Abstract
Natural killer cells are important in innate defense against viral infections. The interplay between stimulatory and inhibitory natural killer cell receptors and their corresponding human leukocyte antigen ligands are known to influence the outcome of acute Hepatitis C virus infection. Frequencies of NK receptor genes (8 inhibitory, 6 activating and 2 pseudogenes) and HLA class II alleles (DRB1, DQB1) were analyzed in 160 Puerto-Rican American drug users with Hepatitis C virus infection; 121 had chronic viremia (CV) and 39 were spontaneous clearance (SC). We further ruled out genetic stratification using short tandem repeats. Interaction between KIR gene receptor 2DL3/2DL3 and its ligand, C1/C1 of HLA-Cw alleles and spontaneous clearance was confirmed (p value = 0.03, OR = 3.05). We also found a new interaction between the KIR receptor gene 2DL3 with HLA-DRB1*1201 (p value = 0.0001 OR: 22) associated with SC, and an association of HLA DQB1*0501 (p value= 0.05 OR 0.30) with CV. Our findings suggested a role for MHC class II alleles in Hepatitis C virus peptide presentation to T cells together with NK ligand interaction involving pathways that will be useful for the development of immunotherapeutic interventions.
Keywords: HCV, KIR, HLA-C, genetic interactions, clinical outcome
Introduction
Natural killer (NK) cells are important in the innate defense against viral infections (Kirwan, 2007). NK cells express surface binding proteins (killer cell immunoglobulin-like receptors or KIRs) that have the capacity to bind ligands on the surface of most cells. The interplay between stimulatory and inhibitory KIRs and their corresponding HLA ligands is likely to play a role in the outcome of acute Hepatitis C viral (HCV) infection, leading to either chronic viremia (CV) or spontaneous viral clearance (SC).KIRs are encoded in the leukocyte receptor complex of chromosome 19q13.4 and recognize HLA-class I molecules as ligands (Vilches and Parham, 2002). Fourteen functional KIR genes have been characterized; eight inhibitory: 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 3DL1, 3DL2, and 3DL3, which encode for receptors that when bound to HLA class I molecules inhibit NK cell function. Six genes are activating: 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 3DS1, and 2 of the loci are pseudogenes: 2DP1, 3DP1 (Uhrberg et al., 1997). The variable content of KIR inhibitory and activating genes within individuals and significant allelic polymorphisms of the known KIR genes has been extensively described (Hsu et al., 2002).
Previous studies demonstrated that KIR receptor genes and major histocompatibility complex (MHC) class I and II loci helped determining the clinical outcome of HCV infection, summarized in table 1. However, the conflicting results found could be due in part to the heterogeneity of the populations within the case-control studies or among ethnic groups and to the mode of transmission (transfusion vs. injection drug users). (Lopez-Vazquez et al., 2005; Thio et al., 2002; Thio et al., 2001; Fanning et al., 2000; Thursz et al., 1999; Alric et al., 1997; Cramp et al., 1998; Fanning et al., 2004; Azocar et al., 2003; Mangia et al., 1999; McKierman et al., 2004; McKierman et al., 2000; Barrett et al., 1999; Lechmann et al., 1999; Yee, 2004; Zavaglia et al., 1998; Minton et al., 1998; Holer et al., 1997; Yenigun and Durunipar, 2002) Indeed, none of the published studies performed analyses while investigating genetic stratification based on population specific DNA markers. We sought to characterize KIR-HLA associations on genetically comparable case- control population. In this report we confirmed a previously defined KIR/ligand interaction in HCV SC (KIR 2DL3/3DL3 with HLA-C1/C1) (Khakoo et al, 2004) and the association of DQB1*0501 with CV (Azocar et al, 2003). In addition we described a new genetic interaction between the KIR gene 2DL3 and HLA-DRB*1201 in spontaneous clearance of HCV.
Table 1.
Summary of Associations of SC and CV with HLA-class I, II and KIR.
| Injection Drug Users (IDU) | Blood Transfusion | IDU and Blood Transfusion | Unknown | |||||
|---|---|---|---|---|---|---|---|---|
| HLA association | SC | CV | SC | CV | SC | CV | SC | CV |
| Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
Reference (Ethnicity) |
|
| HLA-DRB1*0101 | (Mangia, 1999; McKierman 2004, Fanning, 2000; McKierman 2000;Barret,1999) (C) | (Thio, 2001) (AA and C) | ||||||
| HLA-DRB1*0104 | (Cramp, 1998) (C) | |||||||
| HLA-DRB1*0301 | (McKierman 2004) (C) | (Thio, 2001) (C) | (Yenigun, 2002) (C) | |||||
| HLA-DRB1*0401 | (McKierman 2000) (C) | (Cramp, 1998)(C) | ||||||
| HLA-DRB1*0701 | (Thursz, 1999) (C) | |||||||
| HLA-DRB1*1101 | Present report (H) (trend) | (Thursz, 1999) (C) | (Holer, 1997) (T) | |||||
| HLA-DRB1*1201 | Present report (H) | |||||||
| HLA-DRB1*1301 | (Holer, 1997) (C) | |||||||
| HLA-DRB1*1501 | (Lechmann 1999) (C) | (Lechmann, 1999) (C) | ||||||
| HLA-DQA1*0103 | (Holer, 1997) (C) | |||||||
| HLA-DQA1*0201 | (Zavaglia, 1998) (C) | |||||||
| HLA-DQA1*0301 | (Cramp, 1998) (C) | |||||||
| HLA-DQA1*0801 | (Zavaglia 1998) (C) | |||||||
| HLA-DQB1*0201 | (McKierman 2004) (C) | (Zavaglia, 1998) (C) | ||||||
