Abstract
The FoxO family of Forkhead transcription factors functions at the interface of tumor suppression, energy metabolism, and organismal longevity. FoxO factors are key downstream targets of insulin, growth factor, nutrient, and oxidative stress stimuli that coordinate a wide-range of cellular outputs. FoxO-dependent cellular responses include gluconeogenesis, neuropeptide secretion, atrophy, autophagy, apoptosis, cell cycle arrest, and stress resistance. This review will discuss the roles of the mammalian FoxO family in a variety of cell-types, from stem cells to mature cells, in the context of the whole organism. Given the overwhelming evidence that the FoxO factors promote longevity in invertebrates, this review will also discuss the potential role of the FoxO factors in the aging of mammalian organisms.
A traditional view of FoxO regulation and cellular function
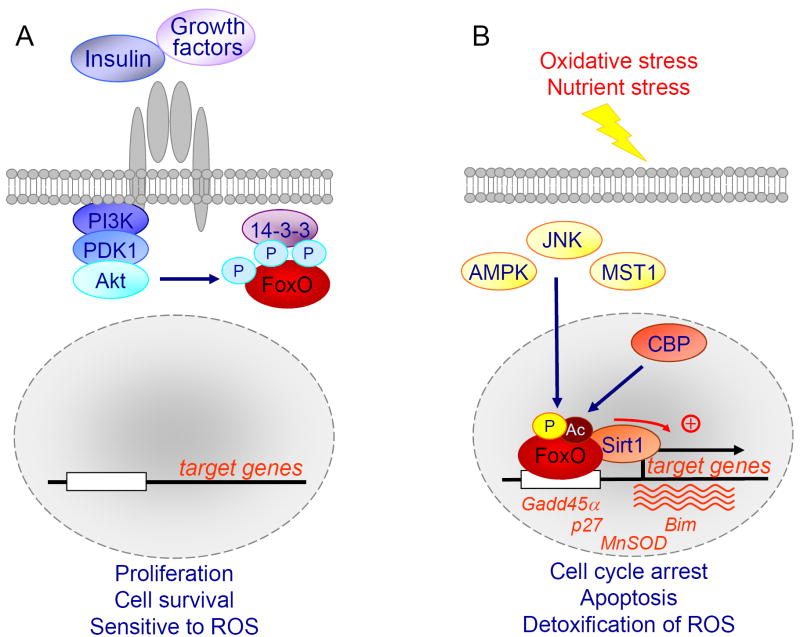
Mammals have four isoforms of the FoxO transcription factor family, FoxO1, FoxO3, FoxO4 and FoxO6. Three of the four FoxO isoforms, FoxO1, FoxO3 and FoxO4, are crucially regulated by Akt-dependent phosphorylation at three specific sites in response to growth factor and insulin stimulation (Thr32, Ser253 and Ser315 for human FoxO3) [1-4]. Akt-dependent phosphorylation of FoxO factors promotes FoxO export from the nucleus to the cytoplasm, thereby repressing FoxO transcriptional function (Fig. 1A). FoxO6 lacks the C-terminal Akt-dependent site and is thus predominantly nuclear, although the phosphorylation of the two remaining Akt-dependent sites inhibits FoxO6 transcriptional activity [5,6]. FoxO factors have emerged as a convergence point of signaling in response to growth factor stimulation and oxidative stress (Fig. 1) [1,7-12]. Insulin and growth factors inhibit FoxO factors through PI3K/Akt, while oxidative stress stimuli activate FoxO factors through a combination of modifications. In addition to the PI3K/Akt pathway, the other major signaling modules that directly regulate the activity of the FoxO factors include the stress-activated Jun-N-terminal kinase (JNK), the mammalian ortholog of the Ste20-like protein kinase (MST1), and the deacetylase Sirt1 (Fig. 1) [9-11,13-15]. The FoxO factors integrate these divergent signals through post-translational modifications, such as phosphorylation, acetylation and mono/poly-ubiquitination, resulting in altered subcellular localization, protein stability, DNA binding properties, and transcriptional activity [1,9,11,12,14,16]. FoxO-dependent transcription plays an important role in a wide variety of cellular outputs, including glucose metabolism, cell cycle arrest, differentiation, detoxification of reactive oxygen species (ROS), repair of damaged DNA, and apoptosis [17-26].
Fig. 1. Negative regulation of FoxO transcription factors by growth factors and positive regulation of FoxO factors by oxidative stress stimuli.
(A) Upon stimulation with growth factors or insulin, Akt directly phosphorylates FoxO factors at three conserved residues, promoting their export from the nucleus by binding with the chaperone 14-3-3, and thus resulting in the inhibition of FoxO-dependent transcription. In the presence of growth factors or insulin FoxO-dependent transcription is inhibited promoting cellular proliferation and survival, but also rendering the cell sensitive to oxidative damage. (B) Oxidative stress induces the phosphorylation, acetylation and monoubiquitination of FoxO factors at a number of regulatory sites by factors such as AMPK (AMP-dependent kinase), JNK (Jun-N-terminal kinase), MST1 (mammalian Ste20-like kinase) and CBP (CREB binding protein). In response to oxidative stress, FoxO factors translocate to the nucleus and bind to the deacetylase Sirt1. Key modification signatures appear to recruit FoxO factors to specific genes involved in cell-cycle arrest and the response to stress. p27, cyclin-dependent kinase inhibitor; MnSOD, manganese superoxide dismutase; Bim, pro-apoptotic Bcl2-interacting mediator of cell death; Gadd45α, growth arrest- and DNA damage-inducible gene 45 α.
Accordingly, a myriad of target genes have been identified that mediate the role of FoxO factors in the various cellular processes (Table 1). The most well defined FoxO-dependent target genes include the cell cycle inhibitors p27 and p21, the stress response genes manganese superoxide dismutase (MnSOD), and growth arrest- and DNA damage-inducible gene 45 (Gadd45), the proapoptotic factors Bcl2-interacting mediator of cell death (Bim) and Fas ligand and the glycogenolytic gene glucose-6-phosphatase (G6pc) (Fig. 1) [17,18,21,22,27-32]. Despite this enormous body of work, the mechanisms by which FoxO factors specify the different cellular responses, and the ensemble of cellular responses and gene targets that are FoxO-dependent have not been fully elucidated.
Table 1.
