Abstract
Background
Microsporidia are well known models of extreme nuclear genome reduction and compaction. The smallest microsporidian genomes have received the most attention, but genomes of different species range in size from 2.3 Mb to 19.5 Mb and the nature of the larger genomes remains unknown.
Results
Here we have undertaken genome sequence surveys of two diverse microsporidia, Brachiola algerae and Edhazardia aedis. In both species we find very large intergenic regions, many transposable elements, and a low gene-density, all in contrast to the small, model microsporidian genomes. We also find no recognizable genes that are not also found in other surveyed or sequenced microsporidian genomes.
Conclusion
Our results demonstrate that microsporidian genome architecture varies greatly between microsporidia. Much of the genome size difference could be accounted for by non-coding material, such as intergenic spaces and retrotransposons, and this suggests that the forces dictating genome size may vary across the phylum.
Background
Microsporidia are obligate intracellular eukaryotic parasites that have been found to infect members of all major animal lineages [1]. The many apparently "primitive" features of microsporidian cells led evolutionary biologists to suggest that they were an early-branching lineage of the eukaryotes [2,3], but molecular phylogeny has since shown that they are instead a derived relative of fungi [4,5]. In light of this, their seemingly primitive features have been re-evaluated as products of reduction and adaptation to life inside another cell [4-8].
One such feature that has attracted considerable attention is their highly reduced genomes. The genome size has been determined for numerous microsporidian species, and they range from 19.5 Mbp in Glugea atherinae to just 2.3 Mbp in Encephalitozoon intestinalis, the smallest eukaryotic genome known [9]. The two best-studied examples are the vertebrate parasite Encephalitozoon cuniculi (2.9 Mbp), the genome of which has been completely sequenced and encodes 1,997 protein-coding genes [10], and the insect parasite Antonospora locustae (5.4 Mbp) for which two sequence surveys are available [11,12]. These two species revealed just how microsporidian genomes had become so small compared with those of other eukaryotes. There has been a severe reduction in the number of genes in the genome, most likely a reflection of the fact that microsporidia are dependent on their hosts for many metabolic processes and import many compounds from their host. Furthermore the genes that remain are packed together very densely: intergenic spaces are minimal. In E. cuniculi there are no selfish elements and just 15 small introns. Genes in E. cuniculi are also shorter than their homologues in Saccharomyces cerevisiae, which is hypothesized to result from the small number of proteins within the cell, and a correspondingly smaller interaction network [10,13]. This extreme compaction appears to have resulted in a high level of gene order conservation between different species of microsporidia [12,14-16] and an unusually high level of overlapping transcription between adjacent genes [17,18].
Because the smallest microsporidian genomes are so unusual, they have garnered the greatest attention, and to date no large-scale survey of any larger genomes is available. This is unfortunate, because the form and content of these larger microsporidian genomes could differ from the smaller ones in many potentially interesting ways. On one hand they may contain a great many more genes, and could therefore reflect a greater cellular or metabolic complexity than the microsporidian parasites we presently know best. On the other hand these genomes may encode a great deal more non-coding DNA, which would have interesting implications for genome evolution within the group, and for why the smaller genomes are so compact. In other eukaryotes, it appears that variation in genome size on a relatively short evolutionary time scale is due to increased or decreased proportions of transposable elements in a genome [19]. There are hints that this may also be at least partially true in some microsporidian genomes where selfish elements have been found [20,21]. Most interestingly, a number of TY3/Gypsy retrotransposons have recently been described from the 15.3 Mbp genome of Nosema bombycis [22]. These elements are apparently flanked by areas of compacted genes that share a high level of synteny with E. cuniculi, perhaps suggesting an invasion of transposable elements into a compacted genome. More recently, the previously unsequenced subtelomeric areas of E. cuniculi have been investigated and found to contain a large family of proteins with at least 30 distinct members. This family of duplicated proteins is also found within other human infecting microsporidia [23], demonstrating that the expansion of gene families might be more common than previously thought.
