Abstract
Arylalkylamine N-acetyltransferase (AANAT) catalyzes the N-acetylation of serotonin, the penultimate step in the synthesis of melatonin. Pineal AANAT activity increases at night in all vertebrates, resulting in increased melatonin production. This increases circulating levels of melatonin, thereby providing a hormonal signal of darkness. Kinetic and structural analysis of AANAT has determined that one element is floppy. This element, termed Loop 1, is one of three loops that comprise the arylalkylamine binding pocket. During the course of chordate evolution, Loop 1 acquired the tripeptide CPL, and the enzyme became highly active. Here we focused on the functional importance of the CPL tripeptide and found that activity was markedly reduced when it was absent. Moreover, increasing the local flexibility of this tripeptide region by P64G and P64A mutations had the counterintuitive effect of reducing activity and reducing the overall movement of Loop 1, as estimated from Langevin dynamics simulations. Binding studies indicate that these mutations increased the off-rate constant of a model substrate without altering the dissociation constant. The structural kink and local rigidity imposed by Pro-64 may enhance activity by favoring configurations of Loop 1 that facilitate catalysis and do not become immobilized by intramolecular interactions.
AANAT3 is a member of the GCN5-related N-acetyltransferase (GNAT) superfamily of acetyltransferases (1–3). It is a 23-kDa cytoplasmic globular protein expressed at high levels in the pineal gland (4) and at variable levels in the retina (5). AANAT has a dual regulatory/synthetic role in melatonin synthesis as the penultimate enzyme in the melatonin pathway (6–8). AANAT activity exhibits a large daily rhythm that is precisely regulated by a complex system. The rhythm is characterized by high values occurring at night and low values during the day. These changes are translated immediately into parallel changes in the production and release of melatonin and in circulating levels of the compound (9–11). The nocturnal increase in melatonin is important to biological timing in vertebrates because it provides a reliable hormonal indication of the duration of the night (6). Hence, AANAT has been referred to as the “Timezyme” (12).
AANAT shares a common Ac-CoA binding fold with members of the GCN5-related N-acetyltransferase (GNAT) acetyltransferase family (13, 14). It also has a unique arylalkylamine binding pocket formed by three protein loops. Acetyl transfer is thought to be initiated after the binding of Ac-CoA followed by binding of the arylalkylamine substrate (15); positioning effects promote the formation of an unstable Ac-CoA:arylalkylamine quaternary complex that then resolves into CoA and the acetylated arylalkylamine.
One of the three loops in the AANAT binding pocket, termed Loop 1, is of special interest because it appears to be floppy, based on different structures (PDB codes 1CJW and 1B6B) obtained with and without a bisubstrate inhibitor of the enzyme, CoA-S-N-acetyltryptamine (CoA-T). CoA-T appears to bind simultaneously to the arylalkylamine and CoA binding pockets, mimicking the transient quaternary structure thought to form during acetyl transfer (15–17). Loop 1 appears to undergo significant changes in organization during substrate binding (14); it is ∼40 residues long and includes flanking helices that are able to partially coil and uncoil. In one configuration Loop 1 occupies the Ac-CoA binding pocket and prevents catalysis. In a second configuration it relocates so as to complete the arylalkylamine binding pocket and to directly contact arylalkylamine substrates and stabilize them in the active center (13, 14). As such, Loop 1 can be seen to convert the arylalkylamine binding pocket from a low affinity open state to a high affinity closed state.
Analysis of the vertebrate AANAT sequence (18) reveals that it has undergone distinct evolutionary changes, including the addition of the tripeptide CPL63–65, which is absent from Loop 1 of the AANAT homologs of lower organisms including the yeast AANAT. Acquisition of this tripeptide is accompanied by a >1000-fold increase in the catalytic rate of the enzyme (19).
Given the relative rigidity of proline residues, Pro-64 might be expected to decrease the motion of Loop 1 and possibly to decrease the catalytic rate of AANAT as a consequence. Here, we studied CPL63–65 and Pro-64 using site-directed mutagenesis and Langevin dynamics simulations. To study the binding characteristics of AANATs, we developed a fluorescent analog of CoA-T, CoA-S-N-acetyl-7-hydroxynaphthylethylamine (CoA-HNE). The results of the studies presented here provide insight into the mechanism through which Pro-64 promotes catalysis.
EXPERIMENTAL PROCEDURES
Preparation of Expression Plasmids—Constructs for expressing mutant proteins were prepared from ovine wild-type AANAT-(2–207) with oligonucleotides containing the desired mutation (QuikChange, Stratagene, La Jolla, CA). The following mutations were made: P64A, P64G, P64W, I57A/V59A, and dCPL (deletion of amino acids CPL). The yeast wild-type construct scAANAT has been described previously (17). All plasmids were sequenced to confirm their identity (Veritas, Rockville, MD). The proteins were expressed fused to GST using the vector pGEX-4T-1 (Amersham Biosciences).