FoxO target genes and functions
| Cellular output | Gene | Function | Regulation in cells | Binding to promoter | Regulation of promoter | References |
|---|---|---|---|---|---|---|
| Metabolism | G6pc | glycogenolysis | + | EMSA | + | [19,29,57] |
| Igfbp1 | regulates IGF activity | + | ChIP | + | [27,57,58,59] | |
| Ppargc1 α | gluconeogenesis | + | [57] | |||
| Pck1 | gluconeogenesis | + | EMSA | + | [57,58] | |
| Food intake | Agrp | orexigenic neuropeptide | + | ChIP; EMSA | + | [61,62] |
| Npy | orexigenic neuropeptide | + | ChIP; EMSA | + | [61] | |
| Pomc | anorexigenic neuropeptide | - | ChIP; EMSA | + | [61,62] | |
| Atrophy | Atrogin-1/MAFbx | muscle-specific ubiquitin ligase | + | EMSA | + | [66,67] |
| MuRF1 | muscle-specific ubiquitin ligase | + | [66] | |||
| Autophagy | Bnip3 | Bcl2-related autophagy regulator | + | ChIP | [21,70] | |
| Gabarapl1 | autophagosome formation | + | ChIP | [72] | ||
| LC3 | autophagosome formation | + | ChIP | + | [70,72] | |
| Atg12I | autophagy-related gene | + | ChIP | [72] | ||
| Apoptosis | Bim | Bcl2-interacting mediator of cell death | + | ChIP | + | [15,28,75] |
| hid | pro-apoptotic | + | ChIP | [76] | ||
| Fas ligand | pro-apoptotic tumor necrosis factor ligand | + | [1] | |||
| Cell cycle arrest | p27Kip1 | binds to and inhibits the cyclin E–CDK2 complex | + | + | [18,82] | |
| p21Cip1 | binds to and inhibits the cyclin E–CDK2 complex | + | ChIP | + | [30,34] | |
| p19INK4d p19Arf | binds to and inhibits cyclin D–CDK4/6 complexes | + | ChIP; EMSA | + | [32,83] | |
| p15INK4b | binds to and inhibits cyclin D–CDK4/6 complexes | + | ChIP; EMSA | + | [32,33,34] | |
| Angiogenesis | Sprouty2 | inhibitor of tyrosine kinase signaling | + | ChIP | [63] | |
| eNOS | endothelial function and neovascularization | - | ChIP | + | [64] | |
| Ang2 | vascular remodelling factor | + | [64] | |||
| Cited2 | CBP/p300-interacting transactivator | + | ChIP | + | [38,63] | |
| Stress resistance | MnSOD | manganese superoxide dismutase | + | ChIP | + | [20,82] |
| Gadd45α | growth arrest- and DNA damage-inducible gene 45 α | + | + | [21] |
Footnotes
The list of target genes is not exhaustive.
Regulation in cells details whether FoxO factors promote (+) or suppress (-) target gene expression in cultured cells by assessment of mRNA or protein levels.
Direct binding of FoxO factors to the promoter was assessed by chromatin immunoprecipitation (ChIP) or electromobility shift assay (EMSA).
Regulation of promoters was tested by the ability of FoxO factors to specifically drive luciferase expression.
Recently discovered signaling pathways regulating FoxO activity
Recent studies are providing further insights into the complexity of FoxO regulatory pathways. FoxO factors have been shown to be regulated by a variety of novel stress stimuli, including DNA damage, nutrient deprivation, cytokines and hypoxia [30,33-38]. For example, DNA damage affects FoxO activity via cyclin-dependent kinase 2 (CDK2) [35]. CDK2 phosphorylates FoxO1 at Ser249, resulting in the sequestration of FoxO1 to the cytoplasm in the absence of DNA damage. In the presence of DNA damage, the repressive effect of CDK2 on FoxO1 sequestration is abolished and FoxO1 translocates to the nucleus to induce apoptosis [35]. In addition, the energy sensor AMP-activated protein kinase (AMPK) has been shown to directly phosphorylate FoxO factors at six regulatory sites that are distinct from the Akt phosphorylation sites, resulting in FoxO activation [36,37]. Activation of FoxO factors by AMPK promotes the preferential expression of a gene expression program that enhances cellular stress resistance [36,37]. While the regulation of FoxO factors is mostly conducted by post-translational modifications, a series of recent studies have highlighted how FoxO factors also integrate extracellular stimuli via alternate mechanisms. For example, the growth-inhibitory cytokine transforming growth factor-β (TGFβ) triggers the formation of a complex between FoxO, Smad and C/EBPβ transcription factors at specific promoters, which results in the expression of cell cycle inhibitor genes, including p15 and p21 [30,33,34,39,40]. The synergy between FoxO and Smad transcription factors has been shown to be essential for the anti-proliferative effects of TGFβ in several cell types, including epithelial and breast cancer cells [30,33]. In addition, an intriguing new study shows that FoxO3 transcription is induced by cellular hypoxia via direct binding of the hypoxia-inducible factor HIF1 to the FoxO3 promoter [38]. The increased expression of FoxO3 resulted in enhanced cellular survival by attenuating HIF-induced apoptosis. These latest studies underscore the intricate regulation of the FoxO factors, by a wide range of diverse stimuli, including DNA damage, glucose availability, cytokines and hypoxia, that may serve to fine tune FoxO activity in different cell-types under different environmental contexts.
Functions of FoxO factors in the context of the whole organism: insights from invertebrates
Studies in invertebrates have shed light on the cellular role of FoxO factors in the context of the entire organism. In contrast to mammals, invertebrate model organisms possess just one isoform of the FoxO transcription factor family denoted DAF-16 for Caenorhabditis elegans and dFOXO for Drosophila melanogaster. The importance of DAF-16 in organismal metabolism and lifespan was revealed in a series of seminal studies on the insulin/FoxO pathway in worms. DAF-16 is necessary for the increase in lifespan provided by mutation of the insulin/insulin-like growth factor receptor daf-2 [41-43]. DAF-16 is also necessary for a distinctive developmental arrest and diapause, called dauer, that is characterized by low metabolic activity and a long lifespan in response to starvation [43]. Interestingly, DAF-16 activity in the cells of specific tissues, particularly the intestine and nervous system, appears to be more pertinent for the promotion of long lifespan than DAF-16 activity in other tissues [44-46]. These results suggest that DAF-16/FoxO regulates the production of secondary signals or hormones that coordinate the metabolism and lifespan of a variety of tissues throughout the organism [47]. In flies, activating dFOXO in the cells of the fat body (the equivalent of mammalian white adipose and liver tissue) also extends lifespan [48,49]. Constitutive dFOXO overexpression and targeted overexpression in neurosecretory insulin-producing cells in flies increases glucose and lipid levels, although a functional dTOR signaling pathway is required to observe the metabolic effects mediated by dFOXO [50]. Thus, work from invertebrates has taught us that the insulin/FoxO pathway is critical to coordinate metabolism and longevity, by acting in both a cell intrinsic and cell extrinsic manner via the production of secondary messengers that act systemically throughout the organism.