Here, we describe low-redundancy genome sequence surveys of two distantly related microsporidia, Brachiola algerae (recently proposed to be renamed Anncaliia algerae [24]) and Edhazardia aedis. Both of these species have mosquitoes as their type hosts with the former having a very broad host range, including man, and the latter restricted to mosquitoes [25]. The genome size of E. aedis is not known, whereas the genome size of B. algerae has been estimated to be 15–20 Mbp [23]. In both species, we show that gene density is very low compared to that of the better-studied species of microsporidia. Specifically, we have found a considerable proportion of repetitive elements in both genomes, large stretches of non-coding DNA, and some evidence that the gene density may vary over the genome. These surveys open the possibility that microsporidian genomes are not universally compacted, large genomes do not necessarily encode significantly more genes than do the smaller genomes, and that dense genomes may sometimes revert to a gene-sparse state.
Results and discussion
General features of the sequence data
Two short-insert genomic libraries were constructed from B. algerae and a total of 219 clones fully or partially sequenced to yield 203,748 bp of non-overlapping sequence. A single E. aedis library was constructed and 290 sequence reads from 182 clones yielded 233,509 bp of non-overlapping sequence. Comparing these with the dense, gene-rich genome of E. cuniculi (Table 1) reveals a sharp contrast in the overall nature of the genomes. From the B. algerae survey, 34 genes with identifiable homologues in other organisms were identified, and a further 33 potential ORFs greater than 100 codons but with no recognizable similarity to any other gene were found, resulting in a protein-coding content of 11% identifiable coding sequence and 20% including putative ORFs. In E. aedis, only 25 identifiable protein-coding genes and 21 ORFs were found, pushing the range of coding sequence still lower, to 8% identifiable coding sequence or 14% if putative ORFs are included (Table 1). In contrast, 52% of the E. cuniculi genome consists of protein-coding sequences with recognizable similarity to other genes, and the proportion of coding sequence is 87% when ORFs are included [10]. The gene density of A. locustae is similar to that of E. cuniculi [12], as are small regions of other microsporidian genomes that have been sampled [20]. Overall, the gene-densities of B. algerae and E. aedis are about an order of magnitude lower than other microsporidia that have been examined to date.
Table 1.
Summary of the data compared to the genome of E. cuniculi
| E. cuniculi [10] | B. algerae | E. aedis | ||||
| bp | % | bp | % | bp | % | |
| Total sequenced | 2,497,519 | 203,748 | 233,509 | |||
| coding | 2,180,498 | 87 | 41153(67) | 20 | 33,617 (46) | 14 |
| hits | 1,320,216 | 61 | 22260 (34) | 54 | 19,099 (25) | 57 |
| ORFS | 860,274 | 39 | 18893 (33) | 46 | 14,518 (21) | 43 |
| Non-coding | 317,029 | 13 | 162595 | 80 | 199,892 | 86 |
Numbers in brackets indicates the number of genes or gene fragments in each sample. The percentages refer to the proportion of each sequenced class relative to the total amount sequenced.
The overall GC content for both B. algerae (24%) and E. aedis (25%) is also significantly lower than that of E. cuniculi (47%). Not surprisingly, the GC content in the coding regions is slightly higher: 28% for B. algerae and 31% for E. aedis. A smaller sequence survey from Spraguea lophii with a genome size of 6.2 Mb has revealed a bias of 28% [20,26]. There is therefore no obvious correlation between genome size and drift towards low GC content in microsporidia, and similarly no pronounced lineage-specific bias.
Presence of transposable elements
In contrast to the E. cuniculi genome, which does not encode any selfish genetic elements, fragments of reverse transcriptase or complete retrotransposons have been reported in the genomes of N. bombycis, V. corneae and S. lophii [20-22], and repeated sequences suggested to be mobile were reported in N. bombycis and N. costelytrae [27]. The V. corneae reverse transcriptase is closely related to a human LINE sequence, and both the N. bombycis and S. lophii retrotransposons have sequence similarity to each other and to Ty3/gypsy retrotransposons.
In both B. algerae and E. aedis surveys we found extensive evidence of numerous transposable elements (Table 2). The E. aedis fragments all share high similarity to Ty3/gypsy retrotransposons from the N. bombycis and S. lophii. Nine of the seventeen fragments of putatively selfish elements identified in B. algerae are also members of the same family, and once more also share a high degree of similarity to the N. bombycis and S. lophii elements. The remaining fragments from B. algerae were similar to hypothetical proteins resembling transposases.
