Abstract
Caliciviruses have a positive strand RNA genome covalently-linked at the 5’-end to a small protein, VPg. This study examined the biochemical modification of VPg by the ProPol form of the polymerase of human norovirus strain MD145 (GII.4). Recombinant norovirus VPg was shown to be nucleotidylylated in the presence of Mn2+ by MD145 ProPol. Phosphodiesterase I treatment of the nucleotidylylated VPg released the incorporated UMP, which was consistent with linkage of RNA to VPg via a phosphodiester bond. Mutagenesis analysis of VPg identified Tyrosine 27 as the target amino acid for this linkage, and suggested that VPg conformation was important for the reaction. Nucleotidylylation was inefficient in the presence of Mg2+; however the addition of full- and subgenomic-length MD145 RNA transcripts led to a marked enhancement of the nucleotidylylation efficiency in the presence of this divalent cation. Furthermore, evidence was found for the presence of an RNA element near the 3’-end of the polyadenylated genome that enhanced the efficiency of nucleotidylylation in the presence of Mg2+.
Keywords: Norovirus, VPg, Polymerase, nucleotidylylation
Introduction
Noroviruses (NoV), a major cause of acute gastroenteritis (Green, Chanock, and Kapikian, 2001), are members of the family Caliciviridae. The genome is composed of an approximately 7.5 kb single-stranded positive sense RNA molecule that is covalently-linked at the 5’-end to a VPg protein, and is polyadenylated at its 3’-end. The genome is organized into three open reading frames (ORFs): ORF1, ORF2 and ORF3. ORF1 encodes the 200 kDa precursor of the nonstructural proteins, and ORF2 and ORF3 encode the major (VP1) and minor (VP2) structural proteins, respectively (Jiang et al., 1993; Lambden et al., 1993). The 200 kDa polyprotein of human norovirus strain MD145 (belonging to genogroup II.4) is cleaved by the viral proteinase (Pro) to release both precursors and fully processed viral proteins with the following gene order: NS1−2 (N-terminal protein):NS3 (NTPase):NS4 (p20):NS5 (VPg): NS6 (Pro): NS7 (RNA-dependent RNA polymerase) (Belliot et al., 2003; Sosnovtsev et al., 2006). The norovirus RNA-dependent RNA polymerase (RdRp) as well as RdRps from several calicivirus genomes have been expressed in bacteria as a mature form (designated Pol) or a ProPol RdRp precursor, purified, and characterized biochemically (Belliot et al., 2005; Fukushi et al., 2004; Ng et al., 2004; Rohayem et al., 2006a; Wei et al., 2001). Polymerase activity was demonstrated by both forms of norovirus RdRp (Belliot et al., 2005). The initiation of RNA synthesis by the calicivirus RdRp on an RNA template has been reported as primer dependent in some studies and primer independent in others (Belliot et al., 2005; Fukushi et al., 2004; Lopez Vazquez, Martin Alonso, and Parra, 2001; Rohayem et al., 2006a; Wei et al., 2001). Various mechanisms by which initiation might occur in the absence of added primer have been proposed. Moreover, a recent study has shown evidence that the uridylylated VPg protein may serve as a primer for the initiation of norovirus RNA synthesis on polyadenylated RNA templates (Rohayem et al., 2006b).
Studies of the role of the calicivirus VPg in replication and the nature of its linkage to the viral RNA have focused on feline calicivirus (FCV, genus Vesivirus) (Herbert, Brierley, and Brown, 1997; Mitra, Sosnovtsev, and Green, 2004), Norwalk virus and other human norovirus strains (genus Norovirus) (Rohayem et al., 2006b), and rabbit hemorrhagic disease virus (RHDV, genus Lagovirus) (Machin, Martin Alonso, and Parra, 2001). Early work showed that genomic RNA purified from FCV and other vesivirus virions was not infectious after digestion of the VPg by proteinase K (Herbert, Brierley, and Brown, 1997), and it was proposed that the VPg was required for translation (Daughenbaugh et al., 2003; Goodfellow et al., 2005). Recent reports provided further evidence for the involvement of the calicivirus VPg in translation. Human and murine norovirus recombinant VPg proteins were shown to interact directly with the eukaryotic initiation factor eIF3 in a yeast two-hybrid system (Daughenbaugh et al., 2003), and the VPg proteins of both FCV and noroviruses were shown to interact with the translation factor eIF4E (Goodfellow et al., 2005). Moreover, certain translation initiation factor inhibitors, such as 4E-BP1, specific for eIF4E, could markedly reduce translation efficiency in vitro for some caliciviruses (Chaudhry et al., 2006).
The discovery that recombinant VPg expressed from RHDV could be uridylylated by the viral polymerase provided initial evidence that the calicivirus VPg might also serve a primer function during RNA replication, similar to that of the picornavirus VPg (Machin, Martin Alonso, and Parra, 2001). As noted above, evidence for such activity has been reported recently for the norovirus VPg (Rohayem et al., 2006b). It is assumed that the first step in a VPg-mediated priming mechanism would be nucleotidylylation at a specific amino acid residue of the VPg. In RHDV, a tyrosine residue located at position 21 of the VPg was shown to be uridylylated in vitro by recombinant Pol in the presence of Mn2+ in a template-independent manner. Mutagenesis of the corresponding conserved tyrosine residue (position 24) in the FCV VPg was lethal for the growth and recovery of the virus (Mitra, Sosnovtsev, and Green, 2004). These studies suggest that a conserved tyrosine residue is the site of nucleotidylylation for the caliciviruses, but this site has not been identified in the noroviruses.
In this study, we examined biochemical modifications of the VPg by the ProPol form of the MD145 RdRp in an effort to gain further insight into the role of the VPg in RNA replication for the noroviruses. One aim was to determine whether the norovirus VPg undergoes nucleotidylylation by the ProPol form of the viral polymerase and to map the site where this modification occurs. We showed that the VPg was nucleotidylylated by the ProPol form of the human norovirus RdRp in a template-independent manner in the presence of Mn2+, and that the linkage between the nucleotide and VPg was covalent. We identified Tyrosine 27 of the MD145 VPg as the target amino acid for this activity, and showed that the linkage was susceptible to treatment with phosphodiesterase. Efficient nucleotidylylation of the VPg could be achieved in the presence of Mg2+ cations, but only in the presence of selected polyadenylated viral RNA templates. In addition, evidence was found for an enhancing element in calicivirus RNA that might function similarly to the cis-acting replication element (CRE) of the picornaviruses.
Results
Synthesis and purification of recombinant VPg proteins
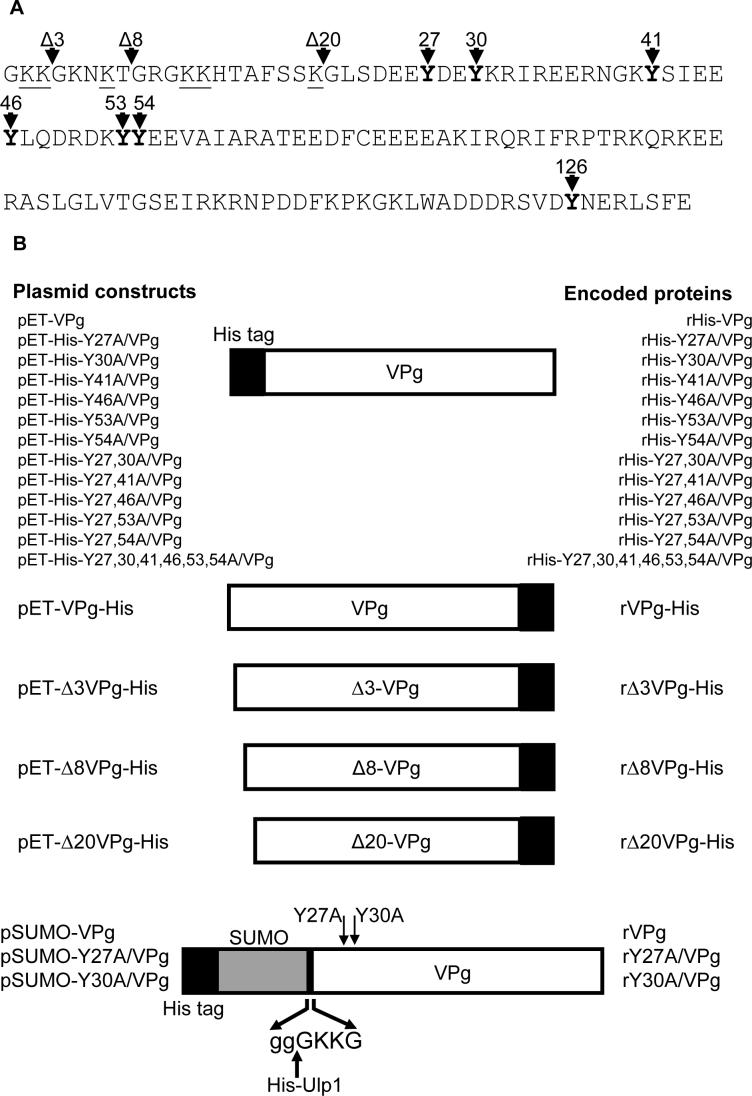
Recombinant VPg proteins were expressed either as N- or C-terminal 6x His-tagged fusions, or as proteins without a His tag (summarized in Figure 1B). The VPg proteins with an N-terminal His-tag were expressed in the pET system and purified on a Ni-NTA (nickel-nitrilotriacetic) column to greater than 85% homogeneity (Figure 1C, lanes 1 to 7). The recombinant proteins rVPg-His, rΔ3VPg-His, rΔ8VPg-His and rΔ20VPg-His, in which the His-tag was engineered at the C-terminus, were purified to over 90% homogeneity (Figure 5A, panel II). The SUMO-VPg recombinant fusion proteins (rHis-SUMO-VPg, rHis-SUMOY27A/VPg, rHis-SUMO-Y30A/VPg) were purified on a Ni-NTA column (as shown for rHis-SUMO-Y27A/VPg in Figure 1C, lane 9) and then incubated with Ulp1 proteinase. The mixture was again passed through a Ni-NTA column to bind the cleaved SUMO-His tag and the His-tag free recombinant VPg protein was collected in the flow-through (as shown for rHis-SUMO-Y27A/VPg in Figure 1C, lane 10). The efficiency varied, but at least 50% of the fusion protein was cleaved with Ulp1 (data not shown). In all purified VPg preparations, minor amounts of a small protein (under 10 kDa) were consistently observed (Figure 1C, lane 10). This 10 kDa protein was not present in recombinant RdRp preparations derived by bacterial expression, and was not immunoprecipitated using VPg-specific immune serum (Figure 6C, lanes 3, 5 and 7).
Figure 1.
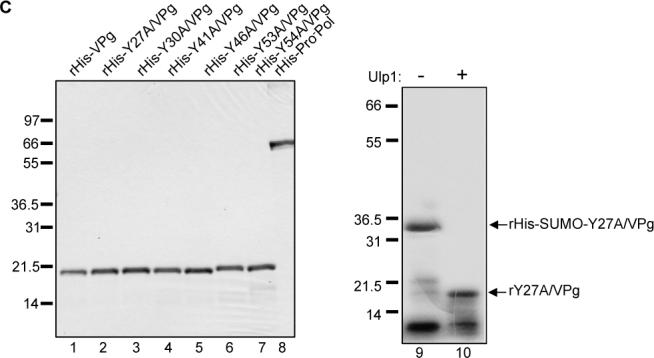
MD145 VPg DNA constructs generated for study. (A) The primary deduced amino acid sequence of the MD145 VPg is shown. The tyrosine residues at positions 27, 30, 41, 46, 53, 54, and 126 are shown in bold type, and the lysine residues in the N-terminal region are underlined. The N-terminus of the VPg encoded in truncated VPg constructs Δ3-, Δ8- and Δ20-VPg-His is indicated by arrows. (B) cDNA clones engineered for expression of the MD145 VPg and its mutated forms in bacteria. His-tag is represented by the black box in the diagram, and is located at the N- or C-terminus. The VPg constructs were designated according to the mutagenized tyrosine (e.g., pET-His-Y27A/VPg). Double tyrosine mutated VPg constructs were designated similarly. VPg proteins with sequential deletions from the N-terminus were designated according to the first amino acid in the truncated VPg (e.g., pET-Δ3VPg-His). For the SUMO-VPg constructs, the SUMO peptide is indicated by a grey box. For the Ulp1 cleavage site, SUMO peptide and VPg amino acids are indicated by lower and lower cases, respectively. The resulting construct was named as indicated above. (C) SDS-PAGE analysis of 1μg bacterially-expressed recombinant VPg proteins (lanes 1−7) and Pro−Pol (lane 8) that were engineered with an N-terminal His tag. The mutated rY27A/VPg (Figure 1C, lanes 9 and 10) was expressed without a His-tag (see Materials and Methods). The rHis-SUMO-Y27A/VPg fusion protein (lane 9) and tag-free rY27A/VPg (lane 10) were resolved by SDS-PAGE. Ulp1 proteinase treatment is indicated above the gel. The migration of the molecular weight marker (Mark XII, Invitrogen) is indicated in kDa here and in the following figures.
