Abstract
Objectives
Development of a user-friendly test alternative to ELISA-based assays to detect IFN-γ by in vitro cultured peripheral blood mononuclear cells (PBMC) stimulated with pathogen-derived antigens.
Design and methods
The molecular components of an operational IFN-γ ELISA-based test were applied in a lateral flow (LF) immuno-sandwich assay using up-converting phosphor (UCP) reporter particles. The analytical sensitivity of the UCP-LF IFN-γ assay (ULIGA) was determined and the assay was qualitatively validated with a selection of 60 supernatants derived from PBMC cultures stimulated with M. leprae derived antigens, mitogen or medium alone.
Results
ULIGA indicated an analytical sensitivity better than 2 pg/mL, and demonstrated four orders of magnitude dynamic range. The assay correlated well with the IFN-γ ELISA.
Conclusions
ULIGA allows detection well below the cutoff value (100 pg/mL) used to define positive responses in the IFN-γ ELISA. The test procedure is less demanding in respect to equipment and labor, and is suited for testing single samples.
Keywords: Cell mediated immunity, IFN-γ, Interferon, Lateral flow, Leprosy, mycobacteria, UCP, Up-converting phosphor, UPT
1. INTRODUCTION
The continued incidence of leprosy in endemic countries is thought to be caused by the perpetuating reservoir of Mycobacterium leprae infected contacts or persons with subclinical leprosy. However, diagnostic tools for early detection of asymptomatic M. leprae infection are not available yet. The cell-mediated immunity (CMI) plays a crucial role in the protection against mycobacterial infections. This immune response is dependent on CD4+ and CD8+ T cells producing Th1-type cytokines (1). Animals lacking functional genes encoding specific cytokines or cytokine receptors (in particular IFN-γ, IFN-γR, TNF-αR, or IL-12p40) are much more susceptible to mycobacterial infections than control littermates (2,3). Similarly, defects in CD4+ Th1 immunity, type-1-cytokine production or type-1 cytokine receptor signaling are strongly associated with progressive infection and bacterial dissemination in humans (4–7). Since IFN-γ is a stable and robust cytokine, IFN-γ secretion is often used as readout for the induction of CMI as a consequence of mycobacterial infection/exposure. It can be measured in cultures of peripheral blood mononuclear cells (PBMC) or in whole blood cultures upon incubation with pathogen specific antigens.
Serology assays have been developed that detect IgM antibodies against the M. leprae specific phenolic glycolipid I (8). These assays allow the identification of multibacilary (MB) leprosy patients; individuals with strong humoral immunity. However, the assays fail to detect most paucibacillary (PB) leprosy patients and leprosy patients' contacts since these individuals have strong cellular immunity but mostly lack humoral immunity (9). MB leprosy is also hard to detect in a preclinical stage when the patient is already an active source of infection. Thus, to complement current clinical methods, improved diagnostic tools are required that detect M. leprae infection before clinical manifestations arise and distinguish M. leprae from infection with other pathogenic and environmental mycobacteria and from routine BCG vaccination (10).
Currently two commercially IFN-γ release assays (IGRAs) are now available for diagnosis of M. tuberculosis infection (see refs in (11)). This has inspired research into the feasibility of developing similar CMI-based assays for the identification of asymptomatic (preclinical) leprosy. In order for these test to be applied in the resource poor settings where most of leprosy cases occur, the assay need to be affordable and easy to perform (low demand on equipment and trained personnel). We therefore attempted to modify an in-house used IFN-γ ELISA to a user friendly format appropriate also for the analysis of single specimens. To achieve this, a lateral flow (LF) IFN-γ immunosandwich assay with highly sensitive up-converting phosphor (UCP) reporter particles (12,13) was developed: the assay is referred to as ULIGA (UCP-LF IFN-Gamma assay). The phosphorescent reporter utilized in ULIGA is excited with infrared light (980 nm) to generate visible light, a process known as up-conversion. As up-conversion does not occur in nature, (interfering) auto-fluorescence of other assay components and materials is virtually absent; this feature makes UCP-based assays highly sensitive. The application of UCP in the detection of other antigens has been recently applied for the detection of Respiratory Syncytial Virus (14), Schistosoma species (15) and Yersinia pestis (16). These three assays all use the rapid LF format thus allowing convenient single sample testing. The LF strips exposed to UCP reporters can be analyzed with a simple and portable reader that allows a full instrument-assisted assay analysis (14) which avoids operator bias in the interpretation of assay results.