Table 2.
List of hits to suspected transposable elements:
| Top Blast hit annotation | Accession no. |
| Brachiola algerae | |
| Pol polyprotein Nosema bombycis | 91176517 |
| Pol polyprotein Nosema bombycis | 91176521 |
| Pol polyprotein Nosema bombycis | 91176521 |
| Pol polyprotein Nosema bombycis | 91176521 |
| Pol polyprotein Nosema bombycis | 91176521 |
| Pol polyprotein Nosema bombycis | 91176523 |
| Pol polyprotein Nosema bombycis | 91176523 |
| Pol polyprotein Nosema bombycis | 91176525 |
| Pol polyprotein Nosema bombycis | 91176525 |
| Transposase, putative Acaryochloris marina | 158337326 |
| Predicted protein, Nematostella vectensis | 156394155 |
| Conserved hypothetical protein Akkermansia muciniphila | 166832600 |
| Neisseria meningitidis IS1016 transposase | 161869234 |
| Caenorhabditis briggsae hypothetical protein CBG18017 | 157775203 |
| Caenorhabditis briggsae hypothetical protein CBG18017 | 157775203 |
| Caenorhabditis briggsae Hypothetical protein CBG00277 | 157771110 |
| Caenorhabditis briggsae Hypothetical protein CBG21915 | 157749299 |
| Edhazardia aedis | |
| Pol polyprotein Nosema bombycis | 91176525 |
| 6 different Pol polyproteins Nosema bombycis | 91176519 |
| Pol polyprotein Nosema bombycis | 91176525 |
The level of similarity between the Ty3/gypsy elements from N. bombycis, S. lophii, E. aedis and B. algerae and the fact that the host groups for these four species are not closely related, is strongly suggestive that an ancestral family of retrotransposons existed in the common ancestor of these microsporidia. In molecular phylogenies of microsporidia, the true Nosema-group is consistently found to be a sister-lineage of the Encephalitozoon-group to the exclusion of lineages that include E. aedis, B. algerae and S. lophii [28-30]. If the Ty3/gypsy retroelements identified here are ancestral to the genomes where they have been found, it means that it was also ancestral to E. cuniculi and must have been completely purged from its genome. This raises some curious questions about the N. bombycis genome. Here, the Ty3/gypsy retrotransposons were found to be nested within blocks of compacted genes that were often conserved in order with homologues from other microsporidian genomes [22]. Based on this it was suggested that the elements could have invaded a compact genome, and perhaps later facilitated some genomic rearrangements [22]. Reconciling the ancient presence of these elements with the nature of the N. bombycis genome is complicated. It is possible that the ancestral genome contained many such elements and had a low gene density. This genome could have subsequently compacted in several lineages, some of which lost the retroelements as part of the compaction process (e.g., E. cuniculi), while others kept them and compacted the genome around them (e.g., N. bombycis). It is also possible that compaction happened in an earlier common ancestor of some of these lineages and that certain genomes have 're-expanded'. In either event, the retention of large numbers of selfish elements in an otherwise compact genome is of interest, as one might expect that compaction would be strongly inclined to lead to the loss of non-coding material such as selfish elements. It serves to illustrate the way compaction affects different aspects of the genome in different lineages, another possible example being the differential loss or retention of introns in relict nucleomorph genomes [31,32].
Gene density, order, and size
The small number of genes identified and the large continuous stretches of non-coding sequence in both surveys lead to the obvious conclusion that the gene-density of these genomes is much lower than those of E. cuniculi or A. locustae. The average intergenic distances in these genomes cannot readily be determined since few have been completely sequenced. In B. algerae four clones encoded two genes and the distances between them are 108, 206, 276, and 552 bp. In E. aedis a single clone encoded two adjacent genes, and the intergenic spaces between them is 1,324 bp. At the same time, the largest continuous stretches of sequence from which we could identify no genes were 2,412 (or 2,943 in a likely subtelomeric region next to an SSU gene) and 2,068 bp in B. algerae and E. aedis, respectively. The average distance between genes in E. cuniculi and A. locustae samples is 129 and 211 bp [10,12]. From the existing data it seems likely that the average distance between genes in B. algerae and E. aedis is much larger than that of either of the well studied microsporidian genomes, and that the density across at least the B. algerae genome may be more heterogeneous.