| HLA-DQB1*0301 | (Thio, 2001), (Cramp,1998) (AA and C) | |||||||
| HLA-DQB1*0501 | (Azocar, 2003) (trend)(H) Present report (H) | (McKierman 2004) (C) | (Thio, 2001) (AA and C) | |||||
| HLADRB1*1101/DQB1*0301 | (Yee, 2004) (C) | (Thursz, 1999; Alric, 1997; Minton 1998) (C) | ||||||
| HLA-DRB1*1104/DQB1*0301 | (Mangia 1999)(C) | (Zavaglia, 1998) (C) | ||||||
| HLA-DRB1*12 | ||||||||
| HLA C1/C1/KIR 2DL3/2DL3 | (Khakoo, 2004)(AA); | (Khakoo, 2004) (C) | ||||||
| HLA C1/C1/KIR2DL3/2DL3 + HLADRB1*1201 | ||||||||
SC: Spontaneous Clearance; CV: Chronic Viremia; C: Caucasian; AA: African American; Hispanic;
Materials and Methods
Human Subjects
160 drug users with evidence of HCV infection were studied. Thirty-nine HCV infected patients (24.4%) underwent spontaneous clearance (SC) with anti-HCV IgG detected by both third-generation enzyme immunoassay (EIA) and recombinant immunoblot assay (RIBA) and were negative for HCV-RNA in serum in 2 successive samples at least 6 months apart. Chronic viremic HCV patients (N = 121) (75.6%) had anti-HCV antibodies and detectable HCV-RNA in serum for more than six months with or without persistent increased ALT levels. HCV RNA was determined by COBAS Amplicor HCV Kit (Roche Diagnostic Systems, Branchburg, NJ, USA; lower limit of detection: 50 IU/mL). HCV genotypes were determined by restriction fragment length polymorphism (RFLP) analysis of the product of the RT-PCR reaction.
All subjects had Puerto Rican ethnic background and were recruited from the HCV Community Screening and Counseling Programs at the Primary Care Clinic of the Northgate Medical Center in Springfield, Massachusetts and gave written informed consent to participate. This study was approved by Institutional Review Boards.
HLA class I and class II typing
Genomic DNA from peripheral blood was obtained using (QIAGEN- DNA Blood, Qiagen Leusden, and The Netherlands). HLA typing was performed by PCR method with published primer sequences to amplify the HLA class I and II region and sequence-specific oligonucleotide probes (SSOP, HLA quick-type kits, Lifecodes, Stamford, CT, USA). To solve ambiguous PCR-SSOP typing, PCR-SSP (sequence-specific primer amplifications were used; Unitray SSP Pel-Freez; Milwaukee, WI, USA) according to the manufacturer’s instructions.
KIR typing
The presence or absence of KIR genes was detected by using PCR-SSP method (Hsu et al., 2002; Gomez-Lozano and Vilches 2002). The final concentration of each KIR-specific primer was approximately 1μM. As an internal control we amplified a 560 bp fragment of the HLA-DRA gene; P1: 5′-ACCTGTCACCACAGG-3′ and P2: 5′-CAGACCCACAGTCAGGCCC-3′). Control primers were used at 0.5 μM of final concentration in all PCR-SSP reactions: 100 ng of genomic DNA were amplified in 10 μl of PCR buffer (67 mM of Tris-HCl, pH 8.8, 16 mM of (NH4)2SO4, 2 mM MgCl2, 0.01 % Tween-20, 100 μM of dNTP’s), 1 μM of each primer and 0.4 U of Taq DNA polymerase (Roche Applied Science, IN, USA). PerkinElmer GeneAmp 9600 system with the following PCR conditions used denaturation for 2 minutes at 92°C, then 30 cycles of 10s at 92°C, 30s at 65°C and 90s at 68°C; and final extension at 68° for 10 min. Annealing temperatures were modified for primers amplifying KIR2DL2 (63°C), KIR2DS4D (2DS4 deletion in exon 5) (63°C), 2DS5 (63°C) and 2DS4 (61°C). Amplification products were electrophoresed on 1.4% agarose gels stained with ethidium bromide.
Short Tandem Repeats Genotyping
Fifteen autosomal Short Tandem Repeats (STR) markers (CSF1PO, FGA, THO1, TPOX, VWA, D3S11358, D5S818, D7S820, D8S1179, D13S317, D16S539, D18S51, D21S11, D19S433, D2S1338 and amelogenin) were typed using the Applied Biosystems AmpFl STR Identifiler Kit. PCR amplification was carried out on a Gene Amp 9600 thermocycler (Applied Biosystems, CA, USA) using 1 ng of DNA according to the manufacturer’s protocol. The PCR conditions were: 95° C for 11 min followed by 28 cycles of 94° C for 1 min, 59° C for 1 min, 72° C for 1 min followed by a hold at 60° C for 60 min. PCR products were diluted 1:15 in Hi- Di formamide and GS500-LIZ internal size standard and analyzed on the ABIPrism 3100 Genetic Analyzer (Applied Biosystems, CA, USA). Allele assignments were made using Genotype 3.7 software by comparison with kit allelic ladders (Applied Biosystems, CA, USA).
Statistical methods
To estimate genetic effects of each separate factor, with or without adjusting for the effects of other factors, we used Chi-square and Fisher’s exact test (if the count in a cell is less than 5) for 2 by 2 tables, and multiple logistic regression implemented in SAS version 9.1.3 for adjusting for other factors. The strength of association was expressed by the Odds Ratio (OR). For these analyses (tables 2–5) the frequency of observations in each row was compared to all other study participants. P values <0.05 were considered significant. In each case, the referent group consisted of those individuals that did not have the risk alleles of interest. To confirm the interaction effects of two risk factors, which were found by chi-square tests, we also used dummy explanatory variables (Suits, 1957) to represent subgroups indicating two independent effects and demonstrating a combined or joint effect in the multiple logistic regression model.