2. METHODS
IFN-γ ELISA
IFN-γ levels were determined by ELISA in 96 well MaxiSorb microtiterplates (NUNC) using a coating anti-IFN-γ mAb (mO-13-43-08) and detecting biotinylated anti-IFN-γ pAb (pB-15-43-07). The IFN-γ standard and the anti-IFN-γ antibodies were obtained from UCytech (Utrecht, The Netherlands). Presence of biotinylated antibody was detected enzymatically using streptavidin-HRP. The cutoff value to define positive cell responses was set beforehand at 100 pg/mL. The assay-sensitivity level was 20–50 pg/mL, values for unstimulated cell cultures were typically <20 pg/mL (10). As a positive control stimulus, 1% PHA (phytoheamagglutinin) and 10 µg/mL PPD (purified protein derivative of M. tuberculosis, Statens Serum Institute, Copenhagen, Denmark) were used.
ULIGA
An IFN-γ specific up-converting phosphor conjugate (reporter conjugate) was prepared by conjugating 25 µg of monoclonal anti IFN-γ antibody (mO-13-43-08, UCytech) per mg carboxylated phosphor particles as described previously (17). The reporter conjugate (1 mg/mL) was stored at 4 °C and had a shelf life of at least 6 months. Prior to use it was diluted to 2.5 µg/mL in LF assay buffer (100 mM Hepes pH 7.4, 270 mM NaCl, 1 % BSA (w/v), 0.5% (v/v) Tween-20), and sonicated for 1 min (100 W). The IFN-γ LF assay comprised three steps (Fig. 1A): 1) 60 min incubation (thermoshaker: 37 °C, 1200 rpm) of 40 µL reporter conjugate with 40 µL of sample; 2) addition of 10 µL (1 ng/µL) of polyclonal biotinylated anti IFN-γ antibody (pB-15-43-07, UCytech, Utrecht, The Netherlands), continued incubation for another 60 min; and, 3) addition of 30 µL LF assay buffer and initiation of LF by addition of the mixture to a microtiterplate well containing a LF strip. The LF strip comprised a sample pad, nitrocellulose membrane with an avidin test line and a rabbit anti-mouse flow control line (12). The assay in summary: a (culture) sample was incubated with an IFN-γ specific reporter to allow binding of IFN-γ to the reporter particle and, upon addition of a biotinylated polyclonal anti IFN-γ antibody an immuno sandwich was formed which could be captured by avidin present on the LF strip. A Packard FluoroCount™ microtiter-plate reader adapted with an infrared laser (980 nm) was used to scan the LF strips. Upon IR excitation, UCP reporter particles emitted green light that was detected with a 550 nm band pass filter. Luminescence was measured, compiled and displayed as relative fluorescence units (RFU) in a 2-D plot as function of the position on the strip.
Fig. 1.