Figure 5.
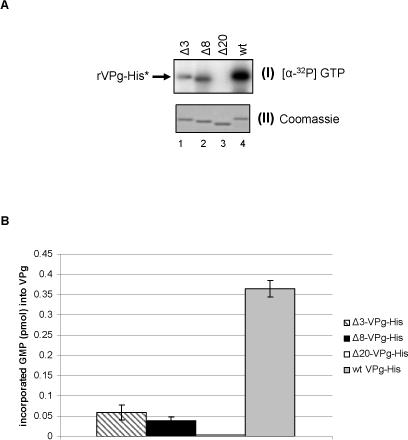
Deletional mutagenesis of the N-terminus of the norovirus VPg. (A) Three μg of wild type rVPg-His (wt, lane 4), rΔ3-VPg-His (Δ3, lane 1), rΔ8-VPg-His (Δ8, lane lane 2) and rΔ20-VPg-His (Δ20, lane 3) were assayed for guanylylation in the presence of 1 μg of rE1189A/ProPol-His, [α32P]-GTP and 1 mM MnCl2. Wild type and truncated VPg proteins are indicated above the gel. The autoradiography and Coomassie blue staining of the modified VPg proteins are shown on panels I, and II, respectively. Panels I and II represent the same gel. (B) Amount of incorporated GMP into wt and truncated VPgs. Three μg of wild type rVPg-His (grey column), rΔ3-VPg-His (Δ3, hatched column), rΔ8-VPg-His (Δ8, black column) and rΔ20-VPg-His (Δ20, white column) were assayed for guanylylation as described above. The assays were conducted in triplicate. The amount of incorporated UMP was calculated as described in Materials and Methods and reported here as pmol per μg of VPg. Columns represent the mean value of three experiments and the standard deviation is indicated by vertical bars.
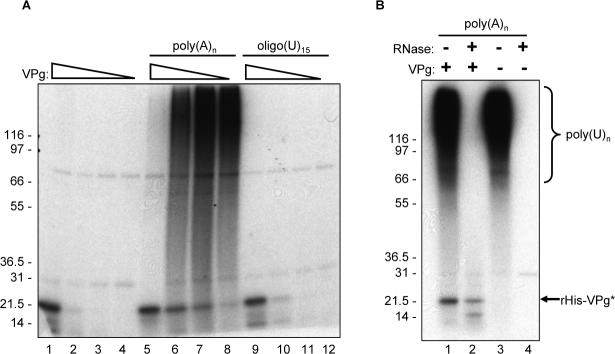
Figure 6.
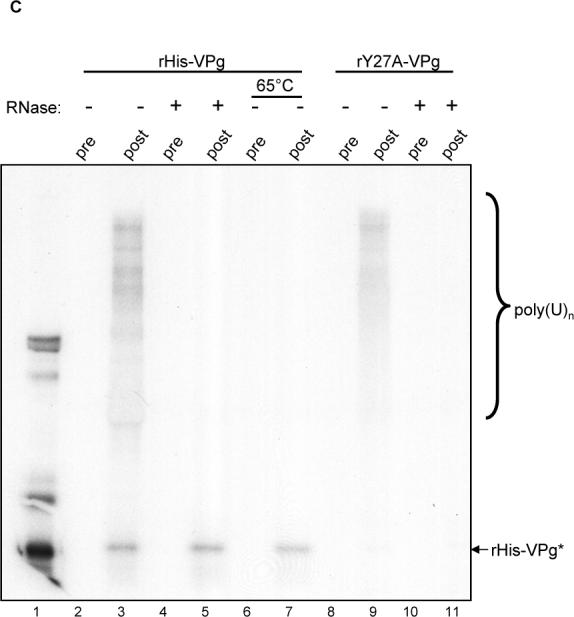
VPg uridylylation in the presence of homopolymeric poly(A)n templates. (A) Three μg (lanes 1, 5 and 9), 1.5 μg (lanes 2, 6 and 10), 0.75 μg (lanes 3, 7 and 11) and 0.375 μg (lanes 4, 8 and 12) of rHis-VPg (corresponding to 180, 90, 45, and 22.5 pmol per assay, respectively) were assayed for uridylylation in the presence of rHis-Pro−Pol (13 pmol), [α32P]-UTP and 1 mM MnCl2. Uridylylation assays were also performed in the presence of 5 μg of homopolymeric poly(A)n templates (Roche) (lanes 5 to 8) or 10 μM oligo(rU)15 primer (Invitrogen) (lanes 9 to 12). The decreasing amounts of rHis-VPg are represented by an open triangle above the gel. The presence of poly(A)n and oligo(U)15 in each reaction is indicated above the gel. (B) One μg of rHis-Pro−Pol was incubated with 5 μg of poly(A)n, 5 μCi [α32P]-UTP, and 1 mM MnCl2 in the presence (lanes 1 and 2) or absence (lanes 3 and 4) of 1.5 μg rHis-VPg). Five μl of each reaction were resolved by SDS-PAGE without treatment (lanes 1 and 3) or following treatment with RNase prior to SDS-PAGE (lanes 2 and 4). The combinations of RNase and VPg are indicated above the gel. Labeled poly(U)n and rHis-VPg are indicated by bracket and arrow, respectively on the right side of the gel. (C) Immunoprecipitation assay of the uridylylation reaction mixture with an antiserum raised against bacterially-expressed rHis-VPg (Belliot et al., 2003). For each immunoprecipitation assay, a uridylylation assay was first performed with rHis-Pro-Pol, rHis-VPg (lanes 2 to 7), and rHis-Y27A/VPg (lanes 8 to 11). The reaction mixture was then assayed by immunoprecipitation without treatment (lanes 2, 3, 8 and 9) or following treatment with RNase (lanes 4, 5, 10 and 11) or heating at 65°C for 10 min (lanes 6 and 7). Labeled materials were resuspended with electrophoresis lysis buffer, released by boiling and directly resolved onto a 12% Tris-glycine polyacrylamide denaturing gel, or used for RIPA. The serum (VPg pre or post-immunization) used in each reaction is indicated. The positive control in lane 1 was 5μl of a VPg uridylylation assay as described above. Labeled poly(U)n and rHis-VPg are indicated on the right with a bracket and arrow, respectively.
Nucleotidylylation of the norovirus VPg by the Pro−Pol form of the polymerase
Our previous studies showed that both Pol and ProPol forms of the norovirus RdRp are active as a polymerase (2). Because previous studies had shown that the Pol form of both the RHDV and norovirus RdRp was able to mediate uridylylation of the VPg (Machin, Martin Alonso, and Parra, 2001; Rohayem et al., 2006b), we decided to investigate whether the ProPol form had this activity. Our preliminary studies showed that both the Pol and ProPol forms of the MD145 norovirus RdRp (Belliot et al., 2005) could uridylylate the MD145 VPg (data not shown). It should be noted that the distantly-related picornavirus 3CDpro protein, which is also a precursor containing a proteinase and polymerase domain, was not active in a VPg nucleotidylylation assay. However, it was able to stimulate uridylylation by the mature 3Dpol form of the polymerase in the presence of a cis-acting replication element, or CRE (Paul et al., 2000). A nucleotidylation reaction was performed in which enzymatically-active MD145 ProPol (rHis-Pro−Pol), which contained an inactivated proteinase domain to prevent autocatalytic cleavage (Belliot et al., 2001; Belliot et al., 2005), was incubated with purified MD145 VPg (rHis-VPg) in the presence of each of the [α-P32]-labeled nucleotides (UTP, GTP, ATP, or CTP) individually and Mn2+, as described in Materials and Methods. Radiolabeling of the recombinant VPg protein (21 kDa) was observed with each of the four nucleotides (Figure 2A, lanes 1−4), with adenylylation appearing least efficient. An additional radiolabeled band was detected with an observed mass of 76 kDa, consistent in size with the ProPol noted above (Figure 1C, lane 10). A similar radiolabeling of the poliovirus 3D pol has been observed in VPg uridylylation reactions, but it is not clear whether this auto-uridylylation of 3Dpol is essential for replication (Richards et al., 2006). A radiolabeled 10 kDa band was also observed in some experiments. This small protein, noted above, was likely a bacterial protein that co-purified with the recombinant VPg or a VPg degradation product. The amount of radioactivity incorporated into the guanylylated and uridylylated 21 kDa forms of the VPg protein was determined as described in the Materials and Methods. The guanylylation reaction was found to be approximately two times more efficient than the uridylylation reaction under these conditions (Figure 2B). The ability of the MD145 ProPol to utilize all four NTPs in the modification of VPg was similar to that reported previously for RHDV Pol (Machin, Martin Alonso, and Parra, 2001).
Figure 2.
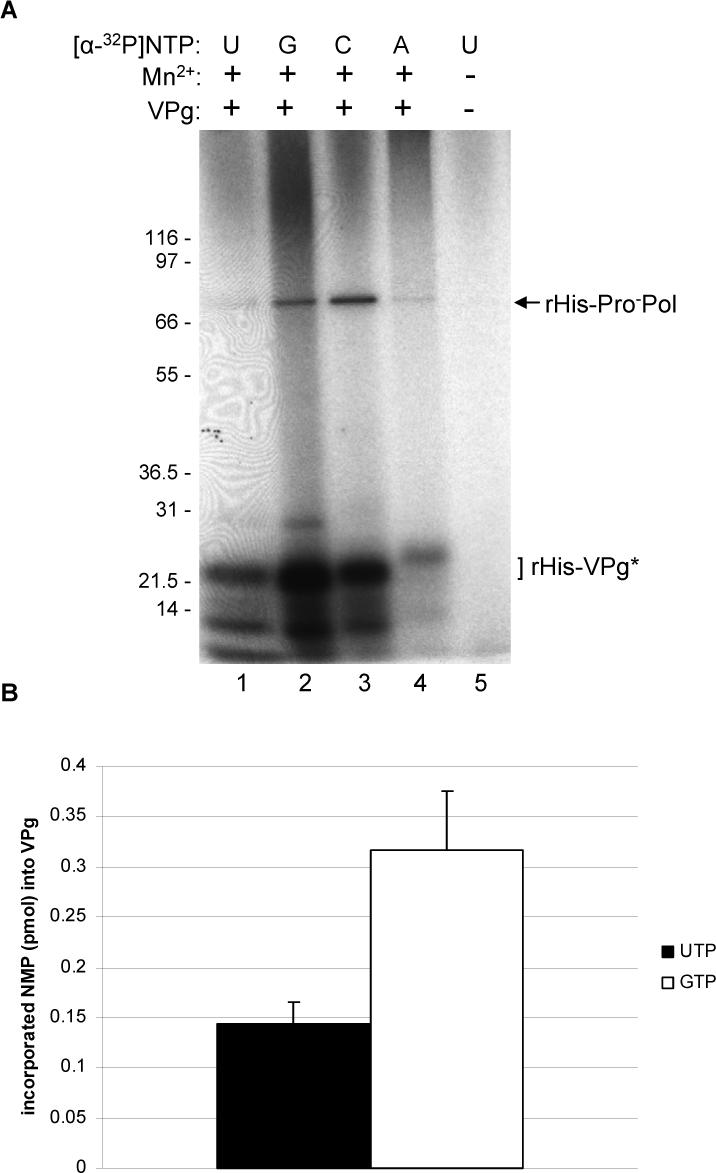
Nucleotidylylation of rHis-VPg by the rHis-Pro−Pol polymerase. (A) Two μg of rHis-VPg were assayed in the presence of [α32P]-NTP (either UTP, lanes 1 and 5; GTP, lane 2; CTP, lane 3 and ATP, lane 4). Lane 5 contains a negative control without VPg and Mn2+. The NTP used in each assay, and the presence or absence of VPg and Mn2+ (0.5 mM final) is indicated above the gel. The identities of the radiolabeled proteins are indicated on the right, and the modified rVPg protein is indicated by an asterisk here and the following figures. (B) In a separate experiment, guanylylation and uridylylation reactions of rHis-VPg were conducted in triplicate. Five μl of each assay was analyzed by SDS-PAGE. Labeled VPg separated by SDS-PAGE, was excised from the gel and the amount of UMP (black column) and GMP (white column) incorporated into VPg was determined. The graph represents the mean value of the triplicate assay for each NTP. Incorporated NMP is given in pmol/μg of rHis-VPg (left side of the graph). Standard deviations are indicated by vertical bars.