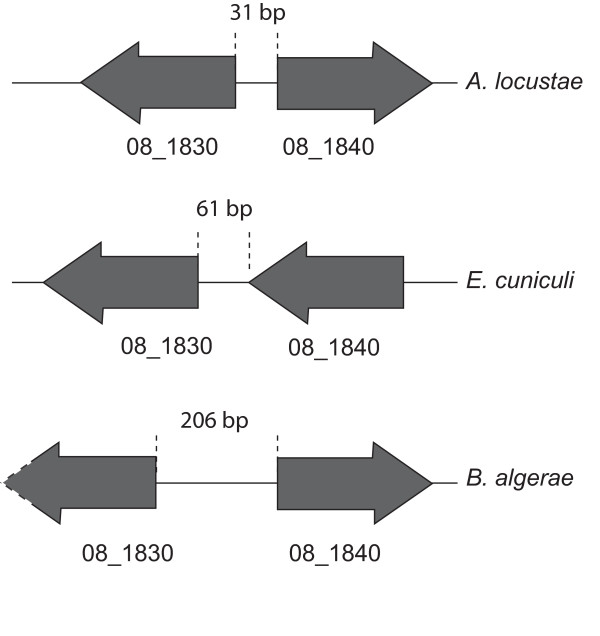
Of the pairs of adjacent genes we identified, one B. algerae pair is also adjacent in both A. locustae and E. cuniculi (Figure 1) (the pair separated by 206 bp in B. algerae). It has previously been shown that the order of gene pairs in A. locustae and E. cuniculi is highly conserved, and this has been hypothesized to be related to the compaction of the genome [12]. The conservation of one of four B. algerae gene pairs suggests that areas of this genome may be under similar constraints. If the conservation of genome order is related to compaction, this also suggests that the compacted state may have existed in the ancestor of B. algerae, E. cuniculi and A. locustae, which is consistent with phylogenies that suggest some relationship between B. algerae and A. locustae [[25] and unpublished data].
Figure 1.
Area of conserved gene synteny between three species of microsporidia. A fragment of the B. algerae genome aligned to corresponding regions from E. cunculi and A. locustae. The gene order, but not orientation, is conserved. Arrowheads indicate gene orientation and dashed white line indicates incomplete gene sequences. Intergenic space lengths are indicated.
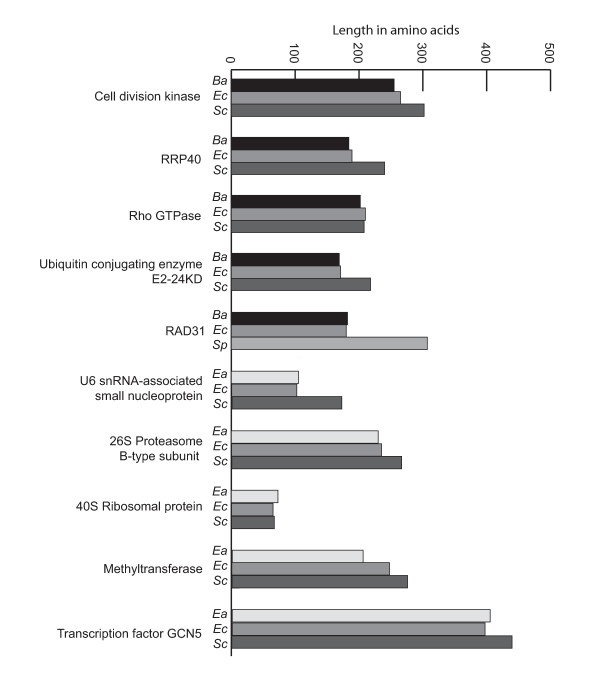
In addition to being densely packed, E. cuniculi genes have also been shown to be shorter on average than homologues in the S. cerevisiae genome [10]. This has been discussed in the context of genome compaction, but also suggested to be the result of a reduction in the number of proteins in the cell, which leads to smaller interaction networks, which in turn allows proteins to reduce their number or complexity of interacting domains [13]. In yeast, it has been shown that there is a correlation between protein size and connectivity, with larger proteins displaying a greater number of interactions [33]. We examined the only five full-length genes identified in the B. algerae survey with homologues in yeast and found that all five were shorter than S. cerevisiae homologues, and more surprisingly most were also shorter than the E. cuniculi homologues (Figure 2). Similarly, from the E. aedis survey, only five full-length genes were found (Figure 2), and four of these were shorter than S. cerevisiae homologues, and comparable in size with the E. cuniculi homologues. The sole E. aedis protein predicted to be larger than the yeast counterpart encodes a ribosomal protein (Figure 2). Given that these genomes are not compacted, it suggests that proteins are either shorter due to a reduced proteome complexity, or that they had an ancestor with a compacted genome, or both.