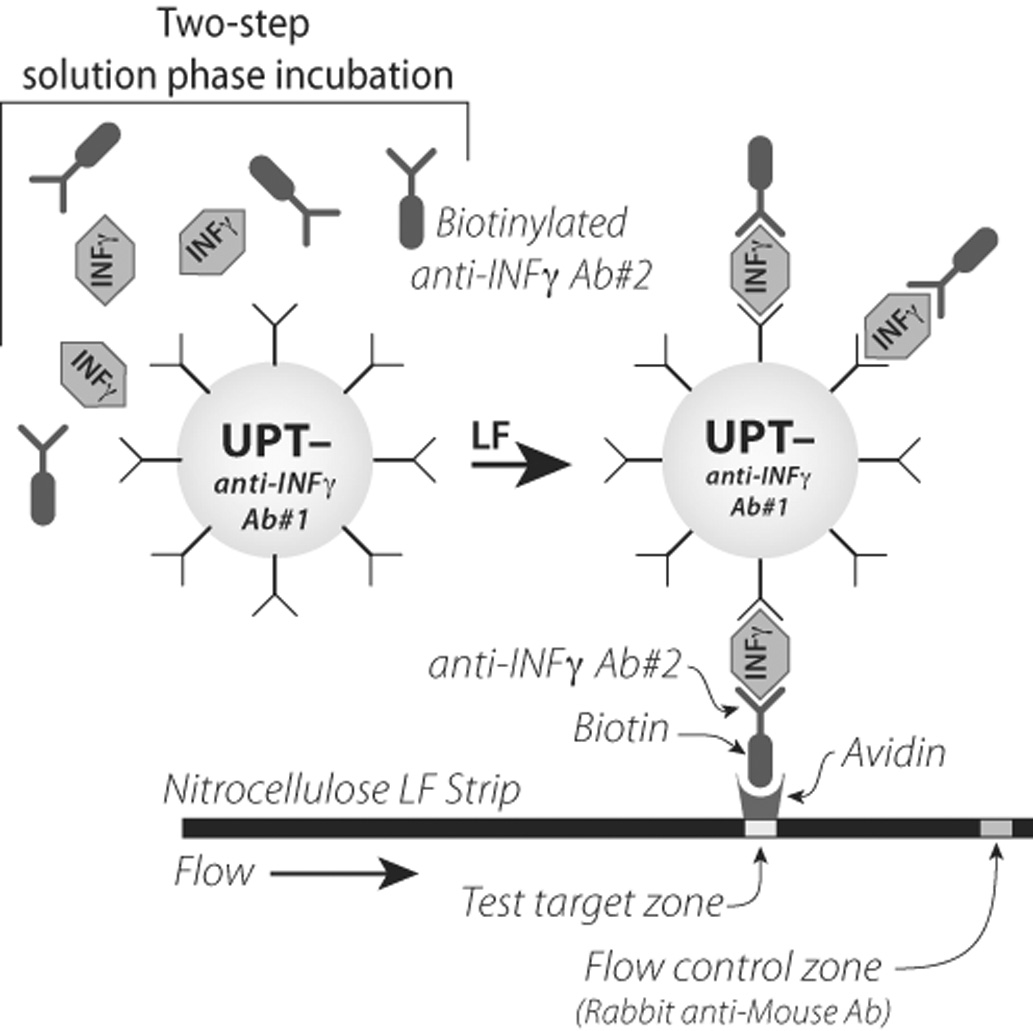
Immuno sandwich combined with lateral flow utilizing up-converting phosphor particles to detect IFN-γ (ULIGA). The solution based incubation is a two step protocol: incubation of the IFN-γ specific reporter with the test sample is followed by incubation with a biotinylated polyclonal anti IFN-γ antibody. Reporter particles containing the IFN-γ antibody sandwich are captured on the test line of a lateral flow strip. Luminescence was measured as relative fluorescence units.
IFN-γ samples
The samples used for development and validation of ULIGA were: 1) human IFN-γ standard in LF assay buffer; 2) IFN-γ in whole blood samples or in culture medium (IMDM; Invitrogen) supplemented with 10% serum (IMDM-serum); and 3) supernatant of in vitro cultures of peripheral blood mononuclear cells stimulated with M. leprae unique recombinant proteins (10). In detail, venous blood was obtained from healthy blood donors in heparinized tubes and used for direct incubation with antigens (WBA) or isolation of peripheral blood mononuclear cells (PBMC) by Ficoll density centrifugation. For WBA 1 mL undiluted blood was plated in 24-well plates. PBMC (2 × 106 cells/mL) were plated in triplicate cultures in 96-well round bottom plates in 200 µL/well of adoptive immunotherapy medium (AIM-V; Invitrogen) supplemented with 100 U/mL penicillin, 100 µg/mL streptomycin and 2 mM L-glutamine (Invitrogen). M. leprae proteins or control antigens were added at final concentrations of 10 µg/mL for both assays. After 6 days of culture at 37°C at 5% CO2, 90% relative humidity, 75 µl supernatants were removed from each well, triplicates were pooled and frozen in aliquots at −20 °C until further analysis by ELISA.