We next established the minimal requirements for the nucleotidylation reaction. As noted above, the uridylylation reaction resulted in the production of three major radiolabeled products (Figure 3A, lanes 1 and 4). The recombinant VPg protein (21kDa) was not radiolabeled in the absence of Mn2+ (Figure 3A, lane 2) or rHis-Pro−Pol (Figure 3A, lane 3), or when incubated with heat-inactivated rHis-Pro−Pol (Figure 3A, lane 5). These data suggested that the modification of the VPg was associated with an active polymerase, and required a divalent cation. The observed efficiency of the VPg nucleotidylylation was similar in the presence of 0.5 to 1 mM MnCl2 (data not shown). No radiolabeled VPg was observed when rHis-Pro−Pol was replaced by rHis-Pro−, indicating that the Pol domain of the ProPol polymerase was responsible for the activity (Figure 3A, Lane 6).
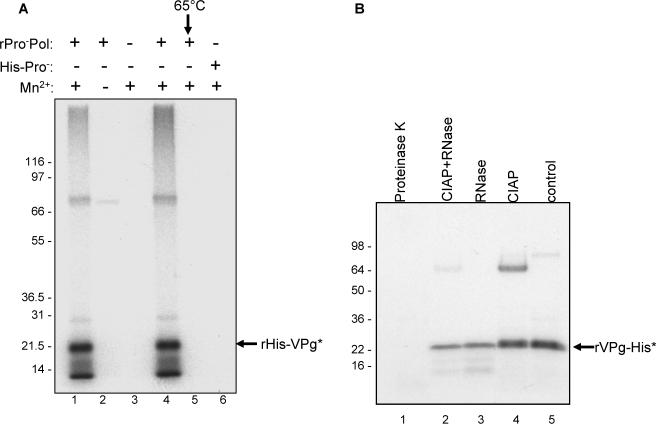
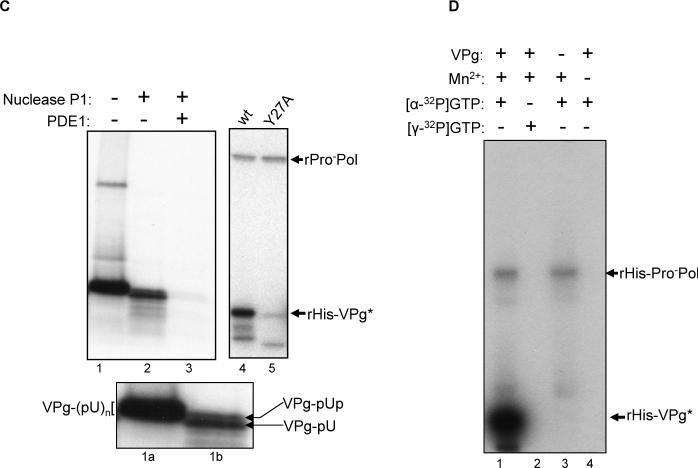
Figure 3.
Characterization of the VPg modification by ProPol. (A) Autoradiography of the VPg-uridylylation assay. Two μg of rHis-VPg was incubated in the presence of [α32P]-UTP and several combinations of rHis-Pro−Pol (1 μg), rHis-Pro− (1 μg) and Mn2+ as indicated above the gel. rHis-Pro−Pol was also heated at 65°C for 10 min prior to addition to the assay (lane 5). (B) Autoradiography of the UTP-labeled VPg after enzymatic treatment. Uridylylation assay and enzymatic treatments are described in the Materials and Methods. The 15 μl reaction was Proteinase K- or RNase-treated (lanes 1 and 3). The 15 μl reaction was also CIAP-treated (Calf Intestinal Alkaline Phosphatase, lane 4) or CIAP- and RNase-treated (lane 2). Enzymatic treatments are indicated above the gel. For the control (lane 5), 5 μl of the uridylylation assay were directly resolved by SDS-PAGE. (C) Nuclease P1 and phosphodiesterase 1 (PDE1) treatment of the uridylylated VPg. For the control (lane 1), 5 μl of the assay was directly resolved by SDS-PAGE. Enzymatic treatments are indicated above the gel. The insert contains a magnified view of the labeled VPg from lane 1 (lane 1a) and 2 (lane 2a). The putative forms of labeled VPg are indicated by arrows (VPg-pUpU and VPg-pU) or bracket (VPg-p(U)n). (D) Two μg of rHis-VPg were assayed for nucleotidylylation in the presence of [α32P]-GTP (lanes 1, 3 and 4) or [γ32P]-GTP (lane 2). The combinations of VPg, Mn2+ (1 mM final), [α32P]-GTP and [γ32P]-GTP are indicated above the gel. rHis-Pro−Pol and rHis-VPg are indicated by arrows.
Various enzymatic treatments of the radiolabeled VPg protein were performed to examine the biochemical nature of the VPg modification. Treatment of the radiolabeled VPg with proteinase K abolished the detection of radiolabeled protein, as expected (compare Figure 3B, lanes 1 and 5). Coomassie blue staining of the proteins in the corresponding gel confirmed proteolytic degradation of the VPg (data not shown). Treatment of the radiolabeled VPg with calf intestinal alkaline phosphatase (CIAP) did not abolish the radioactivity (Figure 3B, lane 4), suggesting that the modification of the VPg was not due to phosphorylation. Further treatment of the radiolabeled VPg with RNase (a mixture of enzymes isolated from bovine pancreas) and RNase in the presence of CIAP (Figure 3B, lane 2), which together would hydrolyze a diverse number of potential covalent bonds, resulted in the weaker intensity of the VPg labeling, presumably by converting VPg-p(U)n to the VPg-pUp form. A similar product was observed when labeled VPg was treated only with the RNase mix (Figure 3B, lane 3). We next examined the activity of nuclease P1, an enzyme that cleaves 3’-phosphodiester bonds and 3’-phosphomonoester bonds, leaving a 3’-end hydroxyl group (Figure 3C, lanes 1 and 2). A magnified view of the uridylylated VPg after P1 treatment showed the presence of a doublet presumably composed of VPg-p(U) and VPg-pUpU, with the latter form consistent with incomplete processing by the nuclease P1 (Figure 3C, lanes 1a and 2a). The intensity of the radiolabeled VPg was reduced but not abolished, suggesting that a radiolabeled UMP moiety remained. The nuclease P1-treated VPg protein was further incubated in the presence of phosphodiesterase 1 (PDE1), which can hydrolyze a 5’-phosphodiester bond in a ribonucleotide with a free 3’-hydroxyl group (Figure 3C, lane 3). This treatment caused a complete loss of radiolabel from the VPg, suggesting that the VPg was linked to the UMP molecule via a 5’-phosphodiester bond. The presence of the intact VPg protein in the products of PDE1 hydrolysis was confirmed by Coomassie blue staining of protein in the corresponding gel (data not shown).
Because all calicivirus genomes begin with a 5’-end guanosine residue, and it is presumed that the VPg is linked to this terminal nucleotide, we further examined the nature of the linkage mediated by ProPol between the norovirus VPg and GTP in a guanylylation reaction. VPg was labeled when [α-32P]-GTP was employed in the reaction (Figure 3D, lane 1), but not [ γ-32P]-GTP (Figure 3D, lane 2). These data suggest that the linkage of the norovirus VPg to RNA utilizes an alpha phosphate group, similar to the picornaviruses, and that NMP is incorporated. The absence of radiolabeling with [γ-32P]-GTP is consistent with the phosphatase data above (Figure 3B), in that a phosphorylation event was not responsible for the radiolabeling of the VPg.
Mapping tyrosine at position 27 in the MD145 VPg protein as the site of nucleotidylylation
Our next experiments addressed the identity of the amino acid residue in the norovirus VPg that undergoes modification by nucleotidylylation. A tyrosine residue previously shown to be conserved among the caliciviruses was identified as the site of uridylylation by the RHDV polymerase (Machin, Martin Alonso, and Parra, 2001). This conserved tyrosine residue is located at position 27 in the VPg of MD145. A mutagenized VPg protein designated rHis-Y27A/VPg was engineered in which Tyr 27 was replaced with Ala. Analysis of the rHis-Y27A/VPg protein in a nucleotidylylation reaction (with UTP) showed a marked reduction in the intensity of the signal (Figure 4A, lane 2) compared to that of the wild type VPg, rHis-VPg (Figure 4A, lane 1). The faint band corresponding to rHis-Y27A/VPg (Figure 4A, lane 2) might be explained by the labeling of an alternative tyrosine residue, and we examined this possibility in a series of mutagenesis experiments. The MD145 VPg amino acid sequence contains seven tyrosine residues (Figure 1B), and we engineered several additional modified VPg mutated proteins in which individual tyrosine residues were replaced by alanine residues. Recombinant VPg (rHis-VPg, rHis-Y27A/VPg, rHis-Y30A/VPg, rHis-Y41A/VPg, rHis-Y46A/VPg, rHis-Y53A/VPg, or rHis-Y54A/VPg) was assayed for uridylylation, and the relative amount of UMP incorporated into VPg was measured with a phosphorimager (Figure 4B). The mutagenesis of Tyr 27 showed the greatest effect, reducing the signal by approximately 95% compared to the wild type VPg control. Mutagenesis of Tyr 53 or Tyr 54 reduced the signal by approximately 60%. Surprisingly, the mutagenesis of Tyr 30, 41 and 46 resulted in a 6.4-fold, 2-fold, and 2.7-fold increase in the labeling of VPg, respectively. In order to determine whether Tyr 27 was the site of enhanced uridylylation observed in the single tyrosine mutants, we engineered and tested double tyrosine-mutated VPg proteins (rHis-Y27,30A/VPg, rHis-Y27,41A/VPg, rHis-Y27,46A/VPg, rHis-Y27,53A/VPg, rHis-Y27,54A/VPg). In addition, a mutagenized VPg protein rHis-Y27,30,41,46,53,54A/VPg in which all tyrosine residues (except for position 126) were replaced with alanine residues, was also assayed. Analysis of the Tyr27-mutagenized VPg proteins in a uridylylation reaction yielded proteins with signals that were 5 to 12% of that obtained with the wild type rHis-VPg control (Figure 4B). This result indicated that the enhanced uridylylation observed with the single tyrosine mutations at positions 30, 41 and 46 was likely due to an increase in the efficiency of uridylylation at Tyr 27. Taken together, the mutagenesis studies identified Tyr 27 as the major site of nucleotidylylation in the MD145 VPg.
Figure 4.