Table 2.
Demographics of SC and CV patients
| SC N= 39 | CV N=121 | P value (OR) | |
|---|---|---|---|
| Age mean | 37.8 | 39.9 | 0.77 (0.92) |
| ALT (U/L) mean | 37.29 | 90.79 | <0.0001 (0.06) |
| BMI mean (kg/m2) | 30 | 29 | 0.08 (1.05) |
| Female/male ratio | 10/29 | 31/90 | 0.9 (1.0) |
| HIV-HCV co-infection | 6 (15.4%) | 10/121 (8.3%) | 0.19 (2.02) |
| HCV genotype 1a* | 63/121 (52.1%) | ||
| HCV genotype 1b* | 18/121 (14.9%) | ||
| HCV genotype 2a* | 1/121 (0.8%) | ||
| HCV genotype 2b* | 14/121 (11.6%) | ||
| HCV genotype 3a* | 8/121 (6.6%) | ||
| HCV genotype 4a* | 7/121 (5.8%) | ||
| HCV genotype not determined | 10/121 (8.2%) |
HCV: Hepatitis C virus, SC: Spontaneous clearance, CV: Chronic viremia; BMI: Body mass Index’; OR: Odds Ratio.
Statistical differences were determined using Student’s unpaired t test.
Table 5.
Genetic interaction between NK genotype receptor and HLA- DR*1201.
| SC | CV | |||
|---|---|---|---|---|
| N= 39 | N=121 | |||
| n (%) | n (%) | OR (CI) | p value | |
| KIR 2DL3/2DL3 (−) + HLA-DRB1*1201 (−) | 16 (41.0) | 72 (59.5) | 1 | - |
| KIR 2DL3/2DL3 + HLA-DRB1*1201 (−) | 17 (43.6) | 45 (37.2) | 1.7 (0.72–3.98) | 0.17 |
| KIR 2DL3/2DL3 (−) + HLA-DRB1*1201 | 2 (5.1) | 2 (1.6) | 4.5 (0.30–64.9) | 0.17 |
| KIR 2DL3/2DL3 + HLA-DRB1*1201 | 4 (10.2) | 1 (0.8) | 18.0 (1.57–898.0) | 0.007 |
| KIR 2DL3 (one or two copies) (−) + HLA- DRB1*1201 (−) | 7 (17.9) | 36 (29.7) | 1 | - |
| KIR 2DL3 (one or two copies) + HLA- DRB1*1201 (−) | 26 (66.7) | 82 (67.8) | 1.63 (0.61–4.85) | 0.29 |
| 2DL3 one or two copies (−) + HLA- DRB1*1201 | 0 | 1 (0.8) | ND | 0.84 |
| 2DL3 one or two copies + HLA- DRB1*1201 | 6 (15.4) | 2 (1.6) | 15.4 (2.04–172.5) | 0.002 |
KIR inhibitory genotypes: 2DL3/2DL3: (2DL3 (+), 2DL2 (−), 2DS2 (−); 2DL3/2DL3 (−): 2DL2 (+), 2DL3 (−) 2DS2 (+) SC: Spontaneous clearance,
Population genetics and population stratification analysis
The ARLEQUIN v2.1 population genetics software was used to calculate Hardy-Weinberg Equilibrium (HWE) of STR in cases and controls. STR markers estimated the admixture of Puerto Ricans by the maximum likelihood method. Spaniards (Caucasians from Europe), Angolans and Amerindians were considered as parental populations and these previously published frequencies were used for the admixture estimation analysis. The STR frequencies of parental populations were downloaded from: http://www.uni-duesseldorf.de/WWW/MedFak/Serology/database.html database. Individual ancestry proportions in HCV infected individuals and in healthy controls were calculated using Structure 2.0 software. We compared STR in HCV infected patients using Chi-Square analysis, as previously described (Pritchard and Rosenberg, 1999) STRs with a frequency greater than 10 % were tested for association with the phenotype (HCV CV or SC), under the null hypothesis that there were no significant differences in the frequency of STR markers between the CV and SC groups. P values < 0.05 were considered significant CI 95%
Results
Baseline characteristics
The baseline characteristics of Puerto Rican American IDUs with positive tests for anti-HCV are shown in Table 2. Patients were divided into two groups according to their HCV-RNA status by RT-PCR, chronic viremia (CV) and spontaneous clearance (SC). CV patients had repeatedly quantifiable HCV-RNA (> 50 IU/mL), while SC patients had repeatedly negative tests for HCV RNA (< 50 IU/mL). There were no significant differences between SC and CV individuals with regard to age, gender, body mass index (BMI) or HIV-1/HCV co-infection. Genotype 1 (1a and 1b) HCV was most frequent, followed by genotypes 3 and 4. Also, CV patients had significantly higher abnormal serum ALT levels than those with SC, analyzed by t student test.
Admixture Estimation and Population Stratification analysis
No significant deviations from Hardy-Weinberg equilibrium in the gene frequencies of KIRs and HLA class I and II alleles in the two HCV infected groups were observed. Differences of the STR’s frequencies between groups, using the Chi-Square association statistic test (p < 0.05), were not detected. Furthermore, admixture estimation analysis using the Structure 2.0 program showed, the two HCV outcome groups have comparable population frequencies, SC: 73.5% Caucasian, 15.4 % African and 11.1% Amerindian and CV: 75.3 % Caucasian, 14.2% African and 10.5% Amerindian (Figure 1).Therefore, Puerto Rican Americans have a large Caucasian ancestry contribution and HCV individuals classified by clinical outcome did not differ in their admixture estimates
Figure 1. Triangle plot showing the ancestry estimates.