3. RESULTS AND DISCUSSION
Description of the ULIGA format
Fig. 1 depicts the novel LF-based assay for IFN-γ detection (ULIGA). The IFN-γ monoclonal antibody (antibody 1, Fig. 1) is conjugated to the UCP reporter, whereas in the IFN-γ ELISA this antibody is immobilized to a microtiter plate. This configuration allows (for both assay formats) an initial incubation of the IFN-γ containing sample with the monoclonal antibody followed by a second incubation with the polyclonal antibody. An alternative ULIGA configuration with the polyclonal antibody immobilized on the LF strip showed to be less sensitive (1–2 orders of magnitude, results not shown). The in-solution formed IFN-γ immunosandwich is then captured during LF utilizing the strong avidin-biotin binding; IFN-γ specific biotinylated polyclonal antibody (antibody 2, Fig. 1) interacts with the avidin test zone on the LF strip. UCP reporters not captured at the avidin test line will bind at the flow control line; for this purpose the flow control line is provided with an anti-mouse antibody that interacts with the monoclonal antibody (antibody 1, Fig. 1) conjugated to UCP.
Effect of cell culture medium
Dilution series of IFN-γ in PBS supplemented with 0.1% w/v BSA (PBS-BSA) indicated a sensitivity better than 4.9 pg/mL (Fig. 2A). As the anticipated sample for the IFN-γ assay consists of supernatant from cell cultures and because ULIGA applies avidin-biotin capture, the assay results may be affected by biotin present in the cell culture medium (IMDM with 10% v/v pooled human serum, IMDM-HS). Dilution series of IFN-γ were therefore also analyzed in IMDM-HS. Results indicated a sensitivity between 18.2 and 1.82 pg/mL (Fig. 2B), in agreement with the results obtained with PBS-BSA. However, the test signals (RFU) obtained with IMDM-HS samples were significantly lower than signals obtained with PBS-BSA samples. This is a consequence of free biotin (approximately 0.05 µM) in IMDM that interferes with the LF avidin-biotin capture. The decrease in signal could be reversed by a 5-fold increase of the amount of avidin on the test line (increase of the standard used 800 ng to 4 µg, results not shown). However, as elevated amounts of avidin also increase non-specific binding (18) the amount of avidin of the LF strip was kept at 800 ng for the series of experiments described here.
Fig. 2.

Detection of IFN-γ with ULIGA. A) Detection of IFN-γ diluted in LF assay buffer. B) Detection of IFN-γ diluted in IMDM with 10% pooled human serum. Experiments were performed in triplicate.
Cutoff-threshold in IMDM-HS
Assay results can be expressed in relative fluorescence units (RFU), the value of the peak area of the scan (Fig. 2). The cutoff threshold above which a sample is designated IFN-γ reactive as determined from the experiment presented in Fig. 2B is 601 RFU; calculated as the average zero signal plus 2 std (standard deviation, n=3). An example of a prototype UCP-LF assay that utilizes the test line signal in RFU to determine infection by respiratory syncytial virus (RSV) has been described recently (14). As actual values may show variation when working with several small batches of manually produced LF strips and as values depend on the type of scanner used (portable versus benchtop (12)), scan results are also expressed as a ratio value. The ratio value for ULIGA is defined as the RFU signal obtained at the test line divided by the RFU signal obtained at the flow control line; peak areas of the test line are thus normalized to the peak area of individual rabbit anti-mouse IgG antibody flow control line (17). The cutoff ratio threshold as determined from the experiment presented in Fig. 2B is 0.021; calculated as the average zero ratio plus 2 std (n=3). Note that both (RFU and ratio) threshold values need to be verified by testing significantly larger number of cell cultures of non-infected individuals. Both, RFU and ratio signal indicate an analytical sensitivity better than 2 pg/mL.