In mammals, FoxO isoforms display differential but overlapping expression throughout the organism
In mammals, the FoxO family have a complementary but overlapping expression pattern both during development and in a variety of adult tissues [5,51-53]. During mouse development, FoxO1 is detected at highest levels in the adipose tissue, FoxO3 is most expressed in the liver, FoxO4 in the skeletal muscle and FoxO6 in the central nervous system. In adult mice, FoxO1 is observed at the highest levels in adipose tissue, uterus and ovaries, with lower levels in most other tissues including the skeletal muscle and spleen. The expression pattern of FoxO3 is more ubiquitous but FoxO3 is particularly highly expressed in the brain, spleen, heart, and ovaries. FoxO4 is expressed most highly in skeletal muscle, cardiac muscle, and adipose tissue. Intriguingly, FoxO6 is expressed almost exclusively in the adult brain. The expression of FoxO1 and FoxO3 in human tissues is similar to the murine expression profiles [54]. The expression pattern of the FoxO family in the adult mouse brain is particularly interesting and illustrates the differential expression of the FoxO isoforms. FoxO1 is most abundant in the striatum, dentate gyrus and the ventral hippocampus, whereas FoxO3 shows highest expression throughout the cortex, hippocampus and cerebellum, and FoxO6 is expressed abundantly in the hippocampus, amygdala and cingulate cortex [53]. The hippocampus, amygdala and cerebellum coordinate distinct organismal behavioral responses, such as spatial learning, emotion and motor coordination respectively. The differential expression of the FoxO family in the adult murine brain may point to different cellular functions or potencies for the different FoxO isoforms in neurons. These observations suggest that during evolution, the FoxO pathway has been co-opted to play either: 1) related roles in response to different tissue-specific environment stimuli, or 2) different tissue-specific roles in response to related environmental stimuli.
An integrative model for FoxO function?
The extreme diversity of the cellular roles of the FoxO factors revealed from mammalian cell culture experiments has created a challenge to integrate these multiple roles into a unified model. In addition, many studies investigating FoxO regulation and function have been performed in immortalized or transformed culture cell lines, which are ‘out of the tissue context.’ Hence it is difficult to extrapolate results to conditions in vivo, which are dependent on specialized cellular niches. Despite these limitations, it is becoming increasingly clear that the regulation of FoxO activity by specific stimuli triggers consistent cellular outputs within the same tissue. As cells and tissues differ with respect to the environmental stimuli that they sense, the remainder of this review will focus on the cellular roles of FoxO proteins in the different tissues of the body in response to tissue-specific stimuli. The classification of FoxO function and signaling by tissue provides an interesting framework to understand why the mammalian organism has evolved to utilize members of the FoxO family of factors to govern a variety of different cellular processes in response to different environmental contexts and aging.
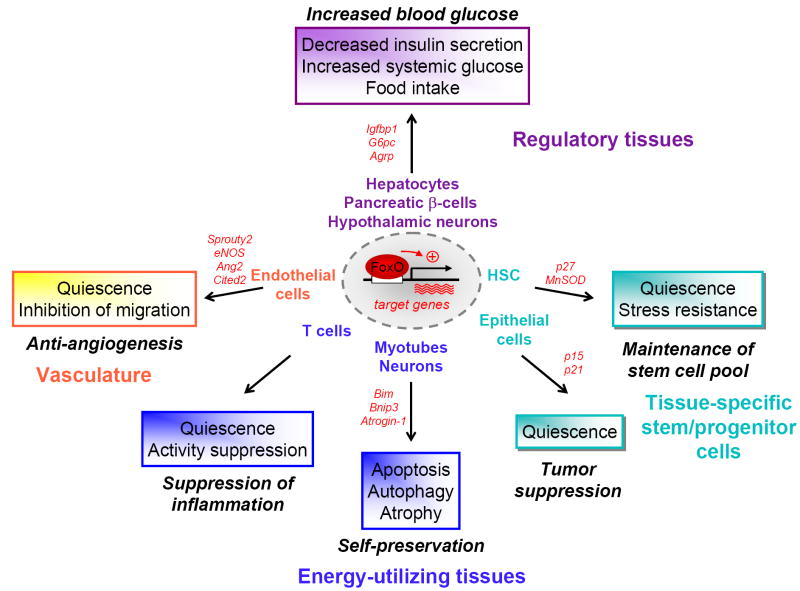
For this review, we have divided mammalian tissues into three broad categories as a framework for our discussions: 1) the ‘regulatory’ tissues, which include the liver, pancreas, and hypothalamus-pituitary axis; 2) the vascular system that allows the connection between tissues; 3) the ‘energy-utilizing tissues’ which include the skeletal muscle, nervous system, and hematopoietic system (Fig. 2). In addition to regulating mature cells, the FoxO factors are also emerging as key regulators of stem cells in adult tissues (Fig. 2). As an example, we will discuss the role of the FoxO family in the stem cells of the hematopoietic system. Given the role of FoxO factors in the longevity of invertebrates and mounting evidence for a role of the insulin signaling pathway in mammalian longevity [55,56], we will also speculate about the possible role of FoxO factors in these tissue categories to regulate mammalian lifespan.
Fig. 2. Role of FoxO transcription factors in tissue homeostasis.
FoxO-dependent transcription serves to increase systemic glucose levels by regulating gluconeogenesis, insulin secretion, and food intake in the regulatory cells of the organism such as the hepatocytes, pancreatic β-cells, and hypothalamic neurons. In energy-utilizing cells, such as myocytes and neurons, the FoxO factors appear to be activated in response to harsh environment conditions such as starvation and oxidative stress to initiate atrophy, autophagy or apoptosis. In T cells, FoxO factors attenuate proliferation and hyperactivation. In the endothelial cells of the vascular system, the FoxO factors regulate genes to suppress proliferation and inhibit migration. FoxO factors also display a crucial role in the maintenance of hematopoietic stem cells (HSC) by coordinating quiescence, stress resistance and terminal differentiation. Overall, FoxO factors appear to promote an organismal metabolic shutdown in response to harsh environmental conditions, such as starvation, to enable the organism to survive in anticipation of improved environmental conditions.
FoxO factors increase organismal glucose levels and food intake by acting on regulatory cell-types
Regulatory cells that control circulatory metabolites and hormones reside in the liver, pancreas, hypothalamus-pituitary axis and adipose tissue. In these regulatory cells, FoxO appears to act at a number of different levels to systemically increase circulating glucose levels (Fig. 2). For example, the ablation of FoxO1 in hepatic cells reduces glucose levels in newborn and adult mice, supporting the notion that FoxO factors promote increased glucose levels in the circulation [57]. FoxO1 regulates glucose levels by acting on target genes such as glucose-6-phosphatase G6pc (glycogenolytic), and phosphoenolpyruvate carboxykinase 1 Pck1, and peroxisome proliferator-activated receptor gamma coactivator 1α Ppargc1α (gluconeogenetic) (Table 1) [29,57,58]. Interestingly, FoxO1 deletion in the hepatocytes protects against excessive glucose production and diabetes in insulin receptor null mice [57]. In cultured β-cells of the pancreas, modest expression of a constitutively nuclear form of FoxO1 leads to a ‘metabolic diapause’ status by repressing glycolysis, reducing insulin secretion, and increasing free fatty acid oxidation [59]. Expression of FoxO1 in the β-cells of transgenic mice also prevents the compensatory proliferation of β-cells that occurs during insulin-resistance [60]. In contrast to the study performed in vitro [59], FoxO1 expression in vivo does not decrease circulating insulin levels [60], suggesting that the FoxO factors can act at multiple levels to increase circulating glucose by either decreasing insulin secretion or by attenuating β-cell division. Thus, FoxO factors act in concert to increase systemic glucose by regulating hepatic gluconeogenesis and glycogenolysis and by reducing the net insulin production by the pancreas. These observations raise the intriguing possibility that FoxO factors would promote diabetes in mammals if they are constitutively active. It is possible that the FoxO factors have evolved to participate in the adaptation to low nutrient availability, and that the observed diabetic phenotype is an overshoot of the adaptative response when food is plentiful.