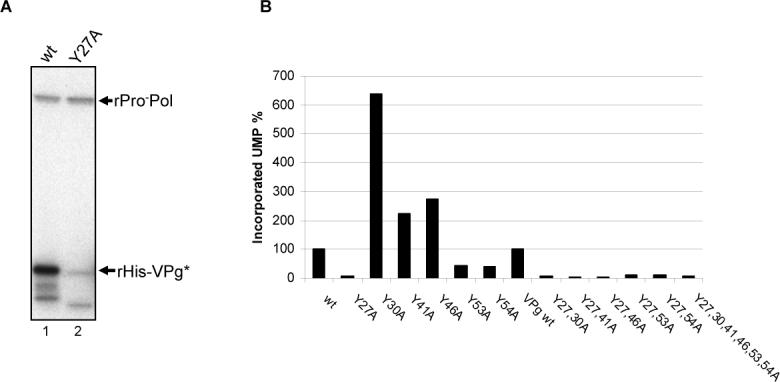
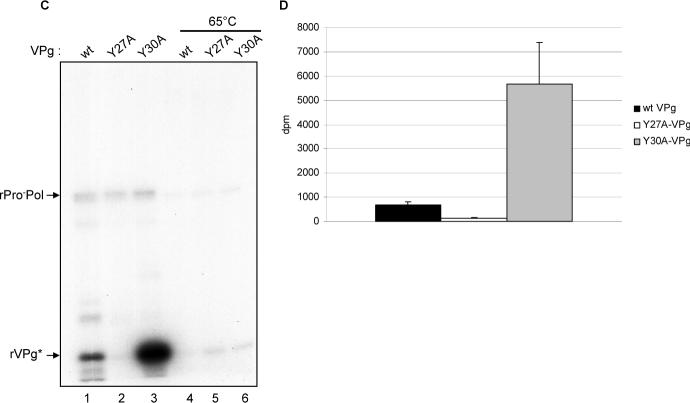
Role of the tyrosine residues in VPg uridylylation. (A) The expression and purification of the His tag-free rPro−Pol has been described previously (Belliot et al., 2005). Two μg of rHis-VPg (lane 1) or rHis-Y27A/VPg (lane 2) were incubated in the presence of rPro−Pol, 1 mM MnCl2 and [α32P]-UTP. Labeled proteins are indicated by arrows on the right side of the gel. (B) Effect of the tyrosine residues at positions 27, 30, 41, 46, 53 and 54 on the uridylylation of the VPg proteins. The VPg mutated proteins are indicated below the graph. rVPg protein (1.5 μg) was incubated with 1 μg of SUMO expressed rPro−Pol in the presence of 5 μCi [α32P]-UTP and 0.5 mM MnCl2. Five μl of each reaction were analyzed by SDS-PAGE. The relative amount of incorporated UMP was determined by using a Phosphorimager. Incorporation values are given in percentage of the level of incorporation for the wt VPg protein (100%). (C) SUMO-expressed wild type (wt, lanes 1 and 4) VPg, rY27A/VPg (lanes 2 and 5) and rY30A/VPg (lanes 3 and 6) were assayed for uridylylation in the presence of tag-free rPro−Pol, 1 mM MnCl2, and [α32P]-UTP. Wild type and mutated VPgs were also heated at 65°C for 10 min prior to incubation with rPro-Pol (lanes 4 to 6). Uridylylation assays were then conducted the same way. The proteins of interest are indicated by arrows. (D) Role of the tyrosine residues at positions 27 and 30 in uridylylation. Tag-free- wt rVPg (black column), rY27A/VPg (white column) and rY30A/VPg (grey column) were assayed for uridylylation in the presence of tag-free rPro−Pol. The uridylylation assays were conducted in triplicate and 5 μl of each assays were analyzed by SDS-PAGE. The amount of incorporated UMP is given in disintegration per min (dpm) per μg of VPg. Columns represent mean values and standard deviations are represented by vertical bars.
In order to address whether the recognition of Tyr27 as the site of nucleotidylylation might be influenced by the presence of a His-tag on the VPg protein, we expressed wt VPg (rVPg) and mutated VPg proteins, rY27A- and rY30A/VPg, in a bacterial expression system (SUMO) that allowed the purification of native VPg without a His-tag. Again, the mutagenesis of Tyr 27 in a native (non-His-tagged) VPg resulted in a marked decrease in incorporation of radiolabel in a uridylylation reaction (Figure 4C, lane 2) compared to the wt VPg control (Figure 4C, lane 1). As observed above, uridylylation of rY30A/VPg (Figure 4C, Lane 3) was enhanced (Figure 4D). These data suggested that the His-tag did not have a major effect on the modification of the VPg, and that conformation likely plays a role in presentation of Tyr27 to the RdRp. Structural studies of the VPg by X-ray crystallography should give further insight.
Influence of the N-terminal region of the VPg protein on nucleotidylylation
A lysine-rich NTP-binding motif (AYTKKGK) in the potyvirus (potato A virus) VPg was found to play a critical role in uridylylation (Puustinen and Makinen, 2004). The presence of a similar lysine-rich region at the N-terminus of the calicivirus VPg (Figure 1A) prompted us to examine its role in nucleotidylylation. A series of mutagenized VPg proteins was engineered in which the first 3, 8, or 20 amino acids were deleted. The recombinant wt VPg protein and the truncated forms of the VPg (rΔ3-, rΔ8- and rΔ20-VPg-His) (Figure 5A, panel I) were assayed in a guanylylation reaction in the presence of [α-32P] -GTP (Figure 5A). The guanylylation of rVPg-His (wt), rΔ3-VPg-His and rΔ8-VPg-His was observed in the presence of 1 mM MnCl and [α-32P]-GTP (Figure 5A, panel I, lanes 1, 2, and 4, respectively), while rΔ20-VPg-His protein was not radiolabeled (Figure 5A, panel I, lane 3). The deletion of 3, 8 and 20 amino acids at the N-terminus of the VPg protein reduced the amount of measured radioactivity in the guanylylation reaction by 83, 90 and 100%, respectively, when compared to the wild type VPg control (Figure 5B). It is likely that the N-terminal region of the VPg may be important in the conformational presentation of Tyr 27 to the RdRp for nucleotidylation. Alternatively, the lysine rich N-terminus might have NTP binding properties similar to the potato A virus (Puustinen and Makinen, 2004). Additional mutagenesis studies will be needed to map the precise amino acids in the N-terminus that facilitate nucleotidylylation of the VPg.
Uridylylation of VPg in the presence of poly (A) templates
It was previously shown that uridylylation of the RHDV VPg by Pol could be achieved in the absence of template RNA, and our next experiments examined whether this feature of nucleotidylylation was shared by the MD145 ProPol (Machin, Martin Alonso, and Parra, 2001). An uridylylation assay was performed in the presence or absence of poly(A) template or oligo(U)15 to determine their effects on the nucleotidylylation of the MD145 VPg in the presence of Mn2+. A constant amount of rHis-Pro−Pol (1 μg) was used in each assay, with decreasing concentrations of rHis-VPg protein (3, 1.5, 0.75 and 0.375 μg per assay). There was no detectable difference in the profile of uridylylation assays performed in the absence (Figure 6A, Lanes 1−4) or the presence (Figure 6A, Lanes 9−12) of oligo(U)15 . In contrast, in the presence of poly(A) template, radiolabeled VPg was detected at all four concentrations of VPg examined (Figure 6A, lanes 5−8). Moreover, a diffuse signal (appearing as a smear on the autoradiograph) representing radioactive material was observed near the top of the gel (Figure 6A, lanes 6−8). Further analysis showed that this diffuse signal was generated in a reaction without VPg (Figure 6B, lane 3), and was sensitive to RNase treatment (Figure 6B, lanes 2 and 4). It was likely that this material represented newly-synthesized poly(U)n products generated by the rHis-Pro−Pol polymerase, which has been reported to have primer-independent RdRp activity in the presence of Mn2+ (Belliot et al., 2005; Fukushi et al., 2004). However, in this experiment, we could not rule out the possibility that a proportion of the products were poly(A) templates that had been terminally labeled by the RdRp (Rohayem et al., 2006a).
To examine whether rHis-VPg molecules might be covalently linked to the newly-synthesized radiolabeled products, rHis-VPg and rHis-Y27A/VPg were immunoprecipitated by VPg-specific antibodies from uridylylation assays that included poly(A)n templates (Figure 6C, lanes 2−3 and lanes 8−9). The larger radiolabeled products co-precipitated with both VPg proteins, including the nonuridylylated VPg with a mutagenized Tyr 27 (Figure 6C, Lane 9). Treatment of the uridylylation reaction with RNase (Figure 6C, lanes 4−5 and 10−11) or heat (65°C for 15 min) (Figure 6C, lanes 6 and 7) prior to immunoprecipitation of the VPg and SDS-PAGE led to the disappearance of the high-MW radiolabeled products, while retaining the radiolabeled precipitated VPg. In order to verify that the high-MW products were radiolabeled RNA products, the material remaining after heat-treatment and immunoprecipitation of the VPg was ethanol-precipitated and analyzed in a denaturing agarose gel. Autoradiography of the dried gel showed the presence of radiolabeled material (data not shown). Taken together, these data suggested that most of the incorporated label in the uridylylation reaction was not covalently linked to the uridylylated VPg. In addition, the synthesis of these products could occur without the involvement of a uridylylated VPg (Figure 6B, lane 3). The efficient co-precipitation of these RNA products with VPg under the conditions of the radioimmunoprecipitation assay (RIPA) suggested that the VPg binds RNA. Additional studies will be needed to establish the RNA-binding properties of VPg and its potential role in replication.
Nucleotidylylation of VPg in the presence of genomic RNA
Uridylylation of the RHDV VPg was shown to be template-independent and optimal in the presence of Mn2+ divalent cations, and weak uridylylation of the VPg was observed also in the presence of Mg2+ (Machin, Martin Alonso, and Parra, 2001). In contrast, uridylylation of the poliovirus VPg was shown to be template-dependent in the presence of Mn2+ (Paul et al., 1998). Moreover, efficient uridylylation of the poliovirus VPg by 3Dpol in the presence of Mg2+ was dependent on the presence of a CRE element, and the reaction was enhanced by the addition of 3CDpro (Paul et al., 2000). We evaluated whether the addition of various RNA templates corresponding to the MD145 genome might improve the efficiency of VPg nucleotidylylation by ProPol in the presence of Mg2+. Plasmid constructs containing either full-length or partial cloned sequences of the MD145 norovirus genome engineered immediately downstream of the T7 RNA polymerase promoter were linearized and used to generate RNA molecules by in vitro transcription with T7 polymerase (Figure 7A). Nucleotidylylation assays were performed similar to those described above, except that all four NTPs were included in the reaction and Mg2+ was used as the divalent cation. In addition, we utilized rHis-E1189A/ProPol (which is similar in RdRp activity to rHis-Pro−Pol, but containing an active proteinase and a mutagenized cleavage site between Pro and Pol) so that the bifunctional enzymatic activity of the precursor protein would remain intact.
Figure 7.