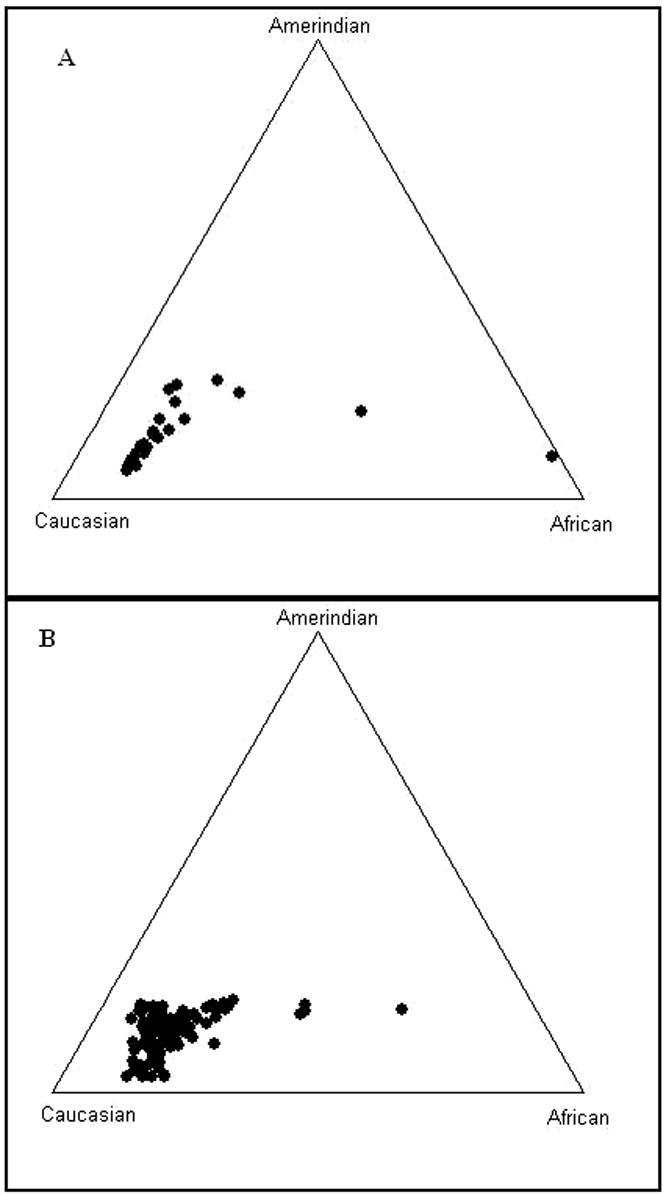
A) HCV spontaneous clearance (SC) Puerto Rican patients (N=39) (73.5% Caucasian, 15.4 % African and 11.1% Amerindian), and B) HCV chronic viremic (CV) Puerto Rican patients (N=121) (75.3% Caucasian, 14.2% African and 10.5% Amerindian) using a panel of 15 STR and 3 SNP unlinked markers with the maximum likelihood method and the computational program Structure 2.0. The parental populations are represented in each corner of the triangle. The circles represent individuals; which were clustered and distributed according to the ancestral proportion of the subject.
HLA class I alleles and activating genotypes and clinical outcome
We next analyzed the frequencies of KIR inhibitory and activating receptors in the two groups, SC and CV (Figure 2). Significant differences in the frequencies of inhibitory or activating KIR receptor genes between SC and CV were not detected. We analyzed the HLA-C groups: C1/C1: 2 copies of C1 alleles; C1/C1 (−): C1/C2 and C2/C2; one or two copies of C1: C1/C1 and C1/C2; and KIR inhibitory genotypes: KIR 2DL3/2DL3: 2DL3 (+), 2DL2 (−), 2DS2 (−); KIR 2DL3/2DL2 (presence of a copy of either KIR) and KIR 2DL2/2DL2: KIR2DL2 (+), KIR2DL3 (−). Different HLA class I alleles act as specific ligands for the different variants of KIRs. KIR2DL2, KIR2DL3 and KIR2DS2 interact with HLA-Cw group 1 alleles (termed C1 ligands) characterized by Ser77/Asn80, and KIR2DL1 and KIR2DS1 interact with HLA-Cw group 2 alleles (C2 ligands; Asn77/Lys80) (Uhrberg et al., 1997). In table 3, we confirmed the report (by Khakoo et al., 2004) of joint effects or biological interaction between HLA-C1/C1 and KIR2DL3/2DL3 homozygous genotypes: (11/39 (28.2%) in SC and 15/121 (12.5 % in CV p value= 0.03 OR = 3.05, 95% CI = 1.00–9.08). This interaction effect was evaluated by a multiple regression analysis in which we studied effects according to three groupings concurrently: C1/C1 + 2DL3/2DL3, C1/C1 (−) + 2DL3/2DL3 (+), and C1/C1 (+) + 2DL3/2DL3(−). In this joint analysis, an interaction effect of C1/C1 + 2DL3/2DL3 was confirmed (p value =0.0243 OR = 3.10 95% CI =1.16, 8.31) The single effects of C1/C1 (−) + 2DL3/2DL3 (p value= 0.66 OR = 1.23 95% CI = 0.48–3.19–9.08) and C1/C1 + 2DL3/2DL3 (−) (p value= 0.67 OR = 1.27, 95% CI = 0.43–3.8) were not significant.
Figure 2. NK receptor gene frequencies.