Protein Expression and Purification—Expression vectors were transformed into Escherichia coli strain BL21Gold (DE3) pLysS (Novagen, Madison, WI). The cells were grown at 37 °C; when the A600 = 0.6, the cultures were cooled to 25 °C, and isopropyl β-d-1-thiogalactopyranoside was added (final concentration = 0.2 mm). Cells were harvested after 12 h (5000 × g, 30 min, 4 °C) and resuspended in 2× phosphate-buffered saline, pH 7.5, containing 10 mm dithiothreitol (Buffer A) and a preparation of protease inhibitors (Complete, Roche Applied Science). Cells were then lysed by sonication, and the resulting lysate was centrifuged (8500 × g, 25 min, 4 °C). The supernatant was mixed with glutathione-Sepharose 4B affinity matrix (Amersham Biosciences) preequilibrated with Buffer A. The suspension was agitated for 1 h and then packed into a glass column. The protein was washed with 5 column volumes of Buffer A followed by 5 column volumes of buffer containing 50 mm Tris-Cl, 0.1 m sodium citrate, 5 mm dithiothreitol, and 10% glycerol, pH 7.8. GST fusion protein was then eluted with 5 column volumes of Buffer A containing 10 mm glutathione. Protein was concentrated and dialyzed against buffer containing 0.1 m ammonium acetate, 50 mm NaCl, and 2 mm TCEP and stored at -80 °C. The typical yield was 15 mg per 1 liter of bacterial culture. For all studies GST fusion proteins were used, consistent with previous studies (15, 16).
SDS-Polyacrylamide Electrophoresis—Proteins were resolved on preformed 14% Tris, glycine (1 mm) gels using the manufacturer's protocol (Invitrogen) (20). Rainbow standard (Amersham Biosciences) was used to determine the molecular mass of the proteins.
AANAT Enzyme Assay—AANAT activity was measured radiochemically. Each assay contained the indicated concentration of tryptamine and [3H]Ac-CoA (0.5 mm, 4mCi/mmol, Moravek Biochemicals, Brea, CA). Assays were performed in 0.1 m sodium phosphate buffer, pH 6.8, containing bovine serum albumin (2 mg/ml) and 1 mm dithiothreitol. 100-μl reactions were incubated for 30 min at 37 °C. N-[3H]Acetyltryptamine was then extracted into 1 ml of chloroform, 0.5-ml aliquots were dried, and radioactivity was determined. The Vmax and Km were calculated by non-linear fitting (SigmaPlot Version 10.0, Systat Software, Inc., Point Richmond, CA) using the equation V = Vmax S/(Km+S), where V is the observed enzyme velocity (activity), Vmax is the maximum enzyme velocity, S is the substrate concentration, and Km is the Michaelis-Menten constant.
Preparation of Bisubstrate Inhibitor CoA-HNE—CoA-HNE was prepared by mixing of 3 mg of CoA dissolved in 400 μl of 1 m Tris, pH 8.0, with 10 mg of N-[2-(7-Hydroxy-1-naphthyl) ethyl]-2-bromoacetamide (NIMH chemical synthesis program, B-804) dissolved in 400 μl of MeOH (supplemental Fig. 1). The reaction mixture was slowly mixed for 1 h at 37 °C, and the product was purified by reverse-phase chromatography using a C18 column. The reaction solution was loaded on the column using a mobile phase of 2 mm NH4Ac, pH 9.0, in water, and CoA-HNE was eluted with a gradient of 0–95% MeOH. The identity of the product was confirmed by mass spectrometry (QTrap LC/MS/MS System, Applied Biosystems, Foster City, CA) (supplemental Fig. 2).
Spectrofluorimetry—Fluorescence measurements were done on an ISS PC1 photon-counting spectrofluorimeter (ISS, Inc., Champaign, IL). Protein unfolding was analyzed by determining changes in the tryptophan fluorescence emission spectra of the proteins. To monitor unfolding as a function of guanidine denaturation, the proteins were excited at 285 nm, and emission was monitored at 339 nm. Fluorescence anisotropy was measured using CoA-HNE to characterize the binding of the probe; the excitation wavelength was 325 nm, and emission at 420 nm was measured. Kd was calculated by non-linear fitting (SigmaPlot Version 10.0) using the equation A = A0 + (Amax - A0)X/(Kd + X), where A is the observed fluorescence anisotropy, A0 is the fluorescence anisotropy of the probe without any added protein, Amax is the fluorescence anisotropy when all probes are bound, X is the concentration of free protein, and Kd is the dissociation constant. The concentration of free protein has been calculated based on the fractional saturation of the probe and the known concentrations of the probe and total protein.
