice—For each strain of C. neoformans tested, eight 4–6-week-old C57Bl/6 female mice were anesthetized with ketamine and intranasally inoculated with 2 to 5 × 104 cryptococcal cells in 50 μl of saline as in Refs. 24 and 25. Three mice were sacrificed at 1 h post-inoculation and the remaining 5 were sacrificed after 1 week. After sacrifice, the lungs were harvested and serial dilutions were plated on YPD agar to quantitate colony forming units.
RESULTS
Generation of CXT1 Deletion Strains—To begin defining the role in cryptococcal biology and capsule synthesis of the β1,2-xylosyltransferase we discovered (10), we deleted the corresponding gene (CXT1) in the acapsular C. neoformans strain CAP67. This strain lacks GXM and has therefore been the primary source for isolation of the less abundant capsule polysaccharide, GalXM (5). We first made a construct containing the nourseothricin drug resistance marker (NAT) flanked by the CXT1 5′- and 3′-untranslated regions (Fig. 2). After biolistic transformation, nourseothricin-resistant colonies were screened by PCR for the desired gene replacement (supplemental Fig. S1). A correct transformant was identified and genetic crossing was used to obtain cxt1Δ and CXT1 isogenic pairs in both encapsulated and acapsular strain backgrounds (see “Experimental Procedures”). Gene replacement and single site insertion was confirmed in these strains by DNA blotting (data not shown).
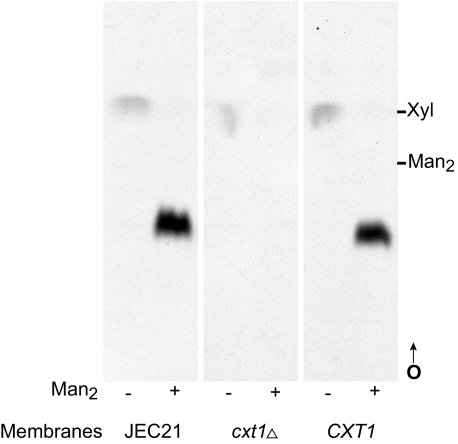
Xylosyltransferase (XT) Activity in cxt1Δ Strains—To confirm that deletion of CXT1 led to loss of XT activity, membranes from the isogenic cxt1Δ/CXT1 pairs were assayed as previously described (11). In this assay wild-type membranes produce a dominant xylosylated product, Xyl-β1,2-Man-α-1,3-Man (10), only in the presence of a Man-α1,3-Man (Man2) substrate (Fig. 3, first two tracks). This activity was dramatically reduced by deletion of CXT1 in both encapsulated and acapsular strain backgrounds (Fig. 3 and data not shown). In some experiments, we did observe minor residual XT activity in cxt1Δ membranes; this formed a product that co-migrated with the wild-type product on TLC plates (data not shown; see “Discussion”).
FIGURE 3.
Xylosyltransferase activity in encapsulated cxt1Δ and wild-type strains derived from JEC21. A standard XT assay of membranes from the indicated strains, incubated without (–) or with (+) the Man-α-1,3-Man (Man2) substrate, was performed; an autoradiogram of a thin layer chromatograph of the products is shown. Migration positions of xylose (Xyl) and Man2 standards are indicated. CXT1, isogenic wild-type strain; O, origin; arrow, direction of migration.
Growth and Capsule Morphology of cxt1Δ—We assessed growth of the encapsulated cxt1Δ strain relative to its isogenic wild-type partner and JEC21. Growth at 30 and 37 °C on YPD agar with and without SDS and in YPD medium at 30 °C was comparable for all strains (data not shown). The mutation therefore does not alter overall viability in standard culture conditions.
We also examined capsule morphology of Δcxt1 and wild-type cells. Cells grown in liquid culture were well separated and excluded India Ink from a region around the cell, both suggesting the presence of capsular material (data not shown). As another approach to detecting potentially subtle capsule alterations, we examined capsule binding with a panel of anticapsular monoclonal antibodies, including two that are thought to require capsular xylose for binding (19). In these experiments, the cxt1Δ strain stained similarly to its isogenic wild-type partner and JEC21 with all antibodies tested (data not shown). This suggested that any capsule defect in the mutant was less drastic than complete loss of xylose.