Figure 2.
Comparison of microsporidian and yeast protein lengths. The number of codons for all full-length proteins found within B. algerae (Ba) and E. aedis (Ea) sequence surveys compared to homologues from S. cerevisiae (Sc) (or Schizosaccahromyces pombe (Sp) in cases where S. cerevisiae does not encode a homologue).
Gene content and coding capacity
There is no experimental estimate of the E. aedis genome size, but the B. algerae genome has been estimated to be between 15–20 Mbp by pulsed-field gel electrophoresis [23]. This is much larger than the genomes of either E. cuniculi (2.9 Mbp) or A. locustae (5.4 Mbp). As we show, much of the genome size difference can be attributed to the significantly lower gene-densities of B. algerae and E. aedis. However, it is still possible that one or both of these genomes is also larger because it contains more genes than E. cuniculi.
The E. cuniculi genome clearly under went a massive gene loss relative to other eukaryotes, but this gene loss may have been ancestral to microsporidia. If this were the case we would expect to find few genes in E. aedis and B. algerae that are present and conserved in other eukaryotes that are not also present in E. cuniculi. That is to say that the B. algerae and E. aedis genomes would not have more conserved genes than the pool remaining in the ancestral microsporidian after this gene loss event.
Our sampling of E. aedis and B. algerae genomes shows that this scenario is quite possible. Of the protein-coding genes with identifiable homologues in some other genome that we found in B. algerae (34 cases) and E. aedis (25 cases), every one is also present in E. cuniculi (Table 3). Given the sample size, it is likely that either genome could contain some genes found in other organisms but not E. cuniculi, but it is unlikely that they are abundant. This lack of excess conserved gene homologues is of interest because it implies that the large-scale gene loss characteristic of E. cuniculi took place relatively early in microsporidian evolution, in the ancestor of E. cuniculi, A. locustae, E. aedis and B. algerae.
Table 3.
List of genes identified by BLAST search
| Brachiola algerae | Edhazardia aedis | ||
| Hit | E. cuniculi locus | Hit | E. cuniculi locus |
| Cell Division Kinase | 08_0230 | 14-3-3 Protein 1 | 03_1010 |
| Coatomer coat delta | 08_0340 | 26S proteasome beta-type subunit | 05_0290 |
| DNA directed RNA pol | 01_0600 | 40S ribosomal protein S28 | 09_1275 |
| DNA ligase | 02_1220 | 60S ribosomal protein L8 | 01_0310 |
| DNA mismatch repair | 11_1260 | Aldose Reductase | 01_0970 |
| Dnm1 | 01_1210 | DNA Mismatch Repair Protein | 05_0300 |
| E. cuniculi hypothetical protein | 01_0390 | Endochitinase | 09_1320 |
| E. cuniculi hypothetical protein | 04_0270 | E. cuniculi hypothetical protein | 03_0870 |
| E. cuniculi hypothetical protein | 05_1460 | E. cuniculi hypothetical protein | 05_1000 |
| E. cuniculi hypothetical protein | 06_0970 | E. cuniculi hypothetical protein | 05_1080 |
| E. cuniculi hypothetical protein | 07_0810 | E. cuniculi hypothetical protein | 09_1690 |
| E. cuniculi hypothetical protein | 08_1830 | Myosin heavy chain | 09_1970 |
| E. cuniculi hypothetical protein | 08_1840 | NIFS-like protein | 11_1770 |
| E. cuniculi hypothetical protein | 09_0300 | Phospholipid-transporting ATPase | 09_1440 |