Off-rates of CoA-HNE were monitored using protein preparations that were 75–85% saturated with CoA-HNE; after a control incubation period, the probe was displaced with a 100-fold excess of CoA-T, which does not generate a 420-nm emission signal when excited at 325 nm. The values of koff were calculated by non-linear fitting (SigmaPlot Version 10.0) using the equation A(t) = A(∞) + (A(0) - A(∞))exp(-koff t), where A(t) is the observed fluorescence anisotropy at time t after the initial displacement, and koff is the rate constant.
Molecular Modeling—Langevin dynamics simulations were done to gain insight into the structural and dynamic properties of uncomplexed AANAT-(2–207) in aqueous solution. The wild-type (PDB code 1B6B), P64A, and P64G mutants were studied. The simulations were performed at 25 °C and at zero ionic strength, protonation states of titratable groups were fixed at standard values compatible with physiological pH, explicit counter ions were not added, the N- and C-terminal residues missing in the crystal structure were not modeled, and the termini were capped with an acetylated and an amidated group. The pre-proline Ω dihedral angles were fixed in the trans conformation; cis-trans proline isomerization was not allowed during the dynamics. The screened Coulomb potentials continuum model was used to represent electrostatic and hydrophobic effects of the solvent (21, 22). The CHARMM program (23), Version c33b1, was used with the CMAP correction (24) applied to the all-atom (PAR22) force field (25). A cutoff of 12 Å was used for the non-bonded interactions as implemented in the screened Coulomb potentials continuum model. A collision frequency of 50/ps was applied to all atoms except hydrogen. All bond lengths were fixed with the SHAKE algorithm, and a time step of 2 fs was used with a Leapfrog Verlet integrator. The simulations were extended up to 120 ns (60 ns for P64G) preceded by an equilibration phase of 0.5 ns.
RESULTS
Mutant Selection and Molecular Dynamics—Mutations were selected in part on the evolutionary differences in Loop 1. As discussed above, the CPL63–65 tripeptide is a notable difference between the Loop 1 sequences of AANAT and AANAT homologs.
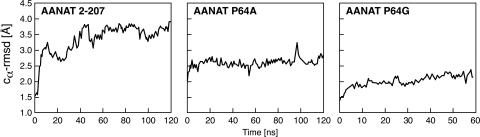
Mutations were also selected in part on the results of molecular modeling. Dynamics simulations showed that elements of secondary structure present in the crystal structure were conserved throughout the simulation. Relatively unstructured regions became ordered as the simulation proceeded. However, substantial conformational changes were observed after ∼20 ns in the sequence Thr-41—Cys-77, which contains Loop 1. The root mean square deviations of superimposed Cα atoms (Cα-rmsd) show variations in the ∼3–4-Å range (Fig. 1). Smaller changes were also observed in Loop 2 and Loop 3 (Cα-rmsd < 2.5 Å). For the wild-type, the structure changes little during equilibration, stabilizing at a root mean square deviation of ∼1.5 Å. The conformational changes develop slowly, essentially within the first ∼60 ns. For the mutants the conformational changes result only from relaxation during equilibration, with no further significant structural changes observed during the dynamics, suggesting a reduced mobility of the loop region. It was also observed during the simulation that two hydrophobic amino acids in Loop 1, Ile-57 and Val-59, interacted partially with the surface of the protein.
FIGURE 1.
The root mean square deviation of superimposed Cα atoms of residues Thr-41—Cys-77, which include Loop 1. Results of dynamics simulated for the wild-type, P64A mutant, and P64G mutant of ovine AANAT are shown. All deviations were calculated relative to the AANAT crystal structure (PDB code 1B6B). rmsd, root mean square deviation.
Based on the above considerations, the following mutations were selected as most likely to be informative regarding the role of Pro-64 and neighboring residues: P64A, P64G, dCPL, and I57A/V59A. In addition, a P64W mutation was selected, with the intention of determining whether an aromatic group at this position would substitute for the P ring.
Langevin dynamics simulation of P64A and P64G mutants suggested an important effect of Pro-64 on the arrangement of Loop 1. Fig. 1 shows the difference in flexibility of Loop 1 between the ovine wild-type and P64A and P64G mutants. The simulations predict that Loop 1 is more rigid in both mutants than in the wild type.
The molecular dynamic analysis of the P64A and P64G mutants and the wild-type enzyme combined with the available structure of the enzyme pointed to the possibility that flexibility was in part inhibited by the interactions of Ile-57 and Val-59 with the surface of the enzyme. According to the simulation and enzyme structures (PDB codes 1CJW and 1IB1), residues Ile-57 and Val-59 remain largely buried in hydrophobic moieties but must become solvent-exposed to allow ligand access to the binding pocket, as suggested by available structures of the active form. Accordingly, we tested the hypothesis that eliminating this interaction with the double-mutant I57A/V59A would increase enzyme activity by reducing the hydrophobic interaction of these residues with the protein, which would destabilize Loop 1 and facilitate exposure of the binding site.