Analysis of Capsule Polysaccharides—The definitive means of establishing the role of an enzyme in capsule polysaccharide synthesis is structural analysis. We therefore purified GXM and GalXM and analyzed these polymers for defects that could not be detected by microscopy or immunofluorescence. GXM was purified from the encapsulated cxt1Δ and the wild-type pair as previously described (20). The results of the NMR analysis of the GXM samples are shown in Table 1. Notably, there was an ∼30% decrease in the amount of xylose in GXM, confirmed by one- or two-dimensional NMR in both native and deacetylated GXM. HSQC analysis of the deacetylated GXM allowed resolution of specific mannose residues. Comparing the mutant to wild-type, there is a decrease of 39% in the anomeric proton signal from the mannose substituted on O-2 with xylose (ManX-2) and a 31.5% increase in the amount of signal from O-2-unsubstituted mannose (Man-2). These changes also support the loss of roughly one-third of the β1,2-linked xylose (see Fig. 1A). Two additional minor xylose residues detected (Xyl-A-1 and Xyl-B-1) were also reduced in the mutant strain. In contrast, there was little change in the abundance of glucuronic acid or of the mannose residues to which glucuronic acid was attached.
TABLE 1.
Integration results of NMR spectra from GXM analysis
| Sample | Method | Residuea,b | CXT1 | cxt1Δ | % Difference | Average Xyl % difference |
|---|---|---|---|---|---|---|
| GXM | HSQC (two-dimensional) | GlcA-1 | 61.0 | 57.4 | -5.9 | |
| Xyl-1 | 60.7 | 42.1 | -30.6 | |||
| GlcA-4 | 83.2 | 76.8 | -7.7 | |||
| Xyl-4 | 147.7 | 98.7 | -33.2 | |||
| GlcA-5 | 101.2 | 104.3 | 3.1 | |||
| GlcA-3 + Xyl-3 | 233.8 | 175.6 | -24.9 | |||
| GlcA-2 + Xyl-2 | 306.2 | 247.6 | -19.2 | |||
| Xyl-5a | 117.2 | 71.2 | -39.2 | |||
| Man-1 | 100.0 | 100.0 | 0 | |||
| -29.4 | ||||||
| Deacetylated GXM | One-dimensional proton | Man | 100.0 | 100.0 | 0 | |
| Xyl | 40.4 | 28.3 | -29.9 | |||
| -29.9 | ||||||
| HSQC (two-dimensional) | Man-1 | 100.0 | 100.0 | 0 | ||
| ManX-2 | 32.5 | 19.8 | -39.1 | |||
| ManG-2 | 32.9 | 32.5 | -1.3 | |||
| Man-2 | 34.9 | 50.9 | 31.5 | |||
| Xyl-1 | 34.7 | 26.6 | -23.5 | |||
| Xyl-5a | 35.9 | 25.3 | -29.6 | |||
| Xyl-5e | 26.0 | 14.6 | -43.8 | |||
| Xyl-A-1 | 7.6 | 4.2 | -44.7 | |||
| Xyl-B-1 | 9.3 | 7.9 | -14.9 | |||
| GlcA-5 | 52.8 | 52.2 | -1.1 | |||
| GlcA-1 | 39.8 | 43.0 | 7.4 | |||
| -33.5 |
GlcA = glucuronic acid; Xyl = xylose; Man = mannose.
Numbers represent the carbon number with which the proton is associated; a = axial, e = equatorial.