| E. cuniculi hypothetical protein | 09_1240 | Putative methyltransferase | 05_0950 |
| E. cuniculi hypothetical protein | 10_1360 | RING-finger-containing ubiquitin ligase | 07_0330 |
| E. cuniculi hypothetical protein | 11_0260 | Similarity to oxidoreductase | 11_1070 |
| GPI Anchor Biosynthesis | 09_1210 | SSU gene | |
| Hsp70 | 02_0100 | Topoisomerase 1 | 06_1520 |
| Isopentyl pyrophosphate δ isomerase | 02_0230 | TPR domain hypothetical protein | 09_1180 |
| LSU gene | Transcriptional activator | 10_1430 | |
| Pelota protein | 03_1380 | Translation initiation factor IF-2P | 09_0070 |
| Phenylalanine tRNA synthase | 07_1660 | Trehalose-6-phosphate synthase | 01_0870 |
| RAD31 DNA damage tolerance | 08_0460 | U6 snRNA-associated small RNP | 05_1310 |
| RAS-like GTP binding protein | 10_0350 | UTP glucose-1-phosphate uridyltransferase | 03_0280 |
| Septin | 09_0820 | Vacuolar protein sorting-associated protein | 03_0900 |
| SER/THR protein kinase | 08_1620 | ||
| Signal Recognition Particle | 04_0980 | ||
| SSU gene | |||
| Syntaxin | 05_0820 | ||
| TFIID 111 KDa | 01_0760 | ||
| TFIID 72/90 KDa | 11_1750 | ||
| TFIID 150 kDa | 09_0090 | ||
| TFIID I | 04_1440 | ||
| U5 Associated snRNP | 11_0870 | ||
| Ubiquitin Conjugating Enzyme E2 | 08_0860 | ||
However, the B. algerae survey consisted of 20% coding sequence, so taking into account the range or estimated genome sizes for B. algerae (15–20 Mbp), this suggests between 2,786 and 3,714 genes in the Brachiola genome (assuming an average gene length of 1,077 as in E. cuniculi). The discrepancy between this predicted coding capacity of B. algerae and the observation that all the recognizable genes we sampled are shared with E. cuniculi could be explained in many ways. First, our sample may be biased to gene-encoding regions, and this would lead to an overestimate of the gene-density. Second, a large number of lineage-specific ORFs could skew the estimate, but the proportion of ORFs we found (47% and 43% for B. algerae and E. aedis, respectively) is similar to that found in E. cuniculi (39%). This issue is also complicated by the fact that we identified several E. cuniculi "ORFs" in our sample, and therefore the proportions of putative ORFs is changing. Third, the genome size estimates may be wrong. Lastly, it is possible that there are many more than 2,000 genes in these organisms, but that the excess is mostly due to recent duplications. We did not sample any duplicates in either genome, though we did find areas of repeats amongst the non-coding areas in both B. algerae and E. aedis. If gene duplications are common, the genome could contain more genes without an increased complexity in the proteome.
Evolution of genome compaction in microsporidia
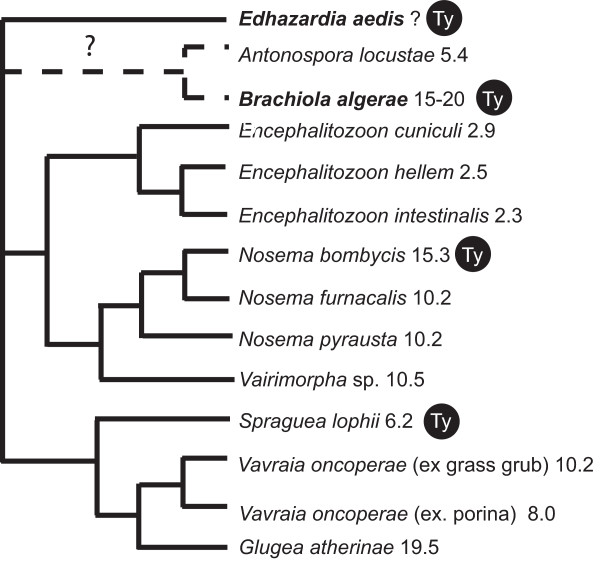
Though the phylogenetic relationships of major microsporidian lineages are not well resolved, phylogenies of rRNA [28] and concatenated tubulin genes (unpublished data) suggest a relationship between B. algerae and A. locustae to the exclusion of E. aedis or E. cuniculi (Figure 3). This raises interesting questions about whether microsporidian genomes have compacted more than once during the diversification of the phylum, or if some have re-expanded from a compacted state.