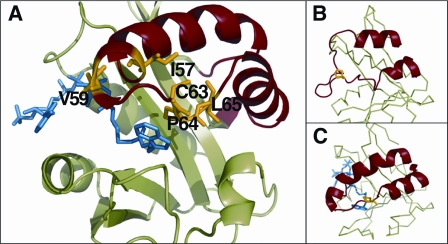
Supplemental Fig. 3 shows the sequence alignment of all proteins mentioned for this investigation. Fig. 2 highlights positions of suggested mutations in the active site of the enzyme and also depicts the two configurations of Loop 1 that were indicated by previous studies (13, 14).
FIGURE 2.
Schema of protein structure. A, position of CoA-T inhibitor (blue) in the active center of AANAT (14). The structure of Loop 1 is highlighted in red, and amino acids mutated during this study are marked in yellow and labeled with symbols. B and C show differences between the organization and position of Loop 1 without (B) and with inhibitor (C). Pictures are rotated several degrees compared with the panel A. Loop 1 is in red, and Pro-64 is highlighted in yellow.
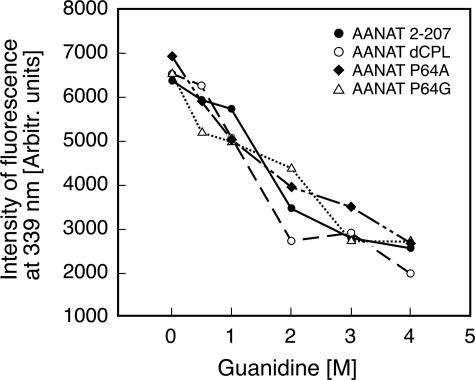
Physical Characterization of the Expressed Proteins—The purified GST fusion preparations were dominated by a single band according to SDS-PAGE analysis; all migrated at the predicted molecular weight (supplemental Fig. 4). To evaluate whether the proteins were well folded, fluorescence intensity was measured as a function of guanidine concentration. Guanidine-dependent unfolding was clearly evident in all cases within 1–2 m (Fig. 3). Based on these observations, it appears that the mutated enzymes were folded normally and that the preparations were homogenous.
FIGURE 3.
Denaturation of proteins in guanidine HCl. Fluorescence of 0.3 μm protein samples in increasing concentrations of guanidine HCl. The buffer used contained 0.1 m ammonium acetate, pH 6.8, 25 mm NaCl, and 2 mm TCEP. The exposure of tryptophans in the structure was monitored by fluorescence (excitation = 285 nm, emission = 339 nm). The similar shape of the denaturation curves indicates the similar and homogenous folding among all prepared protein samples. Curves are representative of several experiments.
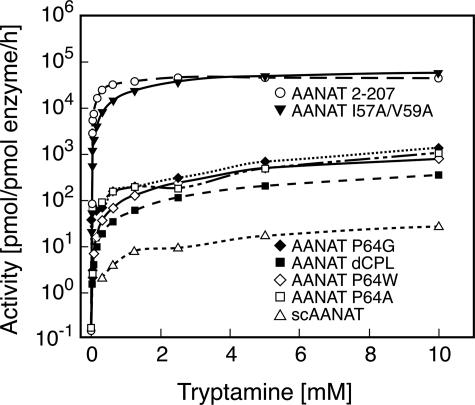
Kinetic Characterization of the Expressed Proteins—The enzyme activities of wild-type and mutant ovine forms of AANAT and of the yeast AANAT homolog were determined (Fig. 4). The I57A/V59A mutation did not alter enzyme activity. In contrast, activities of the other AANAT mutants were ∼0.01 that of the native enzyme, pointing to an important role of Pro-64 in catalysis. In the same series of studies we found that the activities of the P mutants were >10-fold higher than that of the yeast homolog.
FIGURE 4.
Enzyme activity as a function of tryptamine concentration. The enzyme activity of the indicated proteins was measured as a function of the tryptamine concentration. The enzyme assay contained 0.5 mm [3H]Ac-CoA and was performed as described under “Experimental Procedures.” Km and Vmax values appear in Table 1.
Characterization of a Fluorescent Bisubstrate Inhibitor of AANAT as a Probe to Monitor Binding—CoA-HNE was synthesized (see “Experimental Procedures”) and purified by high performance liquid chromatography. The results of liquid chromatography-tandem mass spectroscopy analysis are consistent with the identity of the compound as CoA-HNE because of the presence of a 496.4 m/z peak corresponding to the double-charged ion (supplemental Fig. 2).