We purified GalXM from culture supernatants of the acapsular cxt1Δ and isogenic wild-type strains as described under “Experimental Procedures.” Table 2 shows the integration of peaks corresponding to residues of GalXM in two-dimensional (HSQC) NMR analysis. The structure of GalXM that was derived from NMR analysis of the wild-type strain was identical to that previously published (5) (Fig. 1B) (data not shown). Analysis of cxt1Δ GalXM was complicated by overlap of low intensity signals, as indicated in the footnotes of Table 2. Despite this limitation, we observed almost complete loss of both β-xylose residues that are linked to mannose (Fig. 1B) in the cxt1Δ strain, whereas the amount of β-xylose linked to galactose (β-Xylp-(1→3G)) was unchanged. Signals corresponding to a minor amount of mannose-linked xylose was detected (Table 2), but due to the low abundance of this residue, we could not conclusively determine whether the residual xylose was linked β1,2 or β1,3.
TABLE 2.
HSQC integration results of GaIXM
|
Residue
|
CXT1
|
cxt1Δ
|
||
|---|---|---|---|---|
| Volume | Normalized volumea | Volume | Normalized volumea | |
| 4-β-Galp | 0.37 | 0.51 | 0.52 | 0.63 |
| 3,4-β-Galp | 0.36 | 0.49 | 0.30b | 0.37b |
| α-Manp | 0.2 | 0.27 | 0.71 | 0.87 |
| 3-α-Manp-(1→3M) | 0.33 | 0.45 | NDc | |
| 3-α-Manp-(1→4G) | ND | 0.59 | 0.72 | |
| 2, 3-α-Manp | 0.55 | 0.75 | 0.25d | 0.3d |
| β-Xylp-(1→3G) | 0.25 | 0.34 | 0.30b | 0.37b |
| β-Xylp-(1→3M) | 0.54 | 0.74 | n.d./0.09c,e | -/0.11e |
| β-Xylp-(1→2M) | 0.76 | 1.04 | 0.09/NDc,e | 0.11/-e |
Based on 4-β-Galp + 3,4 β-Galp = 1.
3,4-β-Galp and β-Xylp-(1→3G) overlapped in cxt1Δ. Because these should be equimolar (Fig. 1B), the individual volumes were obtained by dividing the sum by 2.
ND, not detected.
The 2,3-α-Manp peak overlaps with two other peaks in the proton dimension, thus the volume for this residue is an estimated value.
One of these residues was not detected, whereas the other was drastically reduced. However, overlap of the low intensity signals from these residues makes it impossible to resolve which is missing (see text).
To confirm the dramatic loss of xylose from GalXM, and to address the source of the residual xylose we detected, we performed linkage analysis of GalXM. The results of this study (Table 3 and supplemental Figs. S2 and S3) confirm a sharp decrease in both residues, with loss of the β-Xylp-(1→2M) residue causing almost complete loss of 2,3-mannose and loss of the β-Xylp-(1→3M) residue yielding a more than 3-fold increase in terminal mannose (t-Man) (Table 3 and Fig. 1B). Notably, the loss of these two distal xylose residues leads to opposite effects on 3-linked mannose (3-Man): 3-Man increases with loss of the 2-linked xylose and decreases with the loss of the 3-linked xylose (Fig. 1B). For this reason we were still unable to definitively resolve the source of the residual mannose-linked xylose in the mutant GalXM (see “Discussion”).
TABLE 3.
Linkage analysis of GalXM
|
Residuea,b
|
CXT1
|
cxt1Δ
|
||
|---|---|---|---|---|
| Area | Mol% | Area | Mol% | |
| Terminal xylopyranosyl (t-Xyl) | 17,173,422 | 12.4 | NDc | |
| 2,3-Linked mannopyranosyl (2,3-Man) | 21,744,638 | 15.7 | 322,020 | 0.2 |
| 3-Linked mannopyranosyl (3-Man) | 18,164,419 | 13.1 | 44,184,845 | 20.8 |
| Terminal mannopyranosyl (t-Man) | 7,206,471 | 5.2 | 39,962,848 | 18.8 |
| 6-Linked galactopyranosyl (6-Gal) | 19,557,342 | 14.1 | 39,642,211 | 18.6 |
| 3,6-Linked galactopyranosyl (3,6-Gal) | 21,175,731 | 15.3 | 36,949,899 | 17.4 |
| 4-Linked galactopyranosyl (4-Gal) | 17,107,085 | 12.3 | 26,362,816 | 12.4 |
| 3,4-Linked galactopyranosyl (3,4-Gal) | 9,145,202 | 6.6 | 13,039,498 | 6.1 |
| Terminal gluco- or galactofuranosyl (t-Glcf/Galf) | 4,925,987 | 3.6 | 5,903,251 | 2.8 |
| Terminal galactopyranosyl (t-Gal) | 1,276,231 | 0.9 | 2,147,622 | 1.0 |
| 3-Linked galactopyranosyl (3-Gal) | 1,278,349 | 0.9 | 4,229,861 | 2.0 |
| 138,754,877 | 100 | 212,422,851 | 100 | |
GalXM residues in bold differ most drastically between the WT and mutant strains.