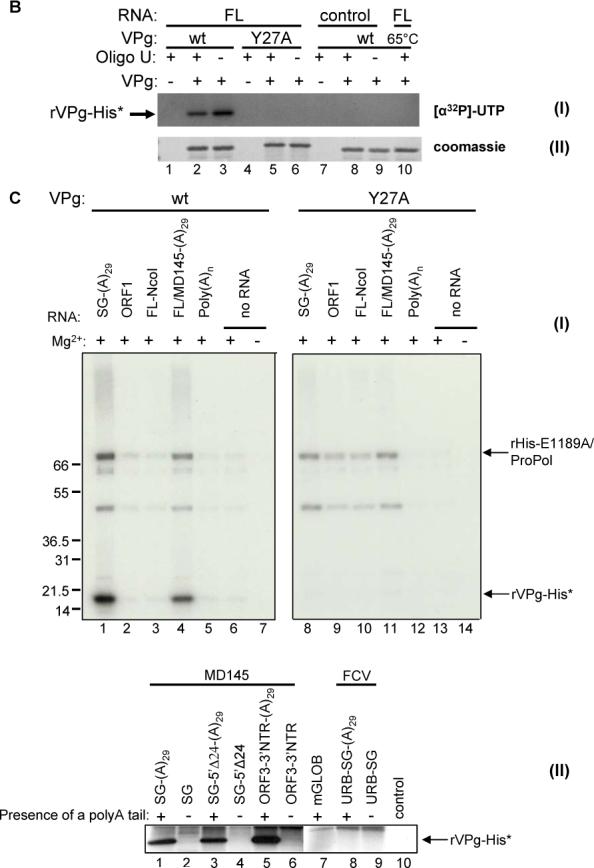
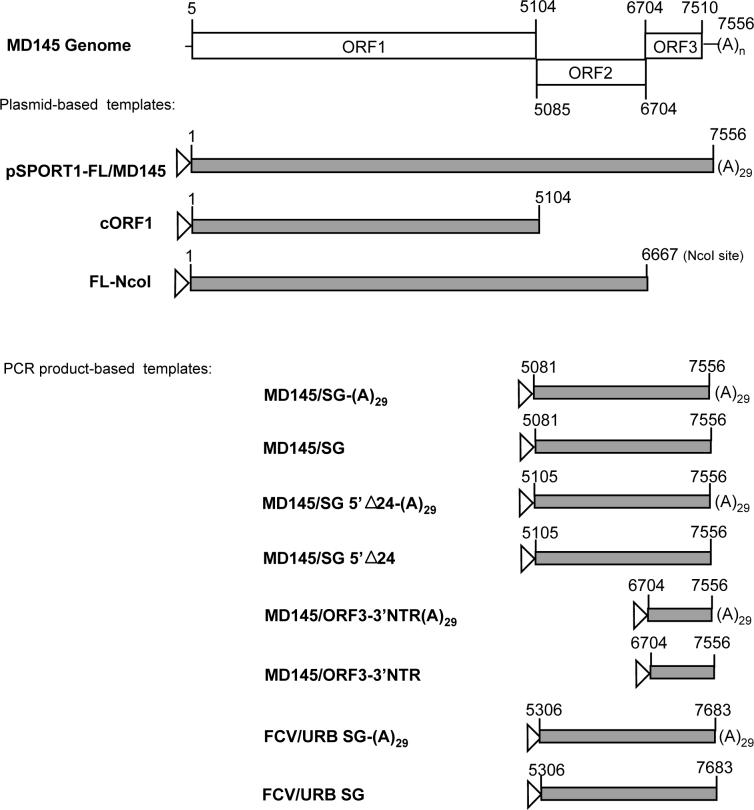
Effect of MD145 genomic and subgenomic RNAs on VPg uridylylation in the presence of Mg2+. (A) Diagram of the constructs used in the experiment. The genomic organization of the MD145 RNA genome is based upon the sequence available in Genbank (AY032605). Positions of the first and last nucleotides for each ORF are indicated. pSPORT1-FL/MD145 and pSPORT1-SG/MD145 were linearized by AatII for RNA production. Both constructs were engineered with a T7 promoter upstream of the 5’-end (open triangle) and a 29 nucleotide length polyA tail at the 3’-end (poly(A)29). To generate genomic RNA lacking ORF3 (named FL-NcoI), the pSPORT1-FL/MD145 plasmid construct was linearized at the NcoI restriction site naturally present at position 6667 of the MD145 genome. Linearized constructs were then used to produce RNA. The cORF1 construct was described previously (linearized by NotI for RNA production) (Belliot et al., 2003). For the generation of PCR DNA templates, a T7 promoter sequence was engineered into the forward primer, and the reverse primer was engineered either with or without a poly(A)29 tail. The pSPORT1-FL/MD145 (above) and pQ14 (Sosnovtsev and Green, 1995) plasmids were used as the templates for generation of the MD145 and Urbana (URB) feline calicivirus PCR products, respectively. (B) VPg uridylylation in the presence of genomic RNA and Mg2+. Five μl of each reaction were directly analyzed by SDS-PAGE (panels I and II). The gel was Coomassie-stained (panel I), dried and subjected to autoradiography (panel II). rVPg-His (lanes 1 to 3 and 7 to 10) and rHis-Y27A/VPg (lanes 4 to 6) used for the assays are indicated above panel I. The uridylylation assays were conducted in the presence of FL genomic RNA (lanes 1 to 6 and 10) or a control RNA without polyA tail (Promega) (lanes 7 to 9). Several combination of VPg and oligo(U) were tested, and they are indicated above panel I. For the negative control, rVPg-His was heated at 65°C for 10 min (lane 10) prior to the uridylylation assay in the presence of FL genomic RNA. Immunoprecipitated rVPg-His is indicated by arrow. (C) Panel I. Wild type rVPg-His (lanes 1 to 7) and rHis-Y27A/VPg (lanes 8 to 14) were assayed for uridylylation in the presence of 1 to 5 μg of subgenomic RNA (SG, lanes 1 and 8), ORF1 RNA (lanes 2 and 9), genomic RNA lacking ORF3 (FLNcoI, lanes 3 and 10), genomic RNA (FL, lanes 4 and 11), poly(A)n template (lane 5 and 12) or in the absence of any RNA (lanes 6, 7, 13 and 14). For the negative control, VPg was assayed in the absence of Mg2+ (lanes 7 and 14). Five μl of each reaction was analyzed by SDS-PAGE. rVPg-His and rHis-E1189A/ProPol are indicated by arrows. Panel II. Wild type rVPg-His was assayed for uridylylation in the presence of Mg2+ and RNA transcripts representing subgenomic regions of either norovirus MD145 (lanes 1−6) or feline calicivirus (FCV) Urbana (lanes 8 and 9) as indicated above the lane and diagrammed in 7A above. A polyadenylated globin messenger RNA (Life Technologies, Gaithersburg, Maryland) was included as a nonviral RNA control (lane 7). The negative control (Lane 10) excluded ProPol and template RNA from the uridylylation reaction mixture.
First, we examined the uridylylation of VPg by rHis-E1189A/ProPol in the presence of full-length (FL) genomic RNA transcripts, with or without oligo(U)15 (Figure 7B). rVPg-His was successfully uridylylated in the presence of Mg2+ and genomic RNA (Figure 7B, lanes 2 and 3 of panel I), and the addition of oligo(U)15 had little effect. The presence of a nonviral control RNA template (nonpolyadenylated) did not enhance uridylylation in the presence of Mg2+ (Figure 7B, lanes 8 and 9, panel I), and again, mutagenesis of Tyr 27 abolished uridylylation (Figure 7B, lanes 5 and 6). An immunoprecipitation assay was performed to confirm the identity of the radiolabeled protein as VPg (data not shown). These data suggested that uridylylation of the VPg through Tyr 27 could be achieved in the presence of Mg2+ and a viral RNA template. As shown above, radiolabeled RNA co-precipitated with the VPg and appeared as high MW products in the gel (Figure 7B, lanes 2−3, 5−6 and 8−9). We ethanol-precipitated the radiolabeled RNA products following heating and immunoprecipitation of the uridylylated VPg, suggesting again that the radiolabeled RNA was not covalently-linked (data not shown). In order to further examine the association of VPg with the RNA, we heated the VPg protein at 65°C for 10 minutes prior to the uridylylation reaction as a negative control. Heating the VPg led to loss of its ability to be uridylylated (Figure 7B, lane 10, panel I and II), and to associate with the radiolabeled RNA derived from the FL template RNA (data not shown), suggesting that conformation was important.
Our data suggested that the FL genomic RNA transcript might contain an element(s) that enhanced the uridylylation of VPg in the presence of Mg2+. To further investigate this possibility, additional RNA transcripts derived from linearized cDNA clones and corresponding to various regions of the MD145 genome (Figure 7A) were each added to a uridylylation reaction containing rVPg-His or rHis-Y27A/VPg and Mg2+ as the divalent cation (Figure 7C). As shown above, the rVPg-His recombinant protein was uridylylated in the presence of a polyadenylated FL genomic RNA transcript (Figure 7C, lane 4, panel I) and of interest, it was efficiently uridylylated in the presence of a polyadenylated MD145 subgenomic RNA that contained ORFs 2 and 3 and the 3’-end nontranslated region (Figure 7C, lane 1, panel I). The VPg was not uridylylated in the presence of RNA corresponding to the FL MD145 ORF1 (Figure 7C, lane 2, panel I), a truncated FL genome transcript lacking 890 nucleotides from the 3’-end and the poly(A) tail (FL-NcoI) (Figure 7C, lane 3, panel I), or a poly(A) template (Figure 7C, lane 5, panel I). These data suggested that the enhanced nucleotidylation activity in the presence of the polyadenylated FL genomic and subgenomic MD145 RNA transcripts was sequence-dependent, and not due to a nonspecific activity in the template preparations or a contaminating nonviral RNA. The VPg was not efficiently uridylylated in the absence of RNA with Mg2+ as the divalent cation (Figure 7C, lane 6, panel I) or when Tyr 27 was mutagenized to alanine (Figure 7C, lanes 8−14, panel I).
To further investigate the location and specificity of a potential CRE-like structure involved in the uridylylation of the VPg protein, we designed and generated T7 promoter- bearing PCR products that would allow the transcription of RNA molecules corresponding to selected regions of the MD145 and feline calicivirus genomes (Table 1 and Figure 7A). In addition, we examined a potential role for the poly(A) tail by engineering viral RNA molecules with and without a poly(A) tract and by including a polyadenylated cellular RNA control (beta-globulin) (Figure 7C, panel II, lane 7). Transcripts corresponding to the predicted full-length MD145 subgenomic RNA (nt 5081−7556 of the genome), a 5-’end truncated subgenomic RNA (nt 5105−7556 of the genome,) and an ORF2-truncated RNA (bearing the ORF3 and 3’-end NTR) were each added to a VPg uridylylation reaction that was performed in the presence of Mg2+ (Figure 7C, panel II, lanes 1 to 6). The addition of each of these three polyadenylated MD145 subgenomic RNA derivatives increased the efficiency of uridylylation in the presence of Mg2+ (Figure 7C, Panel II, lanes 1, 3, and 5), but those lacking a poly(A) tail did not (Figure 7C, panel II, lanes 2, 4, and 6). The addition of a full-length subgenomic RNA (with or without a poly(A) tail) corresponding to the genome of the Urbana (URB) strain of FCV (nt 5306−7683) (Figure 7C, Panel II, lanes 8 and 9) or a polyadenylated cellular RNA (Figure 7C, Panel II, lane 7) did not enhance uridylylation of the VPg. Taken together, these data suggest that an element located near the 3’-end of the MD145 viral RNA was associated with the efficient uridylylation of the VPg protein in the presence of Mg2+. The fine mapping and specificity of this enhancing element will require further study.
Table 1.
Oligonucleotides used for the site directed mutagenesis of the pET constructs and for generating RNA transcripts
| Generated construct |
Template |
Oligonucleotide sequences a,f |
|---|---|---|
| pET-His-Y27A/VPg | pET-His-VPg | GTGATGAAGAGGCCGATGAGTACAAG |
| pET-His-Y30A/VPg | pET-His-VPg | GAGTACGATGAGGCCAAGAGAATCAG |
| pET-His-Y41A/VPg | pET-His-VPg | GAAACGGCAAGGCCTCCATAGAAG |
| pET-His-Y46A/VPg | pET-His-VPg | CCATAGAAGAGGCCCTTCAGGACAG |
| pET-His-Y53A/VPg | pET-His-VPg | GACAGGGACAAGGCCTATGAGGAGGTG |
| pET-His-Y54A/VPg | pET-His-VPg | GGGACAAGTACGCTGAGGAGGTGGC |
| pET-His-Y27,30A/VPg b | pET-His-Y27A/VPg | Y27,30A: GAGgcCGATGAGGCCAAGAGAATCAG |
| pET-His-Y27,41A/VPg | pET-His-Y27A/VPg | GAAACGGCAAGGCCTCCATAGAAG |
| pET-His-Y27,46A/VPg | pET-His-Y27A/VPg | CCATAGAAGAGGCCCTTCAGGACAG |
| pET-His-Y27,53A/VPg | pET-His-Y27A/VPg | GACAGGGACAAGGCCTATGAGGAGGTG |
| pET-His-Y27,54A/VPg | pET-His-Y27A/VPg | GGGACAAGTACGCTGAGGAGGTGGC |
| pET-His-Y27,46,53,54A/VPg c,d | pET-His-Y27,46A/VPg | Y53,54A: GAGgcCCTTCAGGACAGGGACAAGGCCGCTGAGGAGGTGGCCATTGCCAGGGCG |
| pET-His-Y27,30,41,46,53,54A/VPg b,c,e | pET-His-Y27,46,53,54A/VPg | Y41,46A: GAAGAAAGAAACGGCAAGGCCTCCATAGAAGAGgcCCTTCAGGACAGGGAC |
| Y27,30A: GAGgcCGATGAGGCCAAGAGAATCAG | ||
| MD145/SG and MD145/SG-(A)29 | pSPORT1-FL/MD145 | AATTCGTCTCACTGGTAATACGACTCACTATAGGTGAATGAAGATGGCGTCGAGTGACGCCAACCCATC |
| MD145/SG 5′Δ24 and MD145/SG 5′Δ24 -(A)29 | pSPORT1-FL/MD145 | AATTCGTCTCACTGGTAATACGACTCACTATAGCGCCAACCCATCTGATGGGTCCGCAGCCAACCTCGTCCCAGAGG |
| MD145/ORF3−3′NTR and MD145/ORF3−3′NTR-(A)29 | pSPORT1-FL/MD145 | AATTCGTCTCACTGGTAATACGACTCACTATAGATGGCTGGATCTTTCTTTGCTGGATTGGCATCTGATGTCC |
| MD145/SG-(A)29 and MD145/ORF3−3′NTR-(A)29 | pSPORT1-FL/MD145 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTAAAAGACACTAAAGAAGAGAAAGAAAATCAATTTTGTCTTTTC |
| MD145/SG | pSPORT1-FL/MD145 | AAAAGACACTAAAGAAGAGAAAGAAAATCAATTTTGTCTTTTC |
| FCV/URB SG and FCV/URB-SG-(A)29 | pQ14 | AATTCGTCTCACTGGTAATACGACTCACTATAGGTTTGAGCATGTGCTCAACCTGCGCTAACGTGCTTAAATATTATAATTGG |
| FCV/URB SG | pQ14 | CCCTGGGGTTAGGCGCAAATGCGGCAGCCCAAAGGGATACTTGTG |
| FCV/URB SG-(A)29 | pQ14 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTTCCCTGGGGTTAGGCGCAAATGCGGCAGCCCAAAGGGATACTTGTG |
Only the positive-sense oligonucleotide is shown for the pET constructs. Nucleotides substitutions are underlined.