Figure 3.
Schematic consensus of microsporidian phylogenetic relationships. Microsporidian relationships from a consensus of published SSU phylogenies [28, 30, 40] and concatenated tubulin genes (unpublished data). Genome sizes are labeled and the reported presence of Gypsy/Ty transposons is indicated by (Ty).
An obvious factor in the dynamics of genome size is transposable elements. One could imagine a genome expanding due to the invasion of such elements, and indeed many such elements have been found in N. bombycis [22] and now B. algerae and E. aedis. However, the majority of these elements are closely related members of the Ty3/gypsy family. Therefore genome expansion cannot be entirely due to an invasion since the elements must have existed in the common ancestor and been purged from E. cuniculi and possibly other compacted microsporidian genomes. It also remains to be seen if these genomes are substantially heterogeneous. It is possible that many genes do exist in relatively compact regions while other regions are dominated by non-coding sequence. An extreme version of such a situation is seen in the small and compacted genome of the picoplankton Ostreococcus tauri. Here most chromosomes in the genome show a high gene density but 2 chromosomes out of 20 contain 77% of the transposons identified in the genome [34].
A second factor that has been hypothesized to affect genome size is cell size. A correlation between genome size and cell size has been observed in eukaryotes generally [35] and microsporidia specifically [36]. However in the microsporidia, variation in cell size in the different life stages can confound correlations between genome size and cell size. As a rough correlation though, the genome size of B. algerae is estimated at 15–20 Mb and that of A. locustae is reported to be 5.4 Mb [37], whereas the spores of both B. algerae and A. locustae are of comparable sizes [38,39] suggesting, in this case, that cell size is not necessarily a factor. A further consideration is whether the complexity of the life cycles of different microsporidia is reflected in genome size and gene number. Both A. locustae and E. cuniculi have a simple life cycle with monomorphic spores and are restricted to a narrow host range. Edhazardia aedis has a more complex life cycle with multiple spore types and must be adapted to both the larval and adult stage of the mosquito. Brachiola algerae, known to have a larger genome, has a simple life cycle with monomorphic spores, but has a broad host range and can infect both mammals and insects.
Conclusion
The E. cuniculi genome is a model for compacted nuclear genomes, but potentially not a good model for microsporidian genomes generally. We have shown that the genomes of B. algerae and E. aedis are structured very differently: they have large proportions of non-coding sequence and many transposable elements, resulting in a very low and perhaps variable gene-density compared with E. cuniculi. The sample of identifiable genes found in the surveys, and the proportions of these genes shared with E. cuniculi both suggest that the complexity of the proteome is not the major factor contributing to genome size variation. The phylogeny of microsporidia suggests multiple events of compaction and/or expansion, which raises interesting questions about what forces the genomes to compact so severely, and why such a force would then cease to operate on the genome.
Methods
Microsporidia, genomic DNA extraction, and genomic library construction
6.1 × 107 uninucleate Edhazardia aedis spores harvested from Aedes aegypti larvae were ruptured by glass bead-beating, and spores were examined for breakage via light microscopy. E. aedis genomic DNA was purified by the standard phenol-chloroform method and served as template for whole genome rolling-circle amplification using Genomiphi (Amersham). 4.5 μg of amplified E. aedis genomic DNA was sheared, blunt end-repaired, and cloned into pCR4Blunt-TOPO (Invitrogen) according to the manufacturer's specifications. 182 different E. aedis clones with an average length of 1,283 bp were end-sequenced using ABI Big Dye 3.1 chemistry. Six different E. aedis library clones containing coding, non-coding, or transposable segments were checked for chimeric sequence by PCR of non-Genomiphi-treated E. aedis genomic DNA. From this, successful amplification of fragments between 250 and 450 bp did not support the idea of chimeras being present in the Genomiphi-created E. aedis genomic library.