Residues that were <1.5 mol % (n = 9) in both strains and were inconsistent with the structure of GalXM were omitted from this table and calculations.
ND, not detected; see text.
No terminal xylose was detected in the mutant by linkage analysis (Table 3), even though 3,4-linked galactose was still present (corresponding to the presence of a terminal sugar linked β1,3 to galactose; see Fig. 1B). Furthermore, terminal xylose was detected in the mutant by NMR, albeit drastically reduced compared with wild-type. The loss of xylose in the linkage analysis may be due to evaporation of permethylated xylose, which has a low boiling point. Low recovery of terminal xylose was also observed in the wild-type sample, where we detected only 12.4 mol % of terminal xylose (t-Xyl) despite detecting 35.4 mol % of the residues (2,3-Man, 3-Man, and 3,4-Gal) to which terminal xylose is linked (Table 3).
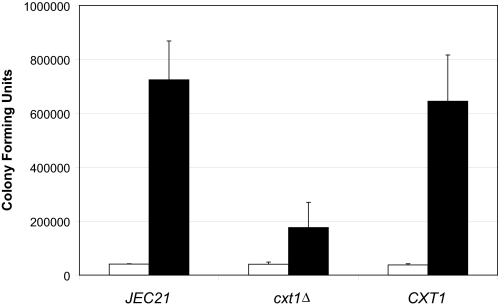
Pulmonary Growth of Encapsulated cxt1Δ Strains in Mice—To assess the impact of CXT1 on virulence, the encapsulated cxt1Δ, isogenic wild-type, and JEC21 strains were each inoculated intranasally into eight C57Bl/6 female mice as previously described (24, 25). As shown in Fig. 4, the number of colony forming units was similar at 1 h after inoculation for all three strains (p > 0.5). However, by 1 week post-inoculation, the number of colony forming units in the lungs of mice infected with the cxt1Δ strain was significantly reduced compared with either of the wild-type strains (p = 0.013 for the isogenic partner and p = 0.002 for JEC21). This shows that loss of CXT1 leads to attenuation of virulence.3 A similar growth defect in mice was also observed in an encapsulated serotype D strain of C. neoformans, which did not express XT activity due to truncation of CXT1 after nucleotide 917 (11) (data not shown).
FIGURE 4.
Growth of cryptococcal strains in murine lungs is attenuated in the absence of CXT1. Growth of each strain indicated was assessed by comparing colony forming units in the lungs of mice at 1 h (open bars) and 7 days (black bars) post-inoculation. The mean ± S.E. are plotted. All strains tested were derived from JEC21.
DISCUSSION
The pathways involved in synthesis of C. neoformans capsule polysaccharides have proven difficult to elucidate, and to date no glycosyltransferases have been clearly implicated in this process (26). We previously discovered a cryptococcal xylosyltransferase (Cxt1p) that transfers xylose to α1,3-dimannoside in a β1,2-linkage (11). We have now demonstrated that this transferase participates in synthesis of both GXM and GalXM.
We performed most of our studies using cells bearing a complete deletion of CXT1. Interestingly, we still detected a small amount (<5% of wild type; not shown) of the normal Cxt1p product in assays performed with membranes from these mutants. We speculate that this product is formed by a second XT enzyme whose similar activity is only detectable in the absence of the more robust CXT1-encoded enzyme.