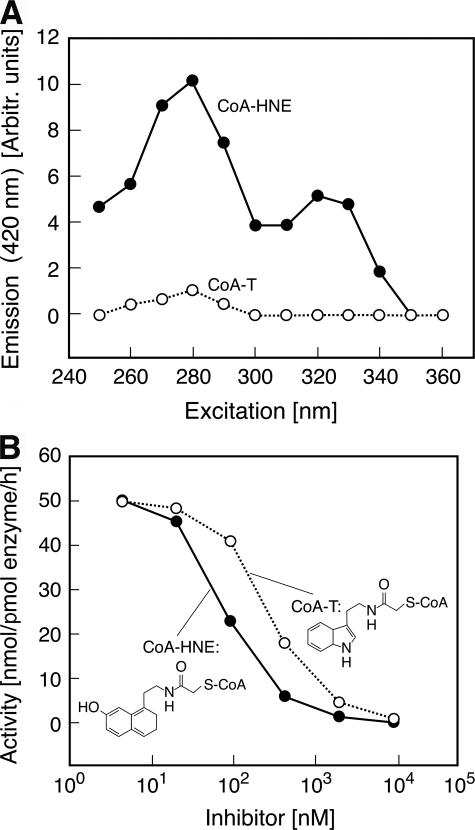
The excitation fluorescence spectrum revealed a peak absorbance at 325 nm not present in the spectra of CoA-T (Fig. 5A). Emission from this excitation peak distinguishes the signal of the probe from those of aromatic groups in the protein itself. Analysis of the inhibitory effects of CoA-HNE indicates that it inhibits AANAT activity with similar potency (IC50 = 115 nm) as does CoA-T (IC50 = 300 nm) (Fig. 5B).
FIGURE 5.
Characterization of bisubstrate inhibitors. A, the fluorescence excitation spectra of 2.8 μm CoA-T and CoA-HNE were determined (excitation wavelength from 250 to 360 nm, emission wavelength = 420 nm). The spectrum of CoA-HNE has a peak between 305 and 350 nm not seen with CoA-T (and also not seen in protein), allowing the former to be used as a fluorescence probe in binding experiments. The buffer used was 0.1 m ammonium acetate, pH 6.8, containing 25 mm NaCl and 2 mm TCEP. Emission is given as arbitrary units. B, inhibition of acetylation activity of ovine wild-type AANAT using the indicated concentrations of CoA-T and CoA-HNE. The assay contained GST-ovine AANAT-(2–207) (4 nm), 10 mm tryptamine, and 0.5 mm [3H]Ac-CoA. Activity was measured as described under “Experimental Procedures.” CoA-T and CoA-HNE had similar inhibitory potency.
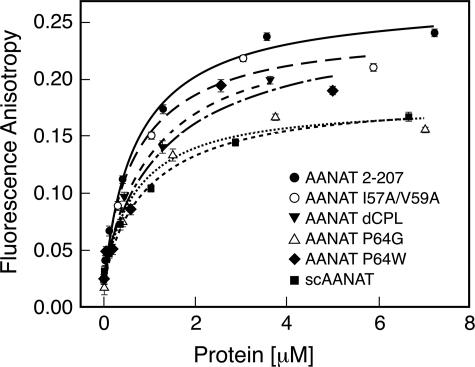
Binding Characteristics of Expressed Proteins—CoA-HNE binding to selected mutants was studied (Fig. 6). Each mutant exhibited generally similar binding properties with Kd values similar to the wild-type, providing further indication that the proteins were correctly folded. Moreover, this indicated that differences in enzyme activity were not associated with differences in Kd values and that the mutated and native enzymes appear to exhibit similar affinity for CoA-HNE. The level of the fluorescence anisotropy for maximum binding was somewhat lower for inactive protein forms. This is likely due to the different flexibility of the hydroxynaphthyl group in the bound state, caused by the absence of the stacking interaction with Pro-64 reported for the wild type (14).
FIGURE 6.
Binding of CoA-HNE to studied proteins. Fluorescence anisotropy as a function of protein concentration added to 3 μm solution of CoA-HNE is shown. The buffer used was 0.1 m ammonium acetate, pH 6.8, containing 25 mm NaCl and 2 mm TCEP. Curves characterize the binding of the probe to the indicated proteins. Derived Kd values are in Table 1.