The hypothalamus regulates food intake and energy homeostasis. FoxO factors exert a further level of control of metabolite homeostasis by coordinating neuropeptide production in hypothalamic neurons [61,62]. Two independent reports have demonstrated that FoxO1 functions in the arcuate nucleus of the hypothalamus to increase food intake and bodyweight by inducing the expression of orexigenic hormones agouti-related protein (Agrp) and/or neuropeptide Y (Npy) (Fig. 2). In addition, FoxO1 appears to have a suppressive effect on the expression of anorexigenic neuropeptides such as pro-opiomelanocortin (Pomc), by inhibiting the induction of this gene by the transcription factor Stat3. The anorexigenic hormones insulin and leptin decrease FoxO1 expression in hypothalamic neurons. The suppressive effects of insulin and leptin on food intake are mediated in part by their ability to inhibit FoxO1 [61]. These studies support the notion that in mammalian regulatory cells, the FoxO transcription factors shift metabolism away from glucose utilization towards a preservation status that is reminiscent of a ‘metabolic diapause.’ By acting at a number of different levels from gluconeogenesis to food intake, FoxO factors have evolved to help the organism adapt to nutrient deprivation. When food is available, FoxO transcription appears to increase organismal food intake and food seeking behaviors via hormonal control. In unfavorable nutrient conditions, FoxO factors alter organismal metabolism to enable the animal to continue functioning by maintaining systemic glucose and lipid levels.
FoxO factors restrict angiogenesis
The blood vessels of the circulatory system connect the ‘regulatory’ and ‘energy-utilizing’ tissues, and are composed of endothelial cells. Emerging evidence suggests that FoxO factors attenuate proliferation and migration of endothelial cells resulting in limited blood vessel formation (Fig. 2). Acute deletion of FoxO1, FoxO3 and FoxO4 in endothelial cells of mice using an inducible Mx-Cre transgene revealed an age-progressive overproliferation of endothelial cells that resulted in hemangiomas and premature death of the animal [63]. Intriguingly, while FoxO1, FoxO3 and FoxO4 deletion was achieved in endothelial cells throughout the body, the hemangiomas were observed in only a subset of tissues, particularly the uterus, liver and skeletal muscle, suggesting that specific extracellular signals --perhaps vascular endothelial growth factor (VEGF)-- also play an important role in tumor progression in the absence of FoxO factors. The factors initiating hemangioma progression in the absence of FoxO factors have not yet been characterized, but Sprouty2, a general receptor tyrosine kinase inhibitor, was identified as an important FoxO target gene involved in the suppression of proliferation and cellular survival. While the ablation of three FoxO family members accentuated the vascular phenotype, FoxO3 ablation in mice enhanced postnatal blood vessel formation in response to hind limb ischemia by abolishing the suppressive effect of FoxO3 on eNOS expression [64]. FoxO1 and FoxO3, but not FoxO4, were demonstrated to bind directly to the eNos promoter in cultured endothelial cells (Table 1). Silencing of FoxO1 and FoxO3 gene expression by siRNA in cultured endothelial cells enhanced vessel formation, migration and vessel sprouting in response to VEGF [64]. These studies are interesting because they provide evidence of a redundant [34,64], but not completely overlapping role for the different isoforms of the FoxO family [63,64]. Indeed, while overexpression of constitutively active forms of FoxO1 or FoxO3 in endothelial cells inhibited vessel formation and migration, overexpression of a constitutively active form of FoxO4 did not inhibit these processes [64]. Furthermore, the program of genes regulated by FoxO1 and FoxO3 in endothelial cells was shown to have overlaps, but also differences [64]. For example, both FoxO1 and FoxO3 regulated eNos, but angiopoietin 2 (Ang2) was exclusively regulated by FoxO1. These studies suggest that FoxO factors limit vascularization, and perhaps even restrict the supply of nutrients to tissues in response to harsh environmental stimuli, although this needs to be formally tested. The possibility that FoxO factors promote a metabolic shutdown is consistent with the observations that FoxO factors restrict vascularization in parallel to maintaining high glucose levels and encouraging food intake in the face of harsh environmental conditions. It is also intriguing that FoxO factors act both as cellular tumor suppressors and regulators of angiogenesis. This intersection provides two mechanisms whereby the FoxO family can limit the development of tumors; by a cell-intrinsic tight control of the cell cycle, as well as by extrinsically limiting the supply of the vasculature to developing tumors.
FoxO factors act to protect undamaged cells in energy-utilizing tissues in response to stress stimuli
The energy-utilizing cell-types of the organism, such as those that reside in the skeletal muscle, nervous system, and immune system are responsive to the regulatory ‘pacemaker’ tissues such as the liver, pancreas, and hypothalamus (Fig. 2). While the entire set of stimuli that regulate FoxO in these energy-utilizing cells is not known yet, particularly for the immune system, it appears that the FoxO family becomes activated when the regulatory tissues and the vasculature are not functioning correctly or are themselves affected by the environment (e.g. during starvation). Under such unfavorable situations, these energy-utilizing cells appear to enter a self-preservation state.
Skeletal muscle
During nutrient deprivation or starvation, the skeletal muscle undergoes protein degradation induced by the activity of two highly conserved processes: 1) ubiquitin-proteosomal atrophy, and 2) lysosomal autophagy. Autophagy is a protective cellular response to nutrient deprivation whereby lysosomal enzymes mediate the recycling of proteins, cytoplasm and cell organelles [65]. Recent studies have revealed that the FoxO factors are key mediators of both atrophy and autophagy in muscle in response to fasting, but also in response to denervation, glucocorticoids and hindlimb suspension [66-72]. During fasting, FoxO1 and FoxO3 induce the transcription of two muscle-specific E3 ubiquitin ligases, atrogin-1/MAFbx and MuRF1, and other components of the ubiquitin-proteasome system (e.g. ZNF216, a novel ubiquitin-binding protein containing a zinc-finger), resulting in skeletal muscle atrophy without apoptosis [66-69]. While atrogin-1 appears to be a direct FoxO target gene, it is not known if other components of the proteasome system, such as ZNF216 are direct FoxO target genes. Interestingly, the induction of atrogin-1 initiates a positive feedback loop to attenuate cardiac hypertrophy by mediating polyubiquination of FoxO1 and FoxO3 to enhance FoxO transcriptional activity [73]. In parallel, FoxO3 has been shown to activate autophagy in response to fasting or denervation by directly controlling the transcription of autophagy-related genes including LC3, Bnip3, Gabarapl1 and Atg12l [70,72]. These autophagy-promoting genes were shown to be direct targets of FoxO factors in myotubes (Table 1). Interestingly, the induction of autophagy by FoxO factors does not seem to be restricted to mammalian muscle, as activating dFOXO in Drosophila in response to starvation also induces autophagy in the fat body [74]. The mammalian studies support the notion that FoxO factors promote an acute atrophy and autophagy in skeletal myotubes in response to starvation or inadequate trophic support by the regulatory ‘pacemaker’ tissues. It is not known if muscular atrophy and autophagy are beneficial to the organism, but atrophy and autophagy may be inducing an acute self-preservation state in anticipation of improved nutritional status. It is likely that prolonged stresses, such as starvation, limb immobilization and chronic glucocorticoids, would negate the transient tissue-preservation provided by FoxO-dependent atrophy and autophagy, and perhaps lead to the induction of FoxO-dependent apoptosis or other FoxO-independent detrimental effects.