Encapsulated strains lacking Cxt1p had normal capsule morphology at the level of light microscopy, and no apparent defect in binding of anticapsular monoclonal antibodies. This is not surprising, because these antibodies were generated against GXM (27–29), and the only defect in the structure of that polysaccharide in cxt1Δ cells is an approximate 30% reduction in β1,2-linked xylose. The nature of the enzyme or enzymes responsible for incorporation of the remaining GXM xylose remains an outstanding question of capsule synthesis.
In contrast to the limited effects of Cxt1p loss on GXM structure, GalXM from the cxt1Δ mutant strain showed an almost complete lack of the two xylose residues linked to mannose (Fig. 1B). The loss of the xylose-linked β1,2 to the reducing mannose of an α1,3-linked dimannose segment is precisely consistent with the in vitro activity of Cxt1p (11). There are two possible explanations for the loss of the more distal β1,3-linked xylose (Fig. 1B): either Cxt1p also has a β1,3-XT activity (which we have not observed in vitro) or, more likely, a separate β1,3-XT enzyme requires the presence of the β1,2-linked xylose before utilizing GalXM as an acceptor. The latter interpretation would also indicate the order of addition of those two xylose residues.
Our analyses showed that a minor amount of mannose-linked xylose remains in GalXM purified from cxt1Δ cells. For technical reasons discussed above, we could not verify whether this xylose is linked β1,2 or β1,3. If this linkage is β1,3, then it is possible that the β1,3-XT can transfer a small amount of xylose even in the complete absence of the β1,2 residue. If this linkage to mannose is β1,2, one intriguing possibility is that its attachment is mediated by the second, less robust XT we detected in our analysis of cxt1Δ enzyme activity. Future studies of the second enzyme will explore this possibility.
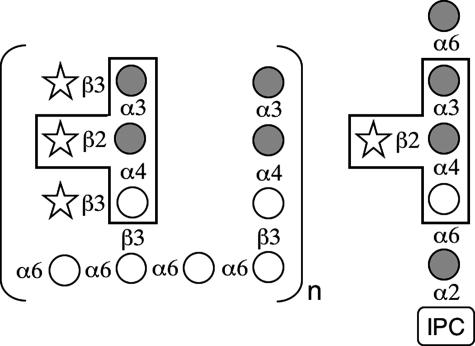
In an interesting biological parallel, cryptococcal glycolipids have structural overlap with capsule polysaccharides. C. neoformans produces glycosylinositol phosphorylceramides; the most abundant of these contains a structural motif (Manα1,3(Xylβ1,2)Manα1,4Gal) that resembles a segment of GalXM (30, 31) (see Fig. 5). Structural analysis of this glycosylinositol phosphorylceramide in the encapsulated cxt1Δ strain revealed complete loss of the β1,2-linked xylose (32) indicating that Cxt1p plays a critical role in β1,2-linked xylose addition to these compounds. Our data thus indicate that Cxt1p is involved in three important pathways: synthesis of GalXM, GXM, and glycosylinositol phosphorylceramides.
FIGURE 5.
Cryptococcal GalXM (left) and glycosylinositol phosphorylceramide (right) share a tetrasaccharide structural motif (boxed). Galactose, open circles; mannose, gray circles; xylose, open stars; IPC, inositol phosphoceramide.
The cxt1Δ mutant is the first strain shown to have a specific defect in GalXM. This altered GalXM may be an important tool for studying the biological role of this polymer, especially in determining the importance of its xylose residues. In addition, the cxt1Δ strain has a partial defect in GXM xylosylation. GXM of other serotypes of C. neoformans is more heavily xylosylated than GXM of the serotype D strains studied here. It will be of further interest to assess the impact of CXT1 deletion on those strains.