Mutagenized sequence corresponding to Y27A is indicated in lower case (primer Y27,30A).
Mutagenized sequence corresponding to Y46A is indicated in lower case (primers Y41,46A and Y53,54A).
Primer set Y53,54A was used to generate the plasmid intermediate construct pET-His-Y27,46,53,54A/VPg (not assayed), using pET-His-Y27,46A/VPg as template.
Primer sets Y41,46A and Y27,30A were used together to mutagenize intermediate template construct pET-His-Y27,46,53,54A/VPg to produce final construct pET-His-Y27,30,41,46,53,54A/VPg.
Nonviral sequences which include a T7 promoter are indicated in bold.
Discussion
VPg proteins are covalently-linked to the RNA genomes of viruses in the families Caliciviridae (Sosnovtsev and Green, 2000), Picornaviridae (Ambros and Baltimore, 1978; Rothberg et al., 1978), Nepoviridae (Daubert, Bruening, and Najarian, 1978; Stanley et al., 1978; Zalloua, Buzayan, and Bruening, 1996), Comoviridae (Jaegle, Wellink, and Goldbach, 1987) and Potyviridae (Murphy et al., 1990; Murphy et al., 1991; Oruetxebarria et al., 2001). The sizes and the functions of VPg proteins vary. Calicivirus genomes possess a 5’-end-linked VPg protein that has been implicated in both translation and replication (Daughenbaugh et al., 2003; Goodfellow et al., 2005; Mitra, Sosnovtsev, and Green, 2004; Rohayem et al., 2006b). This study focused on the characterization of the nucleotidylylation reaction that would link covalently the norovirus VPg to a nucleotide, presumably the first step in the attachment of VPg to the RNA genome and the initiation of RNA synthesis by the RdRp on an RNA template. In this study, we demonstrate that the norovirus VPg can be nucleotidylated by the ProPol form of the norovirus RdRp in a template-independent reaction when Mn2+ is the divalent cation. Our data showed that the presence of a His tag on the VPg protein did not have a deleterious effect on nucleotidylylation, and various forms of recombinant norovirus RdRp including rPro-Pol, rHis-Pro-Pol and rHis-E1189A/ProPol had similar activities. Uridylylation assays, conducted in the presence of poly (A) template, resulted in an increase in the uridylylation of VPg, and in possible de novo synthesis of poly(U) molecules by a possible copy-back mechanism. Our results and a previous study (Rohayem et al., 2006b) suggested that VPg, if functioning as primer, might be covalently linked to nascent poly(U) molecules. However, our RIPA experiments (Figure 6C) indicated that the VPg was uridylylated, but not covalently linked to the newly-synthesized radiolabeled RNA products. Similar observations were made in our analysis of the nucleotidylylation of VPg in the presence of MD145 FL RNA and Mg2+ (Figure 7B), in which we could not show evidence for covalent linkage to the newly-synthesized RNA products generated in vitro. It will be important to verify whether nucleotidylylated VPg is performing a priming function in calicivirus-infected cells as suggested recently (Rohayem et al., 2006b), or is simply part of the mechanism by which the VPg becomes linked to the viral RNA so that it can function in translation.
Similarities of the calicivirus nucleotidylylation reaction with those of other positive strand RNA viruses with VPg proteins such as the picornaviruses and potyviruses include the utilization of tyrosine as the site of RNA linkage in the VPg. We mapped tyrosine 27 of the MD145 norovirus VPg as the site of this linkage, which is conserved among the caliciviruses. The nucleotidylylation reaction of the MD145 VPg was template-independent in the presence of Mn2+, and was not specific for any one nucleotide. Similar observations have been reported in studies of the VPg proteins of RHDV and potyvirus (Machin, Martin Alonso, and Parra, 2001; Puustinen and Makinen, 2004). Although our data showed that the NoV VPg could be linked to any of the four nucleotides, GTP and UTP were the preferred bases. Interestingly, a recent study showed that NoV Pol initiated RNA synthesis with only GTP and UTP in the presence of poly(C) and poly(A) homopolymeric templates, respectively (Rohayem et al., 2006b). In the same study, the authors reported that uridylylation of the VPg protein in the presence of Mn2+ by Pol was strictly template-dependent (Rohayem et al., 2006b), which differed from our findings. Structural differences between Pol and ProPol might explain the variation in the requirement for a poly(A) template in uridylylation when Mn2+ is the divalent cation. The presence of Mn2+ in the reaction might induce a more relaxed conformation of the ProPol polymerase as suggested previously for RHDV Pol (Machin, Martin Alonso, and Parra, 2001).
Our study demonstrated that the ProPol form of the norovirus RdRp can mediate nucleotidylylation of the VPg, consistent with our previous studies showing that the norovirus RdRp is enzymatically active as a precursor protein (ProPol) or in its cleaved form (Pol) (Belliot et al., 2005). The MD145 ProPol and RHDV Pol behaved similarly in in vitro uridylylation assays in that the reaction was template-independent in the presence of Mn2+, the preferred divalent cation (Machin, Martin Alonso, and Parra, 2001). The similarities in nucleotidylation requirements between the ProPol and Pol forms of the RdRp from members of two distinct calicivirus genera suggest that both forms may have the capacity to mediate nucleotidylylation during virus replication. In MNV or in Norwalk virus replicon-bearing cells, the cleaved Pol form of the RdRp is predominant (Chang et al., 2006; Sosnovtsev et al., 2006), whereas in FCV- or RHDV-infected cells the ProPol form is predominant (Konig, Thiel, and Meyers, 1998; Sosnovtsev, Garfield, and Green, 2002). Although the abolition of the cleavage site between the Pro and Pol domains was recently shown to be lethal for MNV (Ward et al., 2007), it does not rule out a role for a transient form of the NoV ProPol precursor in the modification of VPg during replication. It will be of interest in future studies to examine whether the ProPol precursor plays a specific role in nucleotidylation during replication, and whether cellular factors or membranes are involved in this reaction. It should be noted that Pro and Pol domain of 3CDpro were shown to interact with the CRE of human rhinovirus 14 (Yang et al., 2004). It is possible that similar interactions occur in the noroviruses, but additional studies will be needed to map these interacting domains.
The mutagenesis analysis in this study provided evidence for the importance of structural requirements in the norovirus VPg. First, the N-terminal region of the VPg was required for efficient nucleotidylylation of the VPg. This region is rich in lysine residues, and similar to an NTP-binding motif in the potyvirus (potato A virus) VPg. It is possible that the N-terminus of the norovirus VPg might facilitate nucleotidylyation by recruitment of NTPs. Alternatively, the ablation of this region might simply disrupt the conformation of the site of nucleotidylylation at Tyr 27, affecting its presentation to the RdRp. The second observation was an enhancement of nucleotidylylation at Tyr 27 when the nearby tyrosine at residues 30, 41, or 46 was mutated to alanine. Because there is presently no reverse genetics system for the human noroviruses, it is difficult to assess the biological relevance of these mutations. However, a study using the FCV reverse genetics system showed that the mutation of tyrosine at positions other than 24 (residues 12, 76 or 104) were not lethal to the virus (Mitra, Sosnovtsev, and Green, 2004). It will be interesting to examine whether mutations of the tyrosine residues at positions 30, 41 or 46 of the MD145 VPg might actually increase the level of nucleotidylylation of the VPg protein during replication and affect viral growth.
We report in this study the first evidence for the presence of an RNA sequence element in the 3’-end region of the norovirus genome that could specifically and markedly enhance nucleotidylation of the VPg by the RdRp in the presence of Mg2+. The nucleotidylylation was NoV-specific and required the presence of poly (A)-tailed NoV RNA molecules. It is not yet clear whether this element is analogous to the CRE structure of picornaviruses. Of interest, deletional mutagenesis of the FCV genome showed evidence for the importance of sequences near the 3’-end of the genome in viral replication (Sosnovtsev et al., 2005). It is possible that a CRE-based system for nucleotidylylation might help the calicivirus replication machinery distinguish between viral and cellular messenger RNAs. Additional studies will be required to verify whether a CRE-like structure is present in the 3’-end of norovirus genomes, and functions during replication in cells. Replicon (Chang et al., 2006) and rescue systems (Chaudhry, Skinner, and Goodfellow, 2007; Ward et al., 2007) which have been published for human NoV and MNV, respectively, are promising tools to study the replication mechanisms of these viruses.
Materials and Methods
Viruses and materials
Norovirus strain MD145−12 (Hu/NoV/GII.4/MD145−12/1987/US) was described previously (Green et al., 2002), and the sequence of the viral genome was assigned GenBank accession number AY032605. Oligonucleotide primers were purchased from Invitrogen (Carlsbad, CA). [α-32P]-labeled nucleotides UTP, GTP, ATP or CTP at 400 Ci/mmol and [γ-32P]-GTP at 400 Ci/mmol were purchased from Amersham (Piscataway, NJ).
Plasmids and mutagenesis
A cDNA copy of the FL genomic or predicted subgenomic RNA of the MD145 norovirus strain was cloned into the Δlacα/T7-pSPORT1 vector (Belliot et al., 2003) so that the T7 promoter was engineered immediately upstream of the 5’-end. Briefly, two overlapping fragments corresponding to nucleotides (nt) 3398 through 5419 and nt 5419 through the 3’-end of the FL genome were amplified by RT-PCR and cloned into the vector pCR2.1 using the TOPO cloning system (Invitrogen). The fragment corresponding to positions 3393 through 5419 (amplified by RT-PCR with the forward primer 5’-CAAATGGGCATGCTTCTGACAGGATC-3’ and the reverse primer 5’-CGGTGAACGCGTTCCCCGCGAGGATTAC-3’ with an engineered MluI site, underlined) was excised from the pCR2.1 vector with BamHI and MluI and cloned into the corresponding sites of the previously described plasmid cORF1 (Belliot et al., 2003). The resulting construction was named T7-frag-I+II+III+IV+V. The fragment corresponding to the position 5419 through the 3’-end of the viral genome was amplified by RT-PCR using the forward primer 5’-GTAATCCTCGCGGGGAACGCGTTCAC-3’ (MluI site underlined) and the reverse 3’-end primer 5’-GCGGACGTC(T)30AAAAGACAC-3’ with an engineered poly (A) tail of 30 nucleotides (bold type) and an AatII restriction site (underlined) and cloned into pCR2.1 to generate an intermediate construct designated TOPO-frag VI+VII. TOPO-fragVI+VII and the construct T7-frag-I+II+III+IV+V were digested with MluI and AatII and ligated together. The final construction (designated FL/MD145−12) contained the FL genome of the MD145−12 norovirus strain with a poly(A) tail of 30 nucleotides at the 3’-end. For the subgenomic cDNA clone, the fragment corresponding to position 6831 through the poly(A) tract at the 3’-end of the viral genome was amplified by RT-PCR using the forward primer 5’-AACTGCAGTAATACGACTCACTATAGTGAATGAAGATGGCGTCGAGTG-3’ (the PstI site is underlined and the T7 promoter is in bold type) and the reverse 3’-end primer described above. The PCR fragment was cloned into the pCR2.1 vector and the intermediate plasmid construct was named TOPO-SG. The construct TOPO-SG was treated with PstI and HindIII and the fragment of interest containing the first 1778 bp of the 5’-end of the subgenomic cDNA was gel-purified prior to ligation into PstI/HindIII-treated ΔT7-pSPORT1. The resulting plasmid was named pSPORT-5’regSG. For the cloning of the 3’-end of the subgenomic cDNA, the plasmid constructs TOPO-SG and pSPORT-5’regSG were both treated by HindIII and AatII. A fragment of 752 bp representing the 3’-end of the subgenomic RNA was gel purified and ligated with pSPORT-5’regSG to generate the final construct pSPORT1-SG. The plasmid construct pSPORT1-SG contained the cDNA of the FL subgenomic RNA under the control of a T7 promoter, and a poly(A)30 tract at the 3’-end. The selected genomic and subgenomic clones were shown by sequence analysis to represent the consensus sequence of the viral RNA genome.