Nervous system
In the nervous system, the FoxO family appears to be activated in response to various stress stimuli, such as epileptic seizures and oxidative stress, and acts to eliminate damaged neurons by apoptosis. Epileptic brain injury in rats leads to FoxO1 and FoxO3 activation in hippocampal neurons and to the upregulation of the proapoptotic gene Bim leading to neuronal apoptosis [75]. Similarly, ultraviolet damage in the Drosophila retinal nervous tissue induces apoptosis via dFOXO by inducing the pro-apoptotic gene hid [76]. The induction of apoptosis by dFOXO in the fly retina requires activation of the JNK pathway, further supporting the idea that JNK activates FoxO factors [10,77]. Finally, oxidative stress induced by hydrogen peroxide treatment has been shown to promote apoptosis in rat primary cerebellar neurons by the activation of FoxO factors [15]. The induction of neuronal apoptosis in response to oxidative stress is mediated at least in part by direction activation of FoxO factors by MST1 protein kinase, an important effector of cellular apoptosis [15]. It is still unclear why apoptosis is the primarily cellular output to the activation of FoxO factors by environmental stressors in neurons, when FoxO factors clearly have the ability to protect other tissues, such as myotubes, by eliciting autophagy. It is possible that neurons may be more sensitive to apoptosis than other cell-types. While activation of the MST-FoxO pathway causes apoptosis in cultured neurons, the same pathway promotes longevity in C. elegans [15]. Thus, the fact that FoxO factors promote apoptosis in the nervous system, and yet have the ability to elicit a long lifespan in invertebrates creates a conundrum, since apoptosis is a cellular output that is considered terminal and negative. Two explanations may account for this apparent discrepancy: 1) limited apoptosis of damaged neurons may actually be beneficial to help preserve the function of the remaining nervous system; or 2) FoxO may need to be activated in the proper ‘window’ of time/magnitude, otherwise it is detrimental.
Immune system
In the immune system, FoxO transcription factors appear to act at multiple stages to limit the expansion and/or activation of the mature hematopoietic cells, though the specific stimuli that activate FoxO factors in these cells still remain to be established [78,79]. The ablation of FoxO3 in vivo results in T cell proliferation and hyperactivity (e.g. elevated IL-2 expression) due to NF-κB activation, leading to a multi-system inflammatory syndrome [80]. This study indicates that FoxO3 normally suppresses inflammation in mammals. Since the expression of certain genes associated with inflammation is increased during aging (reviewed by [81]), it is tempting to speculate that FoxO factors could also maintain tissue homeostasis by preventing uncontrolled inflammatory responses. However, more work is required to dissect the components of immune function that are beneficial during organismal maintenance versus inflammatory pathways that may accelerate aging.
FoxO factors limit the expansion and regulate the terminal differentiation of stem/precursor cells and proliferative/tumorigenic cells
In addition to their roles in mature cell-types, the FoxO transcription factors also play a role to limit the expansion of stem/progenitor cells of tissues such as the hematopoietic system (Fig. 2). Acute deletion of FoxO1, FoxO3 and FoxO4 in adult murine bone marrow led to the expansion of both the myeloid and lymphoid lineages coupled with increased cell cycling of the long-term hematopoietic stem cells [79], indicating that the FoxO family normally limits the proliferation of these stem cells. Ablation of either FoxO3 alone, or in combination with FoxO1 and FoxO4 resulted in reduced maintenance of the hematopoietic stem cell pool [79,82], as a result of increased ROS and apoptosis, and reduced quiescence, which progressively worsened with age. Interestingly, the cell cycling and survival defects of FoxO-deficient hematopoietic stem cells could be significantly improved by treating mutant mice with the antioxidant N-acetyl-L-cysteine. This finding indicates that oxidative stress is a major factor regulating the proliferation and apoptosis of these stem cells, and also that chemical interventions can recover at least part of the cellular defect. A consequence of the ablation of FoxO factors in hematopoietic cells is increased cell cycle progression and/or resistance to apoptosis leading to lymphoma development via attenuation of p27 and p19 expression [63,83]. In line with these studies, the loss of FoxO transcriptional activity renders progenitor cells or tumor cells resistant to the cytostatic effects of extracellular effectors such as TGFβ, which may contribute to tumor development (Fig. 2) [30,33,34,84,85]. FoxO transcriptional activity is attenuated in progenitor and tumor cells by a variety of mechanisms [30,84,85], including: i) nuclear exclusion upon phosphorylation of FoxO factors by the PI3K/Akt signaling pathway; ii) association of FoxO factors with the transcriptional repressor FoxG; iii) ubiquitin-dependent degradation upon phosphorylation of FoxO factors by IκB kinase β (IKKβ) or extracellular signal-regulated kinase (ERK). Together, these studies frame the FoxO factors as pivotal suppressors of tumorigenesis by coordinating cellular quiescence in stem/progenitor cells. These findings also highlight the importance of the FoxO family in maintaining stem cell pools in adult tissues. Consistent with this possibility, ablation of FoxO3 in female mice leads to the early depletion of the follicular pool and premature infertility [86]. The role of the FoxO family in other tissue-specific stem cells is not known yet, but an intriguing possibility is that the FoxO family may contribute to tissue maintenance and repair during aging by maintaining adult stem cell pools.
Conclusion
Although the regulation and roles of the FoxO family have been well studied, there is still a dearth of knowledge on the mechanisms that specify the decision between different cellular outputs in response to different environmental contexts for these promiscuous transcription factors. Similarly, it is not clear why four FoxO isoforms exist, although there is now evidence to suggest that their roles are not entirely overlapping [63,64]. A model that emerges is that different FoxO isoforms bind to the promoters of target genes with different affinities, perhaps due to differences in structure and post-translational modifications between isoforms resulting in opportunities for differential cellular outputs. Understanding if FoxO factors regulate other cellular responses in addition to those mentioned in this review and elucidating how FoxO factors specify at the molecular level their response to a multitude of environmental stimuli will provide important insights into their organismal function.