Although cxt1Δ cells grow normally in vitro, this strain has an attenuated ability to grow in the lungs of mice. This suggests that the xylose residues missing in the mutant are important for normal host-pathogen interactions. It has been previously shown that xylose is important for virulence, as a strain lacking UDP-xylose does not cause disease in mice, although it grows normally in vitro (33). That strain was mutated in UDP-glucuronic acid decarboxylase, an enzyme that is also present in mammals. The decarboxylase mutant is globally devoid of xylose, which would drastically alter capsule polysaccharides and glycolipids, and may also affect other glycoconjugates. In contrast, the cxt1Δ strain is altered in a fungal-specific enzyme (11) and has a much more restricted xylosylation defect, but still shows attenuated virulence. Together, these observations suggest that fungal-specific glycosyltransferases, such as Cxt1p, may be targets for selective and effective chemotherapeutic agents.
With these studies, we demonstrate for the first time a glycosyltransferase with a direct role in capsule production. This discovery will enable us to address important questions such as the localization of this protein (and others with which it interacts), studies we are now pursuing. Furthermore, Cxt1p defines a new fungal-specific family of large glycosyltransferases (GT Family 90) (11). Other members of this group may also be involved in capsule synthesis, and future studies aimed at this protein family should increase our understanding of the biosynthesis of this fascinating structure.
Supplementary Material
Acknowledgments
We thank Morgann Reilly and Aki Yoneda for helpful discussions and comments on the manuscript, Indrani Bose for productive experimental suggestions, and Joe Heitman, June Kwon-Chung, Tom Kozel, and Gary Cox for strains and reagents. We also thank Hong Liu, Cara Griffith, and Matthew Williams for technical assistance and Parastoo Azadi and Christian Heiss at the Complex Carbohydrate Research Center (CCRC) in Athens, GA, for their efforts and advice on the structural analysis of GXM and GalXM.
This work was supported, in whole or in part, by National Institutes of Health Grants GM R01 071007 (to T. L. D.) and GM F32 072341 (to J. S. K.). This work was also supported by Department of Energy Center for Plant and Microbial Complex Carbohydrates Grant DE-FG09-93ER-20097 and a William Keck Foundation Postdoctoral Fellowship (to J. S. K.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S3.
Footnotes
The abbreviations used are: GXM, glucuronoxylomannan; GalXM, galactoxylomannan; NAT, nourseothricin acetyltransferase; MOPS, 4-morpholinepropanesulfonic acid; HSQC, heteronuclear single quantum coherence; XT, xylosyltransferase.
As part of a large-scale gene deletion project (10), mutants in CXT1 (termed CAP3 in that report) were tested in a different mouse model and showed no phenotypic change; this may be due to the different strain backgrounds, larger inoculum, or intravenous route of infection employed in that study.
References
- 1.Chayakulkeeree, M., and Perfect, J. R. (2006) Infect. Dis. Clin. N. Am. 20 507–544, v–vi [DOI] [PubMed] [Google Scholar]
- 2.Kwon-Chung, K. J., and Rhodes, J. C. (1986) Infect. Immun. 51 218–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bose, I., Reese, A. J., Ory, J. J., Janbon, G., and Doering, T. L. (2003) Eukaryot. Cell 2 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cherniak, R., Valafar, H., Morris, L. C., and Valafar, F. (1998) Clin. Diagn. Lab. Immunol. 5 146–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaishnav, V. V., Bacon, B. E., O'Neill, M., and Cherniak, R. (1998) Carbohydr. Res. 306 315–330 [DOI] [PubMed] [Google Scholar]