Assay validation using cell culture supernatant
A single blinded selection of 60 culture samples was tested with the ULIGA and compared to results obtained using the IFN-γ ELISA. The results are summarized in Tab. 1. Six out of the 60 cultures contained unstimulated samples. The cutoff-threshold determined (as described above) from these six samples was 625 RFU with a ratio value of 0.017. These values are in the same range as the values determined when testing an IFN-γ dilution series in IMDM-serum, respectively 601 and 0.021. Out of the 36 samples that indicated positive cytokine production in ELISA (IFN-γ ≥ 100 pg/mL), 35 samples tested positive using ULIGA applying the ratio cutoff threshold values of 0.17. The one sample (with a ratio value 0.15) that did not test positive with ULIGA indicated an IFN-γ level of 103 pg/mL, just above the 100 pg/mL ELISA cutoff value. Out of the 18 samples (not including the 6 control samples) that produced ELISA-cytokine levels below 100 pg/mL cut-off threshold, 4 samples tested positive using ULIGA. Repetitive testing with ELISA indicated IFN-γ concentrations ranging from 76 to 99 pg/mL. Note here that samples resulting in ELISA values indicating IFN-γ concentrations between 50 and 100 pg/mL are regarded as indecisive. The samples with values indicating concentrations around or below 50 pg/mL are considered non-responders as this level is also found in unstimulated cell cultures. From the remaining 14 samples one resulted in an ELISA IFN-γ concentration of 68 pg/mL whereas the others scored well below 50 pg/mL (ranging from 0–44 pg/mL; average 28 pg/mL). Results are summarized in Table 1. The few minor discrepancies between the ELISA and the ULIGA test are most likely caused by free biotin in the supernatant of the cell culture samples. Whereas variability in cell culture activity does affect the uptake of biotin from the medium, this will affect the concentration of free biotin entering ULIGA. The current assay format is therefore not yet (fully) quantitative when testing supernatant from cell cultures, but it does allow the identification of positive samples that generate responses above the currently maintained threshold of 100 pg/mL.
Tab. 1.
Qualitative analysis of 60 culture samples with ULIGA and ELISA
| ULIGA | ||||||
|---|---|---|---|---|---|---|
| ELISA | Positive | Negative | Discrepant | |||
| ≥100 | 36 | 35 | 1 | 1 | ||
| >50–100 | 5 | 4 | 1 | 1 | ||
| >0–50 | 13 | 0 | 13 | 0 | ||
| 0 | 6 | 0 | 6 | 0 | ||
| Total | 60 | 39 | 21 | 2 | ||
In conclusion
The ULIGA presented here is a general test to measure IFN-γ. In this study the cell mediated IFN-γ response to M. leprae antigens was studied. ULIGA demonstrated analytical sensitivity (cutoff threshold value) of around 2 pg/mL IFN-γ in IMDM-HS; this is 10-fold more sensitive than the sensitivity of the in-house used IFN-γ ELISA. ULIGA also demonstrated a strong correlation with respect to the IFN-γ levels as measured in the IFN-γ ELISA. As the current ULIGA format applies a LF-based avidin-biotin capture, free biotion present in the test samples may affect the quantitative outcome of ULIGA. Nevertheless, the current ULIGA format seems capable of detecting IFN-γ concentrations above the 100 pg/mL in supernatant of cell culture medium, the threshold used to define positive cell responses.