A list of the VPg plasmid constructs and their encoded proteins is shown in Figure 1A. All constructs were generated using the cORF1 construct as template, and verified by sequencing analysis. The constructs pET-VPg, pETPro, pET-His-Pro−Pol and pET-His-E1189A/ProPol were previously described (Belliot et al., 2005; Belliot et al., 2003).
The fragment corresponding to the VPg coding sequence was amplified by PCR with the forward primer 5’-CATGCCatgGGCAAGAAAGGGAAGAACAAGACTGGCCG-3’ which included a NcoI site (underlined), a start codon (lowercase), and the reverse primer 5’-AAGGAAAAAAGCGGCCGCtcaGTGGTGGTGGTGGTGGTGCT CAAAACTGAGTCT-3’ which included a NotI site (underlined), a stop codon (lowercase), and a His tag-encoding sequence (bold type). The fragment was digested with NcoI and NotI and cloned into the corresponding restriction enzyme sites of the pET-28(b) vector (Novagen, Madison, WI) to generate pET-VPg-His. pET-VPg-His was used as template to generate the plasmid constructions pET-Δ3-VPg-His, pET-Δ8-VPg-His and pET-Δ20-VPg-His by using PCR amplicons produced with the forward primers 5’-CATGCCatgGGGAAGAACAAGACTGGCCGTGG-3’ (Δ3), 5’-CATGCCatgGGCCGTGGCAAGAAGCACACAGC-3’ (Δ8), 5’-CATGCCatgGGTCTCAGTGATGAAGAGTACGATGAG-3’ (Δ20) (start codon is in lowercase and NcoI site is underlined), respectively, and the reverse primer used to engineer pET-VPg-His.
The plasmid pET-VPg, described previously (Belliot et al., 2003), was used as template for site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, CA) of the Tyr codons at positions 27, 30, 41, 46, 53 and 54 of the VPg protein (Figure 1A) into Ala residues, to produce the DNA constructs pET-His-Y27A/VPg, pET-His-Y30A/VPg, pET-His-Y41A/VPg, pET-His-Y46A/VPg, pETHis-Y53A/VPg and pET-His-Y54A/VPg, respectively. The plasmid construction pET-His-Y27A/VPg was used as template to produce the double-mutated VPg DNA constructs pET-His-Y27,30A/VPg, pET-His-Y27,41A/VPg, pET-His-Y27,46A/VPg, pET-His-Y27,53A/VPg, and pET-His-Y27,54A/VPg. To produce the DNA construct pET-His-Y27,30,41,46,53,54A/VPg, the construction pET-His-Y27,46A/VPg was sequentially mutagenized to produce the intermediate construct pET-His-Y27,46,53,54A/VPg, which was then used as template to obtain the final construct pET-His-27,30,41,46,53,54A/VPg. Primers used in the VPg mutagenesis experiments are shown in Table 1. All mutagenized VPg constructs were verified by sequencing analysis.
The SUMO expression system (LifeSensors, Malvern, PA.) was used to produce and purify recombinant MD145 VPg that would represent the native protein without a His-tag. The plasmid pET-VPg was employed as template for the PCR amplification of the VPg coding sequence with the forward primer 5’-AAGGAAAAAACGTCTCAAGGTGGCAAGAAAGGGAAGAACAAGACTGG-3’ (the BsmBI site is underlined) and the reverse primer 5’-AAGGAAAAAAGCGGCCGCTCACTCAAAACTGAGTCTCTCATTGTAGTC-3’ (the NotI site is underlined). The resulting DNA fragment was treated with BsmBI and NotI prior to ligation into the BsaI and NotI sites of the pSUMO plasmid. The final construct, designated pSUMO-VPg, was engineered to express the VPg protein fused at its N-terminus to a His-tagged SUMO peptide (Figure 1B). A cleavage site for the SUMO-specific ubiquitin-like protease (Ulp1) was incorporated into the C-terminus of the SUMO peptide. Cleavage of the SUMO-VPg fusion protein by the SUMO-specific Ulp1 proteinase released the VPg with its precise N-terminus and the efficiency of the cleavage was monitored by SDS-PAGE analysis. The plasmid pSUMO-VPg was used as template for site-directed mutagenesis to produce the plasmids pSUMO-Y27A/VPg and pSUMO-Y30A/VPg. The primers used for the mutagenesis are described above for the DNA constructs pET-His-Y27A/VPg and pET-His-Y30A/VPg.
Protein synthesis and purification
pET and pSUMO-related constructs were used to transform E. coli BL21(DE3) cells (Novagen). The protein expression and purification protocols were previously described (Belliot et al., 2005). Briefly, all recombinant VPg proteins were characteristically found in the soluble fraction of E. coli BL21(DE3) cells following induction of expression with 1mM IPTG (Isopropyl β-D-1-thiogalactopyranoside). The recombinant proteins were bound to a Ni-NTA column and eluted with buffer (300 mM NaCl, 50 mM NaH2PO4, 10% glycerol, pH 8) containing 60 mM imidazole. The recombinant proteins His-VPg, VPg-His, His-Y27A/VPg, His-Y30A/VPg, His-Y41A/VPg, His-Y46A/VPg, His-Y453A/VPg, His-Y54A/VPg, His-Y27,30A/VPg, His-Y27,41A/VPg, His-Y27,46A/VPg, His-Y27,53A/VPg, His-Y27,54A/VPg, His-Y27,30,41,46,53,54A/VPg, Δ3-VPg-His, Δ8-VPg-His, Δ20-VPg-His, Pro, His-Pro-Pol and His-E1189A/ProPol were dialyzed overnight at 4°C against 150 mM KCl, 50 mM Tris pH 8, 2 mM DTT and 20% glycerol, aliquoted, and stored at −80°C. The recombinant proteins rVPg-His, rΔ3VPg-His, rΔ8VPg-His and rΔ20VPg-His, in which the His-tag was engineered at the C-terminus, were analyzed by direct N-terminal sequencing using the Edman degradation method. Although an initiating methionine had been engineered at the beginning of each protein, only the rVPg-His retained the methionine residue following expression and purification. The truncated proteins began at the expected amino acid residues 4 (rΔ3VPg-His), 9 (rΔ8VPg-His) and 21 (rΔ20VPg-His). The proteins His-SUMO-VPg, His-SUMO-Y27A/VPg and His-SUMO-Y30A/VPg were dialyzed overnight at 4°C against PBS pH 7.4, 10% glycerol and 2 mM DTT. To remove the SUMO tag, the purified fusion proteins were diluted to 1 mg/ml and incubated with His-tagged Ulp1 (1 unit per 20 μg) (Life Sensors, Inc) at 4°C, overnight. The protein mixture was then diluted to a final volume of 2 ml with a binding buffer containing 300 mM NaCl, 50 mM NaH2PO4 (pH8) and 10% glycerol. Two hundred fifty microliters of Ni-NTA resin slurry (Qiagen, Valencia, Calif) at 50% (vol:vol) were added to the mixture and incubated for 1 h at 4°C. The mixture was centrifuged at 10000x g for 10 min after the incubation and the supernatant containing the purified tag-free recombinant protein (rVPg, rY27A/VPg and rY30A/VPg) was collected. The volume of the supernatant was reduced to 1 ml by using a 10 kDa-cutoff Centricon column (Millipore, Bedford MA) that was centrifuged twice at 3000xg for 45 min. The concentrated proteins were dialyzed overnight at 4°C against 150 mM KCl, 50 mM Tris-HCl (pH 8), 2 mM DTT and 20% glycerol. The dialyzed protein was concentrated further with a 10kDa-cutoff Microcon column (Millipore) that was centrifuged twice at 10000xg for 30 min. The protein quantity was measured by using the Bradford method and the proteins were stored at −80°C.
Nucleotidylylation assay
The nucleotidylylation assay was adapted from Machin et al. (Machin, Martin Alonso, and Parra, 2001) and carried out for 3h at 30°C in a 15 μl reaction mixture containing 50 mM HEPES, 10 mM DTT, 0.5 or 1 mM MnCl2, 67 μM ZnCl2, 10 μM NTP (GTP, UTP, CTP or ATP), 5 μCi [α-32P] NTP (GTP, UTP, CTP or ATP) or [γ-32P]-GTP, 1 μg of rHis-Pro−Pol , rE1189A-ProPol or tag-free rPro−Pol (Belliot et al., 2005) and 0.5 to 3 μg of rHis-VPg. Five μl of the reaction were resolved by SDS-PAGE, unless indicated.
For nucleotidylylation assays in the presence of heteropolymeric RNA templates, RNA transcripts corresponding to the norovirus strain MD145 FL genomic and subgenomic RNAs were synthesized either from AatII-linearized plasmid constructs or PCR fragments using a Ribomax kit (Promega) with T7 polymerase according to the manufacturer's protocol. Following DNAse treatment, transcripts derived from PCR products were further purified with the MEGAclear kit (Ambion). The integrity of all transcribed RNA (following quantification) was monitored by gel electrophoresis. A typical reaction contained 50 mM HEPES, 10 mM DTT, 5 mM MgCl2, 67 μM ZnCl2, 50 mM potassium acetate, 30 μM of each NTP (GTP, UTP, CTP or ATP), 15 μCi [α-32P] UTP, 2.5 μg of rHis-E1189A/ProPol, 2.5 μg of rVPg-His or rHis-Y27A/VPg, and 1 to 5 μg of heteropolymeric RNA or poly r(A)n. The assay was carried out in a 25 μl reaction mixture for 3 h at 30°C. Following the assay, the mixture was heated at 95°C for 5 min, and 5 μl of the reaction were analyzed by SDS-PAGE. In some experiments, the remaining reaction mixture was analyzed in a RIPA (see below).
Enzymatic treatment of VPg
[32P] -labeled uridylylated VPg was dialyzed against 1000 volumes of buffer containing 50 mM NaCl, 50 mM Tris-HCl (pH 8.0) before it was subjected to treatment with proteinase K (0.5 mg/ml, Invitrogen), CIAP (500 U/ml, Invitrogen), or RNase (25 μg/ml, Roche) according to protocols recommended by the manufacturers. For P1 treatment [α32P]–labeled RNase-treated VPg was dialyzed against P1 reaction buffer (0.2 mM ZnCl2 , 2 mM MgCl2, 50 mM Na acetate, pH 5.3) and then was treated with P1 nuclease (MP Biomedicals, Solon, Ohio) at 0.5 u per one μg of VPg for 1 to 3 h at 37°C. For PDE1 treatment, the P1 nuclease-treated VPg was dialyzed against 1000 volumes of buffer containing 50 mM NaCl, 50 mM Tris-HCl (pH8.0) and then against 1000 x volumes of 0.5 x PDE1 reaction buffer (0.05 M TrisHCl pH8.9, 0.05 M NaCl, 7.5 mM MgCl2). The VPg solution was adjusted to 0.11 M TrisHCl (pH8.9), 0.11 M NaCl, 15 mM MgCl2 (1 x PDE1 buffer) using 2x PDE1 buffer and digestion with PDE1 (Worthington, 0.5 u per one μg of VPg) was performed at either 25°C or 37°C for 1 to 3 h.
Following all enzymatic treatments, the VPg was subjected to SDS-PAGE analysis (see below). The integrity of the protein was determined by visualization with Coomassie blue (Gel-code Blue Stain Reagent from Pierce, Rockford, IL), after which the gel was dried and subjected to autoradiography with X-Omat AR film (Kodak, Rochester, NY).
Protein electrophoresis and immunoprecipitation assay
Nucleotidylylation assay products were analyzed by SDS-PAGE on a 4−12% polyacrylamide gel (NUPAGE system from Invitrogen) with 1XMOPS running buffer (Invitrogen), unless indicated. The proteins were stained with Gel-code Blue Stain Reagent, after which the gel was dried and exposed to film. To estimate the amount of NMP incorporated into VPg, the band corresponding to VPg was excised, placed in 5 mls liquid scintillation fluid (ICN, Costa Mesa, CA), and the radioactivity was counted using a Wallac 1409 liquid scintillation counter (Perkin-Elmer). Radiolabeled mutated VPg proteins (rHis-Y/A-VPg) were also analyzed with a Phosphorimager (Molecular Dynamics) and the IP Lab Gel suite (SignalAnalytics, Vienna, VA).