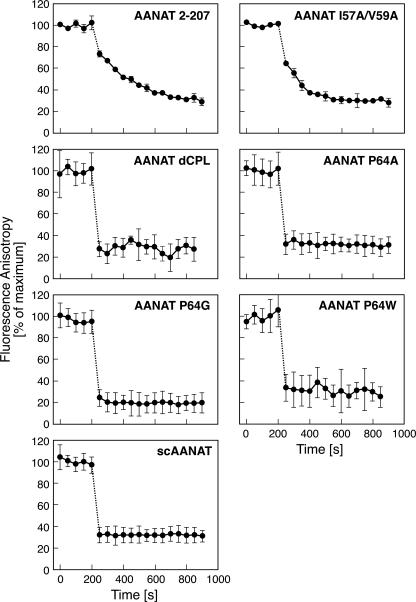
The off-rate of the probe was studied using a displacement strategy in which CoA-HNE was displaced by CoA-T (Fig. 7). The slowest off-rates were observed for ovine wild-type and I57A/V59A mutants. The off-rate of other protein forms was estimated to be at least 10-fold greater than that of wild-type AANAT. Mutant enzymes with a slow off-rate also had high enzyme activity and vice versa. Results of all studies of enzyme activity and binding of the fluorescent probe are summarized in Table 1.
FIGURE 7.
Displacement of inhibitors. Fluorescence anisotropy is shown as a function of time. Samples of proteins (2 μm) were 75–85% saturated with CoA-HNE. After 5 min the fluorescence anisotropy was measured. At t = 200 s, CoA-T was added to a final concentration of 170 μm, and fluorescence anisotropy measurement was resumed. The buffer was 0.1 m ammonium acetate, pH 6.8, containing 25 mm NaCl and 2 mm TCEP. The koff values calculated from the rate of change of fluorescence anisotropy are presented in Table 1.
TABLE 1.
Comprehensive table of results The results of enzyme activity assays measured for 1 mm tryptamine and the results of binding experiments and measurements of the off-rate for the indicated proteins are presented. The wild-type and mutated forms of AANAT are grouped according to enzyme activity.
| Description | Activity | Vmax | Km | koff | Kd | |
|---|---|---|---|---|---|---|
| nmol/pmol enzyme/h | nmol/pmol enzyme/h | mm | s–1 | μm | ||
| High enzyme activity | AANAT-(2–207) | 37 | 48 | 0.33 | 0.006 | 0.77 ± 0.12 |
| AANAT 157A/V59A | 23 | 70 | 2.3 | 0.011 | 0.74 ± 0.24 | |
| Low enzyme activity | AANAT dCPL | 0.2 | <1.0 | >4 | >0.05 | 1.15 ± 0.24 |
| AANAT P64A | 0.06 | <1.0 | >4 | >0.05 | 0.57 ± 0.09 | |
| AANAT P64G | 0.12 | <1.0 | >4 | >0.05 | 0.68 ± 0.19 | |
| AANAT P64W | 0.19 | <1.0 | >4 | >0.05 | 1.34 ± 0.64 | |
| scAANAT | 0.007 | <0.1 | >4 | >0.05 | 1.05 ± 0.15 |
DISCUSSION
The importance of flexible elements for the catalytic and selective characteristics of enzymes is well recognized (26, 27) and is evident from the changes in these elements that occur during adaptive evolution. In cases where floppy loops are movable parts of the catalytic region, optimal enzyme activity appears to reflect a delicate balance between overall flexibility and structural rigidity, resulting in restricted movement that has been well described as “focused motional freedom” (28, 29). Such restricted movement may preferentially result in a conformation that promotes catalysis (30). These characteristics reflect a specific primary amino acid sequence; in some cases proline has been found to determine structure by virtue of the conformational rigidity it imparts (31–33).
The current study revealed that in the case of AANAT, Pro-64 plays a critical role in structure and catalysis. Computer simulations showed that Loop 1 is the most flexible element of the protein. Such flexibility is related to the restricted set of conformations adopted by proline that determine the range of conformations the loop can assume. These computational results are consistent with the results of previous structural studies that first revealed changes in Loop 1 that are associated with binding of the substrate (14).
It is of interest that the results of simulation indicate that the P64A and P64G mutations would render Loop 1 less mobile. We suspect this happened because the presumed increase of local flexibility conferred by A and G permit Loop 1 to assume a greater range of configurations, some of which (arguably, the most favorable) may become involved in intramolecular interactions that stabilize the loop in conformations that do not support catalysis.
The similar Kd values of the AANATs studied indicate that these mutations do not influence the affinity of the enzyme for CoA-HNE. However, marked differences in off-rate were observed. One explanation of these differences is that the CoA-HNE·AANAT complex is more transient in P64A and P64G because Loop 1 is less likely to become positioned so as to stabilize the complex. The prediction that the I57A/V57A mutation would increase enzyme activity was not confirmed, which is consistent with the view that enzyme activity is not simply a function of general flexibility of the loop but that optimal enzyme activity requires more restricted movement.
Likewise, the lower rates of catalysis of the P64A and P64G mutants may reflect the inability of Loop 1 to stabilize the arylalkylamine substrate in the binding pocket for optimal acetyl transfer. The resulting shorter occupancy of the binding pocket, or dwell time in the pocket, reduces the likelihood of acetyl transfer. It would appear that this is an important mechanism responsible for the lower catalytic rate in the Pro-64 mutants and that Pro-64 confers the focused motional freedom required for optimal catalytic activity.