ACKNOWLEDGEMENT
OraSure Technologies Inc. is acknowledged for providing the up-converting phosphor reporter particles (400 nm UPT reporter particles), K. Szuhai (LUMC, Molecular Cell Biology) for his advice during preparation of this document, and W.R. Abrams (NYU College of Dentistry, Basic Sciences) for the artwork in Fig. 1. Part of this work was funded by the U.S. National Institute of Health (NIH) grant UO1DE017855 and the Netherlands Leprosy Relief Foundation (NLR) grant ILEP 702.02.65.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kaufmann SH. Immunity to intracellular bacteria. Annu Rev Immunol. 1993;11:129–163. doi: 10.1146/annurev.iy.11.040193.001021. [DOI] [PubMed] [Google Scholar]
- 2.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178:2249–2254. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper AM, Magram J, Ferrante J, Orme IM. Interleukin 12 (IL-12) is crucial to the development of protective immunity in mice intravenously infected with mycobacterium tuberculosis. J Exp Med. 1997;186:39–45. doi: 10.1084/jem.186.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holland SM, Eisenstein EM, Kuhns DB, et al. Treatment of refractory disseminated nontuberculous mycobacterial infection with interferon gamma. A preliminary report. N Engl J Med. 1994;330:1348–1355. doi: 10.1056/NEJM199405123301904. [DOI] [PubMed] [Google Scholar]
- 5.Newport MJ, C. M. Huxley CM, S. Huston S, et al. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 6.De Jong R, Altare F, Haagen IA, et al. Severe mycobacterial and Salmonella infections in interleukin-12 receptor-deficient patients. Science. 1998;280:1435–1438. doi: 10.1126/science.280.5368.1435. [DOI] [PubMed] [Google Scholar]
- 7.van de Vosse E, Ottenhoff TH. Human host genetic factors in mycobacterial and Salmonella infection: lessons from single gene disorders in IL-12/IL-23-dependent signaling that affect innate and adaptive immunity. Microbes Infect. 2006;8:1167–1173. doi: 10.1016/j.micinf.2005.10.032. [DOI] [PubMed] [Google Scholar]
- 8.Buhrer-Sekula SM, Cunha G, Ferreira WA, Klatser PR. The use of whole blood in a dipstick assay for detection of antibodies to Mycobacterium leprae: a field evaluation. FEMS Immunol Med Microbiol. 1998;21:197–201. doi: 10.1111/j.1574-695X.1998.tb01166.x. [DOI] [PubMed] [Google Scholar]
- 9.Oskam L, Slim E, Buhrer-Sekula S. Serology: recent developments, strengths, limitations and prospects: a state of the art overview. Lepr Rev. 2003;74:196–205. [PubMed] [Google Scholar]
- 10.Geluk A, Klein MR, Franken KL, et al. Postgenomic approach to identify novel Mycobacterium leprae antigens with potential to improve immunodiagnosis of infection. Infect Immun. 2005;73:5636–5644. doi: 10.1128/IAI.73.9.5636-5644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrara G, Losi M, D'Amico R, et al. Use in routine clinical practice of two commercial blood tests for diagnosis of infection with Mycobacterium tuberculosis: a prospective study. Lancet. 2006;367:1328–1334. doi: 10.1016/S0140-6736(06)68579-6. [DOI] [PubMed] [Google Scholar]
- 12.Corstjens PLAM, Li S, Zuiderwijk M, et al. Infrared up-converting phosphors for bioassays; IEE Proc Nanobiotechnol; 2005. pp. 64–72. [DOI] [PubMed] [Google Scholar]
- 13.Zarling DA, Rossi MJ, Peppers NA, et al. Up-converting reporters for biological and other assays using laser excitation techniques. Patent no. 5,674,698. United States Patent. 1997
- 14.Mokkapati VK, Niedbala RS, Kardos K, et al. Evaluation of UPlink-RSV: Prototype rapid antigen assay for detection of respiratory syncytial virus infection. Ann NY Acad Sci. 2007;1098:476–485. doi: 10.1196/annals.1384.021. [DOI] [PubMed] [Google Scholar]
- 15.Corstjens PLAM, van Lieshout L, Zuiderwijk M, et al. Up-converting phosphor technology based lateral flow assay (UPT-LF) for detection of Schistosoma circulating anodic antigen (CAA) in serum. J Clin Microbiol. 2007 doi: 10.1128/JCM.00877-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan ZQ, Zhou L, Zhao YK, et al. Rapid quantitative detection of Yersinia pestis by lateral-flow immunoassay and up-converting phosphor technology-based biosensor. Sensors and Actuators B-Chemical. 2006;119:656–663. doi: 10.1016/j.snb.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Corstjens PLAM, Zuiderwijk M, Brink A, et al. Use of up-converting phosphor reporters in lateral-flow assays to detect specific nucleic acid sequences: a rapid, sensitive dna test to identify human papillomavirus type 16 infection. Clin Chem. 2001;47:1885–1893. [PubMed] [Google Scholar]
- 18.Zuiderwijk M, Tanke HJ, Niedbala RS, Corstjens PLAM. An amplification-free hybridization-based DNA assay to detect Streptococcus pneumoniae utilizing the up-converting phosphor technology. Clin Biochem. 2003;36:401–403. doi: 10.1016/s0009-9120(03)00057-2. [DOI] [PubMed] [Google Scholar]