- 6.McFadden, D. C., De Jesus, M., and Casadevall, A. (2006) J. Biol. Chem. 281 1868–1875 [DOI] [PubMed] [Google Scholar]
- 7.Janbon, G., Himmelreich, U., Moyrand, F., Improvisi, L., and Dromer, F. (2001) Mol. Microbiol. 42 453–467 [DOI] [PubMed] [Google Scholar]
- 8.Vecchiarelli, A. (2005) Curr. Mol. Med. 5 413–420 [DOI] [PubMed] [Google Scholar]
- 9.Pericolini, E., Cenci, E., Monari, C., De Jesus, M., Bistoni, F., Casadevall, A., and Vecchiarelli, A. (2006) Cell. Microbiol. 8 267–275 [DOI] [PubMed] [Google Scholar]
- 10.Moyrand, F., Fontaine, T., and Janbon, G. (2007) Mol. Microbiol. 64 771–781 [DOI] [PubMed] [Google Scholar]
- 11.Klutts, J. S., Levery, S. B., and Doering, T. L. (2007) J. Biol. Chem. 282 17890–17899 [DOI] [PubMed] [Google Scholar]
- 12.Nelson, R. T., Hua, J., Pryor, B., and Lodge, J. K. (2001) Genetics 157 935–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDade, H. C., and Cox, G. M. (2001) Med. Mycol. 39 151–154 [DOI] [PubMed] [Google Scholar]
- 14.Toffaletti, D. L., Rude, T. H., Johnston, S. A., Durack, D. T., and Perfect, J. R. (1993) J. Bacteriol. 175 1405–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goins, C. L., Gerik, K. J., and Lodge, J. K. (2006) Fungal Genet. Biol. 43 531–544 [DOI] [PubMed] [Google Scholar]
- 16.Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A., Struhl, K., Albright, L. M., Coen, D. M., and Varki, A. (2004) in Current Protocols in Molecular Biology (Chanda, V. B., ed) pp. 2.9.1–2.9.15, John Wiley & Sons, New York
- 17.Yan, Z., Li, X., and Xu, J. (2002) J. Clin. Microbiol. 40 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierini, L. M., and Doering, T. L. (2001) Mol. Microbiol. 41 105–115 [DOI] [PubMed] [Google Scholar]
- 19.Kozel, T. R., Levitz, S. M., Dromer, F., Gates, M. A., Thorkildson, P., and Janbon, G. (2003) Infect. Immun. 71 2868–2875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheng, S., and Cherniak, R. (1997) Carbohydr. Res. 301 33–40 [DOI] [PubMed] [Google Scholar]
- 21.Ashwell, G. (1957) Methods Enzymol. 3 73–105 [Google Scholar]
- 22.York, W. S., Darvill, A. G., McNeill, M., Stevenson, T. T., and Albersheim, P. (1986) Methods Enzymol. 118 3–40 [Google Scholar]
- 23.Ciucanu, I., and Kerek, F. (1984) Carbohydr. Res. 131 209–217 [Google Scholar]
- 24.Griffith, C. L., Klutts, J. S., Zhang, L., Levery, S. B., and Doering, T. L. (2004) J. Biol. Chem. 279 51669–51676 [DOI] [PubMed] [Google Scholar]
- 25.Sommer, U., Liu, H., and Doering, T. L. (2003) J. Biol. Chem. 278 47724–47730 [DOI] [PubMed] [Google Scholar]
- 26.Klutts, J. S., Yoneda, A., Reilly, M. C., Bose, I., and Doering, T. L. (2006) FEMS Yeast Res. 6 499–512 [DOI] [PubMed] [Google Scholar]
- 27.Brandt, S., Thorkildson, P., and Kozel, T. R. (2003) Clin. Diagn. Lab. Immunol. 10 903–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckert, T. F., and Kozel, T. R. (1987) Infect. Immun. 55 1895–1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spiropulu, C., Eppard, R. A., Otteson, E., and Kozel, T. R. (1989) Infect. Immun. 57 3240–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez, A. L., Farage, L., Melo, M. N., Mohana-Borges, R. S., Guerardel, Y., Coddeville, B., Wieruszeski, J. M., Mendonca-Previato, L., and Previato, J. O. (2007) Glycobiology 17 1–11C [DOI] [PubMed] [Google Scholar]
- 31.Heise, N., Gutierrez, A. L., Mattos, K. A., Jones, C., Wait, R., Previato, J. O., and Mendonca-Previato, L. (2002) Glycobiology 12 409–420 [DOI] [PubMed] [Google Scholar]
- 32.Owuor, E., Castle, S. A., Thompson, S. H., Garnsey, M. R., Klutts, J. S., Doering, T. L., and Levery, S. B. (2007) Glycobiology 17 1183 [Google Scholar]
- 33.Moyrand, F., Klaproth, B., Himmelreich, U., Dromer, F., and Janbon, G. (2002) Mol. Microbiol. 45 837–849 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.