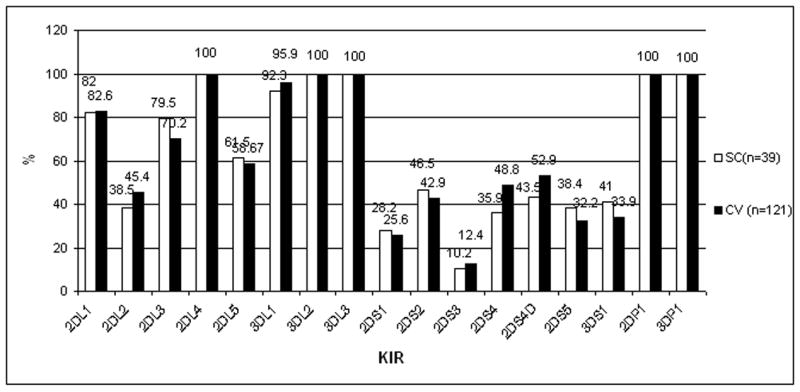
KIR inhibitory and activating genes in Puerto Rican patients with SC and CV. SC: Spontaneous Clearance; CV; Chronic Viremia.
Table 3.
Frequency of HLA-C and inhibitory KIR genotypes in HCV patients with Spontaneous Clearance and Chronic Viremia
| SC | CV | |||
|---|---|---|---|---|
| N = 3* | N = 121 | |||
| n (%) | n (%) | OR (CI) | p value | |
| HLA-C group genotypes | ||||
| C1/C1 (−) | 22 (56.5) | 85 (70.2) | 1 | - |
| C1/C1 | 17 (43.5) | 35 (28.9) | 1.88 (0.82–4.20) | 0.11 |
| Inhibitory genotypes | ||||
| 2DL3/2DL3 (−) | 18 (46.2) | 74 (61.2) | 1 | - |
| 2DL3/2DL3 | 21 (53.8) | 47 (38.8) | 1.84 (0.89–3 79) | 0.1 |
| HLA-C1+inhibitory genotypes | ||||
| C1/C1 (−) + 2DL3/2DL3(−) | 13 (33.3) | 54 (45.0) | 1 | - |
| C1/C1 (−) + 2DL3/2DL3 | 9 (23.1) | 31 (25.8) | 1.20 (0.40–3.46) | 0.8 |
| C1/C1 + 2DL3/2DL3 (−) | 6 (15.4) | 20 (16.7) | 1.25 (0.34–4.12) | 0.78 |
| C1/C1 +2DL3/2DL3 | 11 (28.2) | 15 (12.5) | 3.05 (1.00–9.08) | 0.03 |
C1/C1: 2 copies of C1 alleles; C1/C1 (− ): C1/C2 + C2/C2; KIR inhibitory genotypes: 2DL3/2DL3: (2DL3 (+), 2DL2 (−), 2DS2 (−); 2DL3/2DL3 (−): 2DL2 (+), 2DL3 (−) 2DS2 (+); SC: spontaneous clearance, CV: chronic viremia; OR: Odds Ratio; CI: 95% Confidence Interval.
KIR/HLA class II associations and interaction with HCV outcome
Representative associations between KIR and HLA class II are shown in table 4. We found a significant association between HLA-DRB1*1201 and SC (p value = 0.007, OR = 7.15, 95% CI = 1.48–38.52). This association remained significant after using multiple regression analysis (p =0.007, OR= 7.78 95% CI 1.77–34.1). This association could be due to the presence of this allele in non random association with HLA-DQB1*0301 (p = 0.003 OR 13.71). It also could be due to the joint presence of this allele and 2DL3/2DL3 (p value = 0.007 OR 18.0). We also observed an association between HLA-DQB1*0501 and chronic viremia: SC 7.7% and CV 22.3% (p value = 0.05 OR=0.30, 95% CI = 0.07–1.15);
Table 4.
Association of Class II HLA alleles with Chronic Viremia and Spontaneous Clearance
| Class II Allele | SC N= 39 | CV N=121 | ||
|---|---|---|---|---|
| N (%) | n (%) | OR (CI) | p value | |
| HLA-DQB1*0501 | 3 (7.7) | 26 (22.3) | 0.30 (0.07–1.15) | 0.05 |
| HLA-DRB1*0101 | 4 (10.2) | 21 (17.3) | 0.52 (0.15–1.84) | 0.24 |
| HLA-DQB1*0501 + HLA-DRB1*0101 | 3 (7.7) | 14 (10.7) | 0.64 (0.14–2.56) | 0.49 |
| HLA-DRB1*1101 | 4 (10.2) | 29 (23.9) | 0.36 (0.36–1.19) | 0.07 |
| HLA-DQB1*0301 | 16 (41.0) | 43 (34.7) | 1.27 (0.56–2.81) | 0.53 |
| HLA-DRB1*1101 + HLA-DQB1*0301 | 3 (7.7) | 21 (18.2) | 0.40 (0.40–1.52) | 0.19 |
| HLA-DRB1*1201 | 6 (15.4) | 3 (0.8) | 7.15 (1.48–38.52) | 0.007 |
| HLA-DRB1*1201 + HLA-DQB1*0301 | 4 (10.25) | 0 | 13.71 (ND) | 0.003 |
SC: Spontaneous clearance; CV chronic viremia; OR: Odds Ratio; CI: 95% Confidence Interval; ND: not determined because CI is not accurate.
To model biological interactions we compared two copies of KIR 2DL3 (2DL3/2DL3) or one or two copies of 2DL3 (2DL3/2DL3 + 2DL2/2DL3) and KIR 2DL2 (2DL2/2DL2) or one or two copies of 2DL2 (2DL2/2DL2 + 2DL2/2DL3) with the presence or absence of HLA-DRB1*1201 (Table 4).