The MD145 VPg-specific antiserum (post-immunization), used in the RIPA, was prepared in guinea pigs as described previously (Belliot et al., 2003). Serum from the animal collected before immunization (pre) served as a control for antibody specificity.
Acknowledgments
We thank Tanaji Mitra (NIAID, NIH) for technical assistance. We extend our appreciation to Dr Jerry Keith for helpful discussions and Dr. Albert Z. Kapikian (NIAID, NIH) for support of this project. This work was supported by the Intramural Research Program of NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Stanislav V. Sosnovtsev : SSOSNOVTSE@mail.nih.gov Kyeong-Ok Chang : kchang@vet.k-state.edu Peter McPhie : pmcphie@helix.nih.gov
References
- Ambros V, Baltimore D. Protein is linked to the 5′ end of poliovirus RNA by a phosphodiester linkage to tyrosine. J Biol Chem. 1978;253(15):5263–6. [PubMed] [Google Scholar]
- Belliot G, Noel JS, Li JF, Seto Y, Humphrey CD, Ando T, Glass RI, Monroe SS. Characterization of capsid genes, expressed in the baculovirus system, of three new genetically distinct strains of “Norwalk-like viruses”. J Clin Microbiol. 2001;39(12):4288–95. doi: 10.1128/JCM.39.12.4288-4295.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliot G, Sosnovtsev SV, Chang KO, Babu V, Uche U, Arnold JJ, Cameron CE, Green KY. Norovirus proteinase-polymerase and polymerase are both active forms of RNA-dependent RNA polymerase. J Virol. 2005;79(4):2393–403. doi: 10.1128/JVI.79.4.2393-2403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belliot G, Sosnovtsev SV, Mitra T, Hammer C, Garfield M, Green KY. In vitro proteolytic processing of the MD145 norovirus ORF1 nonstructural polyprotein yields stable precursors and products similar to those detected in calicivirus-infected cells. J Virol. 2003;77(20):10957–74. doi: 10.1128/JVI.77.20.10957-10974.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KO, Sosnovtsev SV, Belliot G, King AD, Green KY. Stable expression of a Norwalk virus RNA replicon in a human hepatoma cell line. Virology. 2006;353(2):463–73. doi: 10.1016/j.virol.2006.06.006. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y, Nayak A, Bordeleau ME, Tanaka J, Pelletier J, Belsham GJ, Roberts LO, Goodfellow IG. Caliciviruses differ in their functional requirements for eIF4F components. J Biol Chem. 2006;281(35):25315–25. doi: 10.1074/jbc.M602230200. [DOI] [PubMed] [Google Scholar]
- Chaudhry Y, Skinner MA, Goodfellow IG. Recovery of genetically defined murine norovirus in tissue culture by using a fowlpox virus expressing T7 RNA polymerase. J Gen Virol. 2007;88(Pt 8):2091–100. doi: 10.1099/vir.0.82940-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubert SD, Bruening G, Najarian RC. Protein bound to the genome RNAs of cowpea mosaic virus. Eur J Biochem. 1978;92(1):45–51. doi: 10.1111/j.1432-1033.1978.tb12721.x. [DOI] [PubMed] [Google Scholar]
- Daughenbaugh KF, Fraser CS, Hershey JW, Hardy ME. The genome-linked protein VPg of the Norwalk virus binds eIF3, suggesting its role in translation initiation complex recruitment. Embo J. 2003;22(11):2852–9. doi: 10.1093/emboj/cdg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushi S, Kojima S, Takai R, Hoshino FB, Oka T, Takeda N, Katayama K, Kageyama T. Poly(A)- and primer-independent RNA polymerase of Norovirus. J Virol. 2004;78(8):3889–96. doi: 10.1128/JVI.78.8.3889-3896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow I, Chaudhry Y, Gioldasi I, Gerondopoulos A, Natoni A, Labrie L, Laliberte JF, Roberts L. Calicivirus translation initiation requires an interaction between VPg and eIF 4 E. EMBO Rep. 2005;6(10):968–72. doi: 10.1038/sj.embor.7400510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY, Belliot G, Taylor JL, Valdesuso J, Lew JF, Kapikian AZ, Lin FY. A predominant role for Norwalk-like viruses as agents of epidemic gastroenteritis in Maryland nursing homes for the elderly. J Infect Dis. 2002;185(2):133–46. doi: 10.1086/338365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green KY, Chanock RM, Kapikian AZ. Human caliciviruses. In: Knipe DM, Howley PM, editors. Fields virology. Lippincott, Williams & Wilkins; Philadelphia, PA: 2001. 1. 2 vols. [Google Scholar]
- Herbert TP, Brierley I, Brown TD. Identification of a protein linked to the genomic and subgenomic mRNAs of feline calicivirus and its role in translation. J Gen Virol. 1997;78(Pt 5):1033–40. doi: 10.1099/0022-1317-78-5-1033. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Wellink J, Goldbach R. The genome-linked protein of Cowpea Mosaic Virus is bound to the 5′ terminus of virus RNA by a phosphodiester linkage to serine. Journal of General Virology. 1987;68:627–632. [Google Scholar]
- Jiang X, Wang M, Wang K, Estes MK. Sequence and genomic organization of Norwalk virus. Virology. 1993;195(1):51–61. doi: 10.1006/viro.1993.1345. [DOI] [PubMed] [Google Scholar]
- Konig M, Thiel HJ, Meyers G. Detection of viral proteins after infection of cultured hepatocytes with rabbit hemorrhagic disease virus. J Virol. 1998;72(5):4492–7. doi: 10.1128/jvi.72.5.4492-4497.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambden PR, Caul EO, Ashley CR, Clarke IN. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science. 1993;259(5094):516–9. doi: 10.1126/science.8380940. [DOI] [PubMed] [Google Scholar]
- Lopez Vazquez AL, Martin Alonso JM, Parra F. Characterisation of the RNA-dependent RNA polymerase from Rabbit hemorrhagic disease virus produced in Escherichia coli. Arch Virol. 2001;146(1):59–69. doi: 10.1007/s007050170191. [DOI] [PubMed] [Google Scholar]
- Machin A, Martin Alonso JM, Parra F. Identification of the amino acid residue involved in rabbit hemorrhagic disease virus VPg uridylylation. J Biol Chem. 2001;276(30):27787–92. doi: 10.1074/jbc.M100707200. [DOI] [PubMed] [Google Scholar]
- Mitra T, Sosnovtsev SV, Green KY. Mutagenesis of tyrosine 24 in the VPg protein is lethal for feline calicivirus. J Virol. 2004;78(9):4931–5. doi: 10.1128/JVI.78.9.4931-4935.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy JF, Rhoads RE, Hunt AG, Shaw JG. The VPg of tobacco etch virus RNA is the 49-kDa proteinase or the N-terminal 24-kDa part of the proteinase. Virology. 1990;178(1):285–8. doi: 10.1016/0042-6822(90)90405-g. [DOI] [PubMed] [Google Scholar]
- Murphy JF, Rychlik W, Rhoads RE, Hunt AG, Shaw JG. A tyrosine residue in the small nuclear inclusion protein of tobacco vein mottling virus links the VPg to the viral RNA. J Virol. 1991;65(1):511–3. doi: 10.1128/jvi.65.1.511-513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KK, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JM, Parra F. Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J Biol Chem. 2004;279(16):16638–45. doi: 10.1074/jbc.M400584200. [DOI] [PubMed] [Google Scholar]
- Oruetxebarria I, Guo D, Merits A, Makinen K, Saarma M, Valkonen JP. Identification of the genome-linked protein in virions of Potato virus A, with comparison to other members in genus Potyvirus. Virus Res. 2001;73(2):103–12. doi: 10.1016/s0168-1702(00)00216-1. [DOI] [PubMed] [Google Scholar]
- Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol. 2000;74(22):10359–70. doi: 10.1128/jvi.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AV, van Boom JH, Filippov D, Wimmer E. Protein-primed RNA synthesis by purified poliovirus RNA polymerase. Nature. 1998;393(6682):280–4. doi: 10.1038/30529. [DOI] [PubMed] [Google Scholar]
- Puustinen P, Makinen K. Uridylylation of the potyvirus VPg by viral replicase NIb correlates with the nucleotide binding capacity of VPg. J Biol Chem. 2004;279(37):38103–10. doi: 10.1074/jbc.M402910200. [DOI] [PubMed] [Google Scholar]
- Richards OC, Spagnolo JF, Lyle JM, Vleck SE, Kuchta RD, Kirkegaard K. Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase. J Virol. 2006;80(15):7405–15. doi: 10.1128/JVI.02533-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohayem J, Jager K, Robel I, Scheffler U, Temme A, Rudolph W. Characterization of norovirus 3Dpol RNA-dependent RNA polymerase activity and initiation of RNA synthesis. J Gen Virol. 2006a;87(Pt 9):2621–30. doi: 10.1099/vir.0.81802-0. [DOI] [PubMed] [Google Scholar]
- Rohayem J, Robel I, Jager K, Scheffler U, Rudolph W. Protein-primed and de novo initiation of RNA synthesis by norovirus 3Dpol. J Virol. 2006b;80(14):7060–9. doi: 10.1128/JVI.02195-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothberg PG, Harris TJ, Nomoto A, Wimmer E. O4-(5′-uridylyl)tyrosine is the bond between the genome-linked protein and the RNA of poliovirus. Proc Natl Acad Sci U S A. 1978;75(10):4868–72. doi: 10.1073/pnas.75.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev S, Green KY. RNA transcripts derived from a cloned full-length copy of the feline calicivirus genome do not require VpG for infectivity. Virology. 1995;210(2):383–90. doi: 10.1006/viro.1995.1354. [DOI] [PubMed] [Google Scholar]
- Sosnovtsev SV, Belliot G, Chang KO, Onwudiwe O, Green KY. Feline calicivirus VP2 is essential for the production of infectious virions. J Virol. 2005;79(7):4012–24. doi: 10.1128/JVI.79.7.4012-4024.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev SV, Belliot G, Chang KO, Prikhodko VG, Thackray LB, Wobus CE, Karst SM, Virgin HW, Green KY. Cleavage map and proteolytic processing of the murine norovirus nonstructural polyprotein in infected cells. J Virol. 2006;80(16):7816–31. doi: 10.1128/JVI.00532-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev SV, Garfield M, Green KY. Processing map and essential cleavage sites of the nonstructural polyprotein encoded by ORF1 of the feline calicivirus genome. J Virol. 2002;76(14):7060–72. doi: 10.1128/JVI.76.14.7060-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnovtsev SV, Green KY. Identification and genomic mapping of the ORF3 and VPg proteins in feline calicivirus virions. Virology. 2000;277(1):193–203. doi: 10.1006/viro.2000.0579. [DOI] [PubMed] [Google Scholar]
- Stanley J, Rottier P, Davies JW, Zabel P, Van Kammen A. A protein linked to the 5′ termini of both RNA components of the cowpea mosaic virus genome. Nucleic Acids Res. 1978;5(12):4505–22. doi: 10.1093/nar/5.12.4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward VK, McCormick CJ, Clarke IN, Salim O, Wobus CE, Thackray LB, Virgin H. W. t., Lambden PR. Recovery of infectious murine norovirus using pol II-driven expression of full-length cDNA. Proc Natl Acad Sci U S A. 2007;104(26):11050–5. doi: 10.1073/pnas.0700336104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L, Huhn JS, Mory A, Pathak HB, Sosnovtsev SV, Green KY, Cameron CE. Proteinase-polymerase precursor as the active form of feline calicivirus RNA-dependent RNA polymerase. J Virol. 2001;75(3):1211–9. doi: 10.1128/JVI.75.3.1211-1219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Rijnbrand R, Watowich S, Lemon SM. Genetic evidence for an interaction between a picornaviral cis-acting RNA replication element and 3CD protein. J Biol Chem. 2004;279(13):12659–67. doi: 10.1074/jbc.M312992200. [DOI] [PubMed] [Google Scholar]
- Zalloua PA, Buzayan JM, Bruening G. Chemical cleavage of 5′-linked protein from tobacco ringspot virus genomic RNAs and characterization of the protein-RNA linkage. Virology. 1996;219(1):1–8. doi: 10.1006/viro.1996.0216. [DOI] [PubMed] [Google Scholar]