Given the evidence discussed here that the FoxO factors coordinate glucose homeostasis, angiogenesis, stem cell maintenance, immune, muscular, and neuronal functions, the implications for diabetes, cancer, autoimmune diseases and neurodegeneration are apparent. Finally, there is overwhelming evidence to support a significant role for FoxO factors in the determination of invertebrate lifespan, although further studies in vivo are required to make conclusions regarding the role of FoxO factors in mammalian longevity. Understanding how FoxO factor cellular functions are integrated into cohesive organismal responses will be significant for therapeutic interventions against age-dependent pathologies such as diabetes, cancer, autoimmune syndromes and neurodegeneration.
Acknowledgments
We apologize for not being able to cite all relevant papers because of space constraints. We thank members of the Brunet lab and in particular Eric L. Greer, Victoria A. Rafalski, Dario R. Valenzano, and Valérie M. Renault for their helpful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
• of special interest
•• of outstanding interest
- 1.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 2.Kops GJ, de Ruiter ND, De Vries-Smits AM, Powell DR, Bos JL, Burgering BM. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 3.Brunet A, Park J, Tran H, Hu LS, Hemmings BA, Greenberg ME. Protein kinase SGK mediates survival signals by phosphorylating the forkhead transcription factor FKHRL1 (FOXO3a) Mol Cell Biol. 2001;21:952–965. doi: 10.1128/MCB.21.3.952-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol. 2002;156:817–828. doi: 10.1083/jcb.200112059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobs FM, van der Heide LP, Wijchers PJ, Burbach JP, Hoekman MF, Smidt MP. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J Biol Chem. 2003;278:35959–35967. doi: 10.1074/jbc.M302804200. [DOI] [PubMed] [Google Scholar]
- 6.van der Heide LP, Jacobs FM, Burbach JP, Hoekman MF, Smidt MP. FoxO6 transcriptional activity is regulated by Thr26 and Ser184, independent of nucleo-cytoplasmic shuttling. Biochem J. 2005;391:623–629. doi: 10.1042/BJ20050525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakae J, Park BC, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 8.Nakae J, Barr V, Accili D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. Embo J. 2000;19:989–996. doi: 10.1093/emboj/19.5.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 10.Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BM. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. Embo J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Horst A, Tertoolen LG, de Vries-Smits LM, Frye RA, Medema RH, Burgering BM. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- 12.van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, Maurice MM, Burgering BM. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 13.Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Regan KM, Wang F, Wang D, Smith DI, van Deursen JM, Tindall DJ. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc Natl Acad Sci U S A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •15.Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villen J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. This study shows that the mammalian Ste20-like kinase MST1 directly phosphorylates FoxO3 at Ser207 in cultured neurons in response to oxidative stress. MST1 promotes FoxO3 nuclear translocation and the induction of proapoptotic genes such as Bim. Overexpression of the MST1 ortholog in C. elegans promotes longevity in a FoxO/daf-16-dependent manner.
- 16.Matsuzaki H, Daitoku H, Hatta M, Aoyama H, Yoshimochi K, Fukamizu A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc Natl Acad Sci U S A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dijkers PF, Medema RH, Pals C, Banerji L, Thomas NS, Lam EW, Burgering BM, Raaijmakers JA, Lammers JW, Koenderman L, et al. Forkhead transcription factor FKHR-L1 modulates cytokine-dependent transcriptional regulation of p27(KIP1) Mol Cell Biol. 2000;20:9138–9148. doi: 10.1128/mcb.20.24.9138-9148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782–787. doi: 10.1038/35008115. [DOI] [PubMed] [Google Scholar]
- 19.Nakae J, Kitamura T, Silver DL, Accili D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J Clin Invest. 2001;108:1359–1367. doi: 10.1172/JCI12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kops GJ, Dansen TB, Polderman PE, Saarloos I, Wirtz KW, Coffer PJ, Huang TT, Bos JL, Medema RH, Burgering BM. Forkhead transcription factor FOXO3a protects quiescent cells from oxidative stress. Nature. 2002;419:316–321. doi: 10.1038/nature01036. [DOI] [PubMed] [Google Scholar]
- 21.Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, Jr, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- 22.Gilley J, Coffer PJ, Ham J. FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J Cell Biol. 2003;162:613–622. doi: 10.1083/jcb.200303026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hribal ML, Nakae J, Kitamura T, Shutter JR, Accili D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J Cell Biol. 2003;162:535–541. doi: 10.1083/jcb.200212107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakae J, Kitamura T, Kitamura Y, Biggs WH, 3rd, Arden KC, Accili D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev Cell. 2003;4:119–129. doi: 10.1016/s1534-5807(02)00401-x. [DOI] [PubMed] [Google Scholar]
- 25.Liu ZP, Wang Z, Yanagisawa H, Olson EN. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev Cell. 2005;9:261–270. doi: 10.1016/j.devcel.2005.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Kitamura T, Kitamura YI, Funahashi Y, Shawber CJ, Castrillon DH, Kollipara R, DePinho RA, Kitajewski J, Accili D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J Clin Invest. 2007;117:2477–2485. doi: 10.1172/JCI32054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 28.Dijkers PF, Medema RH, Lammers JW, Koenderman L, Coffer PJ. Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr Biol. 2000;10:1201–1204. doi: 10.1016/s0960-9822(00)00728-4. [DOI] [PubMed] [Google Scholar]
- 29.Schmoll D, Walker KS, Alessi DR, Grempler R, Burchell A, Guo S, Walther R, Unterman TG. Regulation of glucose-6-phosphatase gene expression by protein kinase Balpha and the forkhead transcription factor FKHR. Evidence for insulin response unit-dependent and -independent effects of insulin on promoter activity. J Biol Chem. 2000;275:36324–36333. doi: 10.1074/jbc.M003616200. [DOI] [PubMed] [Google Scholar]
- 30.Seoane J, Le HV, Shen L, Anderson SA, Massague J. Integration of Smad and forkhead pathways in the control of neuroepithelial and glioblastoma cell proliferation. Cell. 2004;117:211–223. doi: 10.1016/s0092-8674(04)00298-3. [DOI] [PubMed] [Google Scholar]
- 31.Wook Oh S, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nat Genet. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- 32.Katayama K, Nakamura A, Sugimoto Y, Tsuruo T, Fujita N. FOXO transcription factor-dependent p15(INK4b) and p19(INK4d) expression. Oncogene. 2008 doi: 10.1038/sj.onc.1210813. in press. [DOI] [PubMed] [Google Scholar]
- •33.Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massague J. C/EBPbeta at the core of the TGFbeta cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell. 2006;10:203–214. doi: 10.1016/j.ccr.2006.07.019. See Note for 34.
- •34.Gomis RR, Alarcon C, He W, Wang Q, Seoane J, Lash A, Massague J. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci U S A. 2006;103:12747–12752. doi: 10.1073/pnas.0605333103. 33 and 34 reveal that the growth-inhibitory cytokine transforming growth factor β (TGFβ) induces the formation of a transcriptionally active complex between Smad, FoxO and other transcription factors, such as C/EBPβ, to result in cytostasis. TGFβ elicits the expression of a cytostatic gene program in a number of progenitor and tumor cell types, including epithelial and metastatic breast cancer cells.