There was an interaction between KIR 2DL3/2DL3 with the presence of HLA-DRB1*1201 (p value= 0.007 OR 18.0 CI= 1.57–898.0) but KIR 2DL3/2DL3 in the absence of HLA- DRB1*1201 or KIR 2DL3/2DL3 (−) in the presence of HLA-DRB1*1201 were not significant. Although the presence of one or two copies of KIR 2DL3 in presence of HLA-DRB1*1201 was also significant (p value= 0.002, OR: 15.4 CI: 2.04–172.5), the odds ratio is reduced. Showing that the interaction between KIR 2DL3 and DRB1*1201 is mainly from two copies of 2DL3.
This association for the joint effect of HLA-DRB1*1201 and KIR 2DL3/2DL3 remained significant (p value =0.01, OR = 17.18, 95% CI = 1.80–163.6) using multiple logistic regression where we used three dummy variables to represent HLA-DRB1*1201(−) + KIR 2DL3/2DL3; HLA-DRB1*1201 + KIR 2DL3/2DL3(−), and HLA-DRB1*1201 + KIR 2DL3/2DL3. The separate effects of HLA-DRB1*1201 only (p value =0.16 OR 4.29 95%CI: 0.56–32.6) and 2DL3/2DL3 only (p value =0.29 OR 1.53 95%CI: 0.702–3.3) remained not significant.
Also, the association of DQB1*0501 in chronic viremia was independent from any interactions with NK receptors (data not shown).
Discussion
We described KIR and HLA-C ligand genetic interactions together with class II HLA alleles in the outcome of HCV infection consistent with a report of interaction between murine Klra (Ly49P) and MHC class I alleles, and NK cell-mediated innate resistance to cytomegalovirus infection (Desrosiers at al., 2005). The recognition of an H2 molecule presenting a viral peptide resulting from murine CMV infection involves a different MHC interaction than that seen in human HCV infection due to the fact that KIRs are immunoglobulin-like receptors and Ly49P is a lectin-like receptor. Different HLA class I alleles act as specific ligands for the different variants of KIRs. KIR2DL2, KIR2DL3 and KIR2DS2 interact with HLA-Cw group 1 alleles (termed C1 ligands) characterized by Ser77/Asn80, and KIR2DL1 and KIR2DS1 interact with HLA-Cw group 2 alleles (C2 ligands; Asn77/Lys80) (Uhrberg et al., 1997). It is likely that in both cases, recognition was specific for peptides in infected cells that activate receptors. If these peptides are of viral origin, then the implicated HCV product awaits identification. In our study we confirmed the association between C1/C1 and 2DL3/2DL3 homozygous genotypes and spontaneous clearance of the HCV infection consistent with previous observations in a large cohort of patients without history of transfusion with blood products (Khakoo et al, 2004). Of interest, in a recent report these interactions were not found (Rauch A et al, 2007) perhaps because a major difference with those reported previously (Khakoo et al., 2004) was that in the cohort of Europeans (Rauch A et al, 2007) the HCV infected individuals were co-infected with HIV.
We used STRs to rule out genetic stratification in HCV-infected and non-infected Puerto Rican individuals supporting previous studies of admixture estimates using blood groups (Salzano and Bortolini., 2002) and other ancestry informative markers (Bonilla et al., 2004; Zuniga et al., 2006), of important contribution of Caucasian genes in Puerto Ricans and did not detect population stratification in our HCV-infected SC and CV groups.
We confirmed the association of HLA-DQB1*0501 with chronic viremia (Azocar J., 2003). contrasting with those reported in Caucasians (Thio et al., 2001; McKierman et al., 2004) and African Americans (Thio et al., 2001); our study was performed in IDUs, whereas other studies were predominantly transfusion-related HCV (Table 1). The conflicting reports regarding associations between HLA alleles and HCV clinical outcome could have resulted from population stratification (Cardon and Palmer, 2003; Freedman et al., 2004). In all these studies NK cells may be activated not only by HCV-infected hepatocytes and dendritic cells via KIR receptor ligands, but also by pro-inflammatory and regulatory cytokines (Doherty and O’Farelly, 2000, Mailliard et al., 2003). In this regard, reduced frequency of NK cells, altered subset distribution, impaired functional capacity of CD56 positive CD3 negative NK cells and the subsets, CD56dim and CD56bright, as well as significant reduction in serum IL-15 levels of HCV infection associated with more apoptotic changes and defects in the proliferation of NK cells were described (Nattermann et al., 2006). However, contradictory results suggested that circulating NK cell frequency and cytolytic activity, tested ex vivo, is similar between HCV-infected and uninfected groups (Morishima etal. 2006). Therefore, it would be important to investigate the interaction between HLA-Cw1 and KIR2DL3 expressed on NK cells and the outcome of HCV infection using HCV peptides, in particular to explore whether the loss of inhibition mediated by KIR2DL3 together with MHC class II alleles could be associated with strengthened innate immunity against HCV infection (Wang et al 2003; Parham, 2004).
Taken together our results can be explained by a possible mechanism of loss of inhibition of KIR2DL3 (Khakoo, 2004); the interaction between inhibitory KIR2DL3 or KIR2DL2 and the HLA-C1 involves direct contact between KIR and an interfering peptide, producing loss of inhibition (Parham, 2004). Our report of interaction between DRB1*1201 with KIR2DL3/2DL3 in SC suggests that both peptide presentation to T cells via class II alleles and KIRs are involved the outcome of HCV infection. Also, the association of DQB1*0501 with chronic viremia independent from NK genes and the association of HLA-DRB1*1201 with KIR2DL3/2DL3 suggest that peptide presentation to T cells could be involved by production of TH cytokines (Wang et al, 2003). Also, It is important to mention that although our findings are statistically significant, the small sample size was a limitation of our study.. Obtaining large sample sizes in the study population of drug abusers is difficult and confirmation in another population would be beneficial.