DNA was extracted from 2 × 107germinated Brachiola algerae spores (a strain originally isolated the mosquito Anopheles stephensi). Spores were cultivated in vitro in RK13 rabbit kidney cells at 30C in 5% CO2, purified and germinated by incubation in 0.3% H2O2 for 16 hours at 25°C. The germinated spores were concentrated by centrifugation suspended in 100 μg/ml Proteinase K-PBS and incubated for 15 min at 65°C. DNA was extracted using phenol-chloroform, followed by ethanol precipitation. DNA was then dissolved in TE buffer and stored at -70°C until use. A sample of purified Brachiola algerae DNA was amplified using Genomphi (Amersham) to produce 10 μg of DNA and another sample of 1.7 μg of purified DNA was processed directly to make two separate libraries. DNA was sheared, blunt-ended and cloned as described for E. aedis above. A respective 64,433 and 140,051 bases from the Genomiphi and non-Genomiphi treated libraries were sequenced with ABI Big Dye 3.1. This gave a total of 203,748 non-overlapping bases of sequence in 181 contigs with an average length of 1,125 bp.
Areas of six representative B. algerae clones from the Genomiphi-amplified DNA library were reamplified by PCR from genomic DNA to confirm that the Genomiphi process had not amplified chimeric sequences. Primers were designed to areas of 6 clones. These fragments were between 525 and 1000 base pairs and included non-coding areas, putative transposases, transposons, protein-coding genes, and an SSU gene area.
Contigs were analysed by BlastX and BlastN to sequences in GenBank. Open reading frames were considered significantly similar if E values were less than 0.00001. Contigs were further searched for stretches of nucleotides coding for sequences of at least 100 amino acids, and these were considered ORFs.
New sequences were deposited in GenBank under accession numbers ET437577–ET437812 and ET437979–ET437981 (E. aedis) and ET223031–ET223211 (B. algerae).
Authors' contributions
BAPW and RCHL constructed libraries, sequenced clones, analysed data and drafted the paper, JJB and LMW contributed microsporidian material and to the writing of the paper. NMF and PJK conceived of the study, analysed data and drafted the paper.
Acknowledgments
Acknowledgements
This work was supported by a grant from the Canadian Institutes for Health Research to PJK (MOP-42517) and a grant from the Natural Sciences and Engineering Research Council of Canada to NMF. PJK is a Fellow of the Canadian Institute for Advanced Research and senior investigator awards from the Michael Smith Foundation for Health Research.
Contributor Information
Bryony AP Williams, Email: B.A.P.Williams@exeter.ac.uk.
Renny CH Lee, Email: renny@interchange.ubc.ca.
James J Becnel, Email: James.Becnel@ars.usda.gov.
Louis M Weiss, Email: lmweiss@aecom.yu.edu.
Naomi M Fast, Email: nfast@interchange.ubc.ca.
Patrick J Keeling, Email: pkeeling@interchange.ubc.ca.
References
- Didier ES. Microsporidiosis: an emerging and opportunistic infection in humans and animals. Acta Trop. 2005;94:61–76. doi: 10.1016/j.actatropica.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Woese CR. Eukaryotic ribosomes that lack a 5.8S RNA. Nature. 1986;320:287–288. doi: 10.1038/320287a0. [DOI] [PubMed] [Google Scholar]
- Hirt RP, Logsdon JM, Jr., Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: evidence from the largest subunit of RNA polymerase II and other proteins. Proc Natl Acad Sci USA. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlind TD, Li J, Visversvara GS, Vodkin MH, McLaughlin GL, Katiyar SK. Phylogenetic analysis of the b-tubulin sequences from amitochondriate protozoa. Mol Phylogenet Evol. 1996;5:359–367. doi: 10.1006/mpev.1996.0031. [DOI] [PubMed] [Google Scholar]
- Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol Biol Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- Van de Peer Y, Ben Ali A, Meyer A. Microsporidia: accumulating molecular evidence that a group of amitochondriate and suspectedly primitive eukaryotes are just curious fungi. Gene. 2000;246:1–8. doi: 10.1016/S0378-1119(00)00063-9. [DOI] [PubMed] [Google Scholar]
- Gill EE, Fast NM. Assessing the microsporidia-fungi relationship: Combined phylogenetic analysis of eight genes. Gene. 2006;375:103–109. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- Peyretaillade E, Biderre C