- •35.Huang H, Regan KM, Lou Z, Chen J, Tindall DJ. CDK2-dependent phosphorylation of FOXO1 as an apoptotic response to DNA damage. Science. 2006;314:294–297. doi: 10.1126/science.1130512. This study demonstrates that the protein kinase CDK2 phosphorylates FoxO1 at serine 249, which contributes to the sequestration of FoxO1 in the cytoplasm. DNA damage abrogates this phosphorylation, thereby triggering FoxO1 activation and apoptotic cell death.
- •36.Greer EL, Dowlatshahi D, Banko MR, Villen J, Hoang K, Blanchard D, Gygi SP, Brunet A. An AMPK-FOXO pathway mediates longevity induced by a novel method of dietary restriction in C. elegans. Curr Biol. 2007;17:1646–1656. doi: 10.1016/j.cub.2007.08.047. 36 and 37 show that the energy-sensing AMPK is required for the beneficial effects of one dietary restriction method on C. elegans lifespan. AMPK directly phosphorylates DAF-16 and mammalian FoxO3 at six residues. Phosphorylation of mammalian FoxO3 by AMPK is necessary for the upregulation of genes involved in stress resistance and energy metabolism.
- •37.Greer EL, Oskoui PR, Banko MR, Maniar JM, Gygi MP, Gygi SP, Brunet A. The energy sensor AMP-activated protein kinase directly regulates the mammalian FOXO3 transcription factor. J Biol Chem. 2007;282:30107–30119. doi: 10.1074/jbc.M705325200. See Note for 36.
- •38.Bakker WJ, Harris IS, Mak TW. FOXO3a Is Activated in Response to Hypoxic Stress and Inhibits HIF1-Induced Apoptosis via Regulation of CITED2. Mol Cell. 2007;28:941–953. doi: 10.1016/j.molcel.2007.10.035. This study shows that hypoxic stress triggers the activation of FoxO3 by upregulating FoxO3 mRNA. In response to hypoxia, FoxO3 regulates the expression of the transcriptional cofactor Cited 2, which results in the inhibition of HIF1-dependent apoptosis in both normal and cancer cells.
- 39.Shaw WM, Luo S, Landis J, Ashraf J, Murphy CT. The C. elegans TGF-beta Dauer pathway regulates longevity via insulin signaling. Curr Biol. 2007;17:1635–1645. doi: 10.1016/j.cub.2007.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siegenthaler JA, Miller MW. Generation of Cajal-Retzius neurons in mouse forebrain is regulated by transforming growth factor beta-Fox signaling pathways. Dev Biol. 2008;313:35–46. doi: 10.1016/j.ydbio.2007.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 42.Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- 43.Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 44.Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- 45.Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- 46.Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. 2007;303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- 48.Giannakou ME, Goss M, Junger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- 49.Hwangbo DS, Gersham B, Tu MP, Palmer M, Tatar M. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 2004;429:562–566. doi: 10.1038/nature02549. [DOI] [PubMed] [Google Scholar]
- 50.Luong N, Davies CR, Wessells RJ, Graham SM, King MT, Veech R, Bodmer R, Oldham SM. Activated FOXO-mediated insulin resistance is blocked by reduction of TOR activity. Cell Metab. 2006;4:133–142. doi: 10.1016/j.cmet.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 51.Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biggs WH, 3rd, Cavenee WK, Arden KC. Identification and characterization of members of the FKHR (FOX O) subclass of winged-helix transcription factors in the mouse. Mamm Genome. 2001;12:416–425. doi: 10.1007/s003350020002. [DOI] [PubMed] [Google Scholar]
- 53.Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 54.Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich A, et al. Large-scale analysis of the human and mouse transcriptomes. Proc Natl Acad Sci U S A. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature. 2003;421:182–187. doi: 10.1038/nature01298. [DOI] [PubMed] [Google Scholar]
- 56.Selman C, Lingard S, Choudhury AI, Batterham RL, Claret M, Clements M, Ramadani F, Okkenhaug K, Schuster E, Blanc E, et al. Evidence for lifespan extension and delayed age-related biomarkers in insulin receptor substrate 1 null mice. Faseb J. 2008 doi: 10.1096/fj.07-9261com. in press. [DOI] [PubMed] [Google Scholar]
- •57.Matsumoto M, Pocai A, Rossetti L, Depinho RA, Accili D. Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor foxo1 in liver. Cell Metab. 2007;6:208–216. doi: 10.1016/j.cmet.2007.08.006. This study demonstrates that FoxO1 ablation in the hepatocytes of mice results in reduced glucose levels at birth, and in adult mice in response to fasting. FoxO1 ablation attenuates fasting-induced glycogenolysis and gluconeogenesis. FoxO1 ablation in the liver of insulin receptor Insr null mice significantly improves the hyperglycemia of the Insr null mice, suggesting that inhibiting FoxO1 activity would be beneficial to treat diabetes.
- 58.Hall RK, Yamasaki T, Kucera T, Waltner-Law M, O’Brien R, Granner DK. Regulation of phosphoenolpyruvate carboxykinase and insulin-like growth factor-binding protein-1 gene expression by insulin. The role of winged helix/forkhead proteins. J Biol Chem. 2000;275:30169–30175. doi: 10.1074/jbc.M004898200. [DOI] [PubMed] [Google Scholar]
- 59.Buteau J, Shlien A, Foisy S, Accili D. Metabolic diapause in pancreatic beta-cells expressing a gain-of-function mutant of the forkhead protein Foxo1. J Biol Chem. 2007;282:287–293. doi: 10.1074/jbc.M606118200. [DOI] [PubMed] [Google Scholar]
- 60.Okamoto H, Hribal ML, Lin HV, Bennett WR, Ward A, Accili D. Role of the forkhead protein FoxO1 in beta cell compensation to insulin resistance. J Clin Invest. 2006;116:775–782. doi: 10.1172/JCI24967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••61.Kim MS, Pak YK, Jang PG, Namkoong C, Choi YS, Won JC, Kim KS, Kim SW, Kim HS, Park JY, et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nat Neurosci. 2006;9:901–906. doi: 10.1038/nn1731. See Note for 62.
- ••62.Kitamura T, Feng Y, Kitamura YI, Chua SC, Jr, Xu AW, Barsh GS, Rossetti L, Accili D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat Med. 2006;12:534–540. doi: 10.1038/nm1392. 61 and 62 show that a constitutively active form of FoxO1 expressed in the arcuate nucleus of the hypothalamus increases food intake and body weight by directly inducing the transcription of orexigenic peptides agouti-related protein (Agrp) and neuropeptide Y (Npy). The anorexigenic hormones insulin and leptin suppress feeding behaviors by inhibiting FoxO1. FoxO1 also suppresses the expression of the anorexigenic peptide pro-opiomelanocortin (Pomc).