Acknowledgments
This work was supported by NIH grants AI49213, HL29583 and HL59838 (EJY, VR), and AI69939 (RTC). J.Z was supported by grants from the Instituto Nacional de Enfermedades Respiratorias, Mexico City and Fundación Mexico at Harvard, Boston MA. J.A was supported by Northgate Medical Center, Springfield, MA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alric L, Fort M, Izopet J, Vinel JP, Charlet JP, Selves J, et al. Genes of the major histocompatibility complex class II influence the outcome of hepatitis C virus infection. Gastroenterology. 1997;113:1675–1681. doi: 10.1053/gast.1997.v113.pm9352872. [DOI] [PubMed] [Google Scholar]
- 2.Azocar J, Clavijo OP, Yunis E. MHC Class II genes in HCV viral clearance of Hepatitis C infected Hispanic Patients. Hum Immunol. 2003;65:99–102. doi: 10.1016/s0198-8859(02)00722-x. [DOI] [PubMed] [Google Scholar]
- 3.Barrett S, Ryan E, Crowe J. Association of the HLA-DRB1*01 allele with spontaneous clearance in an Irish cohort infected with hepatitis C virus via contaminated anti-D immunoglobulin. J Hepatol. 1999;30:979–983. doi: 10.1016/s0168-8278(99)80249-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonilla C, Shriver MD, Parra EJ, Jones A, Fernández JR. Ancestral proportions and their association with skin pigmentation and bone mineral density in Puerto Rican women from New York City. Hum Genet. 2004;115:57–68. doi: 10.1007/s00439-004-1125-7. [DOI] [PubMed] [Google Scholar]
- 5.Cardon LR, Palmer LJ. Population stratification and spurious allelic association. Lancet. 2003;361:598–604. doi: 10.1016/S0140-6736(03)12520-2. [DOI] [PubMed] [Google Scholar]
- 6.Cramp ME, Carucci P, Underhill J, Naoumov NV, Williams R, Donaldson PT. Association of Class II genotype and spontaneous clearance of hepatitis C viremia. J Hepatol. 1998;29:207–213. doi: 10.1016/s0168-8278(98)80005-6. [DOI] [PubMed] [Google Scholar]
- 7.Desrosiers MP, Kielczewska A, Loredo-Osti JC, Adam SG, Makrigiannis AP, Lemieux S, et al. Epistasis between mouse Klra and major histocompatibility complex class I loci is associated with a new mechanism of natural killer cell-mediated innate resistance to cytomegalovirus infection. Nat Genet. 2005;37:593–599. doi: 10.1038/ng1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doherty DG, O’Farrelly C. Innate and adaptive lymphoid cells in the human liver. Immunol Rev. 2000;174:5–20. doi: 10.1034/j.1600-0528.2002.017416.x. [DOI] [PubMed] [Google Scholar]
- 9.Fanning LJ, Kenny-Walsh E, Shanahan F. Persistence of hepatitis C virus in a white population: associations with human leukocyte antigen class 1. Hum Immunol. 2004;65:745–751. doi: 10.1016/j.humimm.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Fanning LJ, Levis J, Kenny-Walsh E, Wynne F, Whelton M, Shanahan F. Viral clearance in Hepatitis C (1b) infection relationship with human leukocyte antigen class II in an homogeneous population. Hepatology. 2000;31:1334–7. doi: 10.1053/jhep.2000.7437. [DOI] [PubMed] [Google Scholar]
- 11.Freedman ML, Reich D, Penney KL, McDonald GJ, Mignault AA, Patterson N, et al. Assessing the impact of population stratification on genetic association studies. Nat Genet. 2004;36:388–393. doi: 10.1038/ng1333. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Lozano N, Vilches C. Genotyping of human killer-cell immunoglobulin-like receptor genes by polymerase chain reaction with sequence-specific primers: an update. Tissue Antigens. 2002;59:184–193. doi: 10.1034/j.1399-0039.2002.590302.x. [DOI] [PubMed] [Google Scholar]
- 13.Holer T, Gerken G, Notghi A, Knolle P, Lubjuhn R, Taqheri H, et al. MHC class II genes influence the susceptibility to chronic active hepatitis C. J Hepatol. 1997;27:259–264. doi: 10.1016/s0168-8278(97)80169-9. [DOI] [PubMed] [Google Scholar]
- 14.Hsu KC, Liu XR, Selvakumar A, Mickelson E, O’Reilly RJ, Dupont B. Killer Ig-like receptor haplotype analysis by gene content: evidence for genomic diversity with a minimum of six basic framework haplotypes, each with multiple subsets. J Immunol. 2002;169:5118–5129. doi: 10.4049/jimmunol.169.9.5118. [DOI] [PubMed] [Google Scholar]
- 15.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 16.Kirwan SE, Burshtyn DN. Regulation of natural killer cell activity. Curr Opin Immunol. 2007;19 (1):46–54. doi: 10.1016/j.coi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 17.Lechmann M, Schneider EM, Giers G, Kaiser R, Dumoulin FL, Sauerbruch T. Increased frequency of HLA –DR15 (B1*15011) allele in german patients with self limited hepatitis C virus infection. Eur. J. Clin Invest. 1999;29:337–43. doi: 10.1046/j.1365-2362.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Vazquez A, Rodrigo L, Martinez-Borra J, Perez R, Rodriguez M, Fdez-Morera JL, et al. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192:162–165. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 19.Mailliard RB, Son YI, Redlinger R, Coates PT, Giermasz A, Morel PA. Dendritic cells mediate NK cell help for Th1 and CTL responses: two-signal requirement for induction of NK cell helper function. J Immunol. 2003;171:2366–2373. doi: 10.4049/jimmunol.171.5.2366. [DOI] [PubMed] [Google Scholar]