In addition to conferring a physical limitation on the conformation of Loop 1, it is also possible that Pro-64 contributes to catalysis through direct interactions with the aromatic ring of arylalkylamine substrates. This is supported by the results of structural analyses (14), which have determined that the aromatic ring of tryptamine is positioned between Pro-64 and the aromatic group of Phe-188 in the binding pocket. We tested whether an aromatic structure at this site would be active, using a P64W mutation. However, this mutant was not found to be more active than other Pro-64 mutations examined. This could be explained by the fact that the aromatic ring of Trp cannot occupy the same position as did Pro-64 because of steric constrains and differences in the physical features of proline and tryptophan. Together, these findings favor the hypothesis that the structural rigidity conferred by Pro-64 focuses movement of Loop 1.
These studies point to the evolutionary acquisition of Pro-64 as a critical step leading to the high level of activity of AANAT, which is >1000 fold higher than that in AANAT homologs. The increase in activity can be seen as being directly linked to the evolution of the melatonin signal, because this would support high levels of production. Other important changes that have occurred during the course of evolution also enhance the function of AANAT, including the acquisition of two histidines in the core of the enzyme. These appear to be essential for optimal catalysis (34) because they facilitate the ejection of protons from the core to the surface via a distinct proton wire, with the overall purpose of maintaining optimal pH in the active site. The acquisition of C- and N-terminal flanking sequences that contain cyclic AMP-dependent protein kinase phosphorylation sites nested within 14-3-3 binding motifs are essential for precise regulation of the stability and degradation of the enzyme, which govern melatonin signaling (35, 36).
These studies add to the literature which has revealed novel roles of proline, reflecting the importance of structural rigidity of this residue, especially in loops (28). The acquisition of Pro-64 can be seen to play a critical role in biological timekeeping among vertebrates by enhancing the activity of the Timezyme.
Supplementary Material
Acknowledgments
We are grateful to Dr. P. J. Steinbach for helpful discussions. This study utilized the high performance computer capabilities of the Biowulf PC/Linux cluster at the National Institutes of Health.
This work was supported, in whole or in part, by the NIH Intramural Research Program through the NICHD, National Institutes of Health and the Center for Information Technology. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
Footnotes
The abbreviations used are: AANAT, arylalkylamine N-acetyltransferase; CoA-HNE, hydroxynaphthylethylamine coenzyme A; CoA-T, tryptamine-CoA; GST, glutathione S-transferase; TCEP, tris[2-carboxyethyl] phosphine.
References
- 1.Neuwald, A. F., and Landsman, D. (1997) Trends Biochem. Sci. 22 154-155 [DOI] [PubMed] [Google Scholar]
- 2.Vetting, M. W. S., de Carvalho, L., Yu, M., Hegde, S. S., Magnet, S., Roderick, S. L., and Blanchard, J. S. (2005) Arch. Biochem. Biophys. 433 212-226 [DOI] [PubMed] [Google Scholar]
- 3.Dyda, F., Klein, D. C., and Hickman, A. B. (2000) Annu. Rev. Biophys. Biomol. Struct. 29 81-103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coon, S. L., Roseboom, P., Baler, R., Weller, J. L., Namboodiri, M. A., Koonin, E. V., and Klein, D. C. (1995) Science 270 1681-1683 [DOI] [PubMed] [Google Scholar]
- 5.Klein, D. C., Coon, S. L., Roseboom, P. H., Weller, J. L., Bernard, M., Gastel, J. A., Zatz, M., Iuvone, P. M., Rodriguez, I. R., Begay, V., Falcon, J., Cahill, G. M., Cassone, V. M., and Baler, R. (1997) Recent Prog. Horm. Res. 52 307-357 [PubMed] [Google Scholar]
- 6.Arendt, J. (1995) Melatonin and the Mammalian Pineal Gland, Chapman & Hall, London