- ••63.Paik JH, Kollipara R, Chu G, Ji H, Xiao Y, Ding Z, Miao L, Tothova Z, Horner JW, Carrasco DR, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. This is the first study to ablate multiple members of the FoxO family in vivo and demonstrate that the FoxO family are bona fide tumor suppressors. Acute deletion of FoxO1, FoxO3 and FoxO4 in the thymus and endothelial cells resulted in age-dependent progression of thymic lymphomas and hemangiomas. This study also performs a comprehensive transcriptome and promoter analysis to identify many novel FoxO target genes in the endothelium and shows that Sprouty2 is a key target gene of FoxO factors that promotes endothelial cell quiescence.
- •64.Potente M, Urbich C, Sasaki K, Hofmann WK, Heeschen C, Aicher A, Kollipara R, DePinho RA, Zeiher AM, Dimmeler S. Involvement of Foxo transcription factors in angiogenesis and postnatal neovascularization. J Clin Invest. 2005;115:2382–2392. doi: 10.1172/JCI23126. This is a comprehensive study showing that FoxO1 and FoxO3, but not FoxO4, inhibit endothelial cell migration and vessel formation, both in cultured cells and in vivo. This study also demonstrates that while the isoforms of the FoxO family do have overlapping functions, they also display differential preferences for target genes such as angiopoietin 2 (Ang2).
- 65.Massey AC, Zhang C, Cuervo AM. Chaperone-mediated autophagy in aging and disease. Curr Top Dev Biol. 2006;73:205–235. doi: 10.1016/S0070-2153(05)73007-6. [DOI] [PubMed] [Google Scholar]
- •66.Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. This study and 67 show that FoxO1 and FoxO3 promote the atrophy of both cultured myotubes and muscle fibers by inducing the ubiquitin ligases atrogin-1/MAFbx and MuRF1 in response to starvation, glucocorticoids and denervation.
- •67.Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. See Note for 66.
- 68.Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. Embo J. 2006;25:554–564. doi: 10.1038/sj.emboj.7600945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, Goldberg AL, Spiegelman BM. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–16265. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••70.Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, Burden SJ, Di Lisi R, Sandri C, Zhao J, et al. FoxO3 Controls Autophagy in Skeletal Muscle In Vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. This study and 72 reveals that FoxO3 is necessary and sufficient to induce autophagy in skeletal muscle fibers by inducing autophagy-related genes such as LC3 and the Bcl2-related autophagy regulator Bnip3. The gene program induced by FoxO3 is similar to the profile of genes induced in the mouse skeletal muscle in response to denervation and fasting.
- 71.Suzuki N, Motohashi N, Uezumi A, Fukada S, Yoshimura T, Itoyama Y, Aoki M, Miyagoe-Suzuki Y, Takeda S. NO production results in suspension-induced muscle atrophy through dislocation of neuronal NOS. J Clin Invest. 2007;117:2468–2476. doi: 10.1172/JCI30654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ••72.Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, Lecker SH, Goldberg AL. FoxO3 Coordinately Activates Protein Degradation by the Autophagic/Lysosomal and Proteasomal Pathways in Atrophying Muscle Cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. See Note for 70.
- 73.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of Forkhead proteins. J Clin Invest. 2007;117:3211–3223. doi: 10.1172/JCI31757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Juhasz G, Puskas LG, Komonyi O, Erdi B, Maroy P, Neufeld TP, Sass M. Gene expression profiling identifies FKBP39 as an inhibitor of autophagy in larval Drosophila fat body. Cell Death Differ. 2007;14:1181–1190. doi: 10.1038/sj.cdd.4402123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •75.Shinoda S, Schindler CK, Meller R, So NK, Araki T, Yamamoto A, Lan JQ, Taki W, Simon RP, Henshall DC. Bim regulation may determine hippocampal vulnerability after injurious seizures and in temporal lobe epilepsy. J Clin Invest. 2004;113:1059–1068. doi: 10.1172/JCI19971. This study is the first to demonstrate that induction of epilepsy in rats results in the activation of FoxO1 and FoxO3, the upregulation of Bim and the induction of neuronal cell death in vivo. A general phosphatase inhibitor blocks dephosphorylation of FoxO1 and FoxO3 and protects against neuronal death in response to seizures.
- 76.Luo X, Puig O, Hyun J, Bohmann D, Jasper H. Foxo and Fos regulate the decision between cell death and survival in response to UV irradiation. Embo J. 2007;26:380–390. doi: 10.1038/sj.emboj.7601484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- 78.Peng SL. Immune regulation by Foxo transcription factors. Autoimmunity. 2007;40:462–469. doi: 10.1080/08916930701464913. [DOI] [PubMed] [Google Scholar]
- ••79.Tothova Z, Kollipara R, Huntly BJ, Lee BH, Castrillon DH, Cullen DE, McDowell EP, Lazo-Kallanian S, Williams IR, Sears C, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. This is the first study to identify the critical role of the FoxO family in the maintenance of the hematopoietic stem cell (HSC) pool by ablating FoxO1, FoxO3 and FoxO4 in the bone marrow. FoxO-deficient HSCs displayed increased cell cycling, increased apoptosis, increased production of ROS and an impaired ability to repopulate the bone marrow long-term. Administration of the anti-oxidant N-acetyl-L-cysteine in vivo resulted in significant recovery of the FoxO-deficient HSC phenotype.
- •80.Lin L, Hron JD, Peng SL. Regulation of NF-kappaB, Th activation, and autoinflammation by the forkhead transcription factor Foxo3a. Immunity. 2004;21:203–213. doi: 10.1016/j.immuni.2004.06.016. This study reveals that FoxO3 ablation in vivo results in lymphoproliferation, and the inflammation of several organs. The loss of FoxO3 was associated with hyperactivated helper T cells with increased proliferation, and increased Th1 and Th2 cytokine production due to NF-κB activation.
- 81.Wyss-Coray T. Inflammation in Alzheimer disease: driving force, bystander or beneficial response? Nat Med. 2006;12:1005–1015. doi: 10.1038/nm1484. [DOI] [PubMed] [Google Scholar]
- ••82.Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, et al. FoxO3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. This study reports that FoxO3 alone plays a crucial role in the maintenance of the hematopoietic stem cell (HSC) pool by promoting quiescence and detoxifying ROS. Interestingly, the phenotype was progressive with age since the HSC pool was significantly decreased in aged FoxO3 null mice.
- 83.Bouchard C, Lee S, Paulus-Hock V, Loddenkemper C, Eilers M, Schmitt CA. FoxO transcription factors suppress Myc-driven lymphomagenesis via direct activation of Arf. Genes Dev. 2007;21:2775–2787. doi: 10.1101/gad.453107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hu MC, Lee DF, Xia W, Golfman LS, Ou-Yang F, Yang JY, Zou Y, Bao S, Hanada N, Saso H, et al. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 2004;117:225–237. doi: 10.1016/s0092-8674(04)00302-2. [DOI] [PubMed] [Google Scholar]
- 85.Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC, et al. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138–148. doi: 10.1038/ncb1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Castrillon DH, Miao L, Kollipara R, Horner JW, DePinho RA. Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science. 2003;301:215–218. doi: 10.1126/science.1086336. [DOI] [PubMed] [Google Scholar]