- 20.Mangia A, Gentile R, Cascavilla I, Margaglione M, Villani MR, Stella F, et al. HLA class II favors clearance of HCV infection and progression of the chronic liver damage. J. Hepatology. 1999;30:984–989. doi: 10.1016/s0168-8278(99)80250-5. [DOI] [PubMed] [Google Scholar]
- 21.McKierman SM, Hagan R, Curry M, McDonald GS, Kelly A, Nolan N, et al. Distinct MHC class I and II alleles are associated with Hepatitis viral clearance originating from a single source. Hepatology. 2004;40:108–114. doi: 10.1002/hep.20261. [DOI] [PubMed] [Google Scholar]
- 22.McKierman SM, Hagan R, Curry M, McDonald GS, Nolan N, Crowley J, et al. The MHC is a major determinant of viral status, but not fibrotic stage, in individuals infected infected with hepatitis C. Gastroenterology. 2000;118:1124–1130. doi: 10.1016/s0016-5085(00)70365-9. [DOI] [PubMed] [Google Scholar]
- 23.Minton EJ, Smillie D, Neal KR, Irving WL, Underwood JCE, James V. Association between MHC class II alleles and clearance of circulating hepatitis C virus. J. Infect Dis. 1998;178:39–44. doi: 10.1086/515599. [DOI] [PubMed] [Google Scholar]
- 24.Morishima C, Paschal DM, Wang CC, Yoshihara CS, Wood BL, Yeo AE, et al. Decreased NK cell frequency in chronic hepatitis C does not affect ex vivo cytolytic killing. Hepatology. 2006;43:573–580. doi: 10.1002/hep.21073. [DOI] [PubMed] [Google Scholar]
- 25.Nattermann J, Feldmann G, Ahlenstiel G, Langhans B, Sauerbruch T, Spengler U. Surface expression and cytolytic function of natural killer cell receptors is altered in chronic hepatitis C. Gut. 2006;55:869–877. doi: 10.1136/gut.2005.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parham P. Immunology NK cells lose their inhibition. Science. 2004;305:786–7. doi: 10.1126/science.1102025. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard JK, Rosenberg N. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rauch A, Laird R, McKinnon E, Telenti A, Furrer H, Weber R, et al. Influence of inhibitory killer immunoglobulin-like receptors and their HLA-C ligands on resolving hepatitis C virus infection. Tissue antigens. 69(suppl):237–240. doi: 10.1111/j.1399-0039.2006.773_4.x. [DOI] [PubMed] [Google Scholar]
- 29.Salzano FM, Bortolini MC. The evolution and genetics of Latin American populations, Cambridge studies in biological and evolutionary anthropology. Cambridge; New York: Cambridge University Press; 2002. pp. 512–522. [Google Scholar]
- 30.Suits Daniel B. Use of Dummy Variables in Regression Equations. Journal of the American Statistical Association. 1957;52(280):548–551. [Google Scholar]; Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgarther MW, et al. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thio CL, Thomas DL, Goedert JJ, Vlahov D, Nelson KE, Hilgarther MW, et al. Racial differences in HLA class II associations with hepatitis C virus outcomes. J Infect Dis. 2001;184:16–21. doi: 10.1086/321005. [DOI] [PubMed] [Google Scholar]
- 32.Thursz M, Yallop R, Goldin R, Trepo C, Thomas HC. Influence of the MHC Class II genotype on outcome of infection with hepatitis C. Lancet. 1999;354:18–25. doi: 10.1016/s0140-6736(99)91443-5. [DOI] [PubMed] [Google Scholar]
- 33.Uhrberg M, Valiante NM, Shum BP, Shilling HG, Lienert-Weidenbach K, Corliss B, et al. Human diversity in killer cell inhibitory receptor genes. Immunity. 1997;7:753–763. doi: 10.1016/s1074-7613(00)80394-5. [DOI] [PubMed] [Google Scholar]
- 34.Vilches C, Parham P. KIR: diverse, rapidly evolving receptors of innate and adaptative immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 35.Yee LJ. Host genetic determinants in hepatitis C virus infections. Genes and Immunity. 2004;5:237–245. doi: 10.1038/sj.gene.6364090. [DOI] [PubMed] [Google Scholar]
- 36.Yenigun A, Durupinar B. Decreased frequency of the HLA-DRB1*11 allele in patients with chronic hepatitis C virus infection. J Virol. 2002;76:1787–1789. doi: 10.1128/JVI.76.4.1787-1789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zavaglia C, Martinetti M, Silini E, Botteli R, Daielli C, Asti M, et al. Association between HLA class II alleles and protection from or susceptibility to chronic hepatitis C. J Hepatol. 1998;28:1–6. doi: 10.1016/s0168-8278(98)80195-5. [DOI] [PubMed] [Google Scholar]
- 38.Zuniga J, Ilzarbe M, Acunha-Alonzo V, Rosetti F, Herbert Z, Romero V, et al. Allele frequencies for 15 autosomal STR loci and admixture estimates in Puerto Rican Americans. Forensic Sci Int. 2006;164:266–70. doi: 10.1016/j.forsciint.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 39.Wang JH, Layden TJ, Eckels DD. Modulation of the peripheral T-Cell response by CD4 mutants of hepatitis C virus: transition from a Th1 to a Th2 response. Hum Immunol. 2003;64(7):662–73. doi: 10.1016/s0198-8859(03)00070-3. [DOI] [PubMed] [Google Scholar]