- 7.Klein, D. C., and Weller, J. (1970) Science 169 1093-1095 [DOI] [PubMed] [Google Scholar]
- 8.Klein, D. C., and Weller, J. (1972) Science 177 532-533 [DOI] [PubMed] [Google Scholar]
- 9.Reppert, S. M. (1998) Neuron 21 1-4 [DOI] [PubMed] [Google Scholar]
- 10.Dunlap, J. C. (1999) Cell 96 271-290 [DOI] [PubMed] [Google Scholar]
- 11.Roseboom, P. H., Coon, S., Baler, R., McCune, S. K., Weller, J. L., and Klein, D. C. (1996) Endocrinology 137 3033-3045 [DOI] [PubMed] [Google Scholar]
- 12.Klein, D. C. (2007) J. Biol. Chem. 282 4233-4237 [DOI] [PubMed] [Google Scholar]
- 13.Hickman, A. B., Klein, D. C., and Dyda, F. (1999) Mol. Cell 3 23-32 [DOI] [PubMed] [Google Scholar]
- 14.Hickman, A. B., Namboodiri, M., Klein, D. C., and Dyda, F. (1999) Cell 97 361-369 [DOI] [PubMed] [Google Scholar]
- 15.De Angelis, J., Gastel, J., Klein, D. C., and Cole, P. A. (1998) J. Biol. Chem. 273 3045-3050 [DOI] [PubMed] [Google Scholar]
- 16.Khalil, E. M., and Cole, P. (1998) J. Am. Chem. Soc. 120 6195-6196 [Google Scholar]
- 17.Wolf, E., De Angelis, J., Khalil, E. M., Cole, P. A., and Burley, S. K. (2002) J. Mol. Biol. 317 215-224 [DOI] [PubMed] [Google Scholar]
- 18.Coon, S. L., and Klein, D. C. (2006) Mol. Cell. Endocrinol. 252 2-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ganguly, S., Mummaneni, P., Steinbach, P. J., Klein, D. C., and Coon, S. L. (2001) J. Biol. Chem. 276 47239-47247 [DOI] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 21.Hassan, S. A., Guarnieri, F., and Mehler, F. L. (2000) J. Phys. Chem. B 104 6478-6489 [Google Scholar]
- 22.Hassan, S. A., Mehler, E., Zhang, D., and Weinstein, H. (2003) Proteins Struct. Funct. Genet. 51 109-125 [DOI] [PubMed] [Google Scholar]
- 23.Brooks, B. R., Bruccoleri, R., Olafson, B. D., States, D. J., Swaminathan, S., and Karplus, M. (1983) J. Comput. Chem. 4 187-217 [Google Scholar]
- 24.MacKerell, A. D., Feig, M., and Brooks, C. L., III (2004) J. Am. Chem. Soc. 126 698-699 [DOI] [PubMed] [Google Scholar]
- 25.MacKerell, A. D., Bashford, D., Bellott, M., Dunbrack, R. L., Evanseck, J. D., Field, M. J., Fischer, Gao, J., Guo, H., Ha, S., Joseph-McCarthy, D., Kuchnir, L., Kuczera, K., Lau, F. T. K., Mattos, C., Michnick, S., Ngo, T., Nguyen, D. T., Prodhom, B., Reiher, W. E., Roux, B., Schlenkrich, M., Smith, J. C., Stote, R., Straub, J., Watanabe, M., Wiorkiewicz-Kuczera, J., Yin, D., and Karplus, M. (1998) J. Phys. Chem. B 102 3586-3616 [DOI] [PubMed] [Google Scholar]
- 26.Yazer, M. H., and Palcic, M. (2005) Transfus. Med. Rev. 19 210-216 [DOI] [PubMed] [Google Scholar]
- 27.Pizzitutti, F., Giansanti, A., Ballario, P., Ornaghi, P., Torreri, P., Ciccotti, G., and Filetici, P. (2006) J. Mol. Recognit. 19 1-9 [DOI] [PubMed] [Google Scholar]
- 28.Kempf, J. G., Jung, J., Ragain, C., Sampson, N. S., and Loria, J. P. (2007) J. Mol. Biol. 368 131-149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shan, L., Tong, Y., Xie, T., Wang, M., and Wang, J. (2007) Biochemistry 46 11504-11513 [DOI] [PubMed] [Google Scholar]
- 30.Henzler-Wildman, K. A., Lei, M., Thai, V., Kerns, S. J., Karplus, M., and Kern, D. (2007) Nature 450 913-916 [DOI] [PubMed] [Google Scholar]
- 31.Nam, G. H., Cha, S. S., Yun, Y. S., Oh, Y. H., Hong, B. H., Lee, H. S., and Choi, K. Y. (2003) Biochem. J. 375 297-305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramaswamy, S., Park, D., and Plapp, B. V. (1999) Biochemistry 38 13951-13959 [DOI] [PubMed] [Google Scholar]
- 33.Sun, Z., and Liu, J. (2005) Proteins 61 870-877 [DOI] [PubMed] [Google Scholar]
- 34.Scheibner, K. A., De Angelis, J., Burley, S. K., and Cole, P. A. (2002) J. Biol. Chem. 277 18118-18126 [DOI] [PubMed] [Google Scholar]
- 35.Obsil, T., Ghirlando, R., Klein, D. C., Ganguly, S., and Dyda, F. (2001) Cell 105 256-267 [DOI] [PubMed] [Google Scholar]
- 36.Klein, D. C., Ganguly, S., Coon, S. L., Weller, J. L., Obsil, T., Hickman, A. B., and Dyda, F. (2002) Biochem. Soc. Trans. 30 365-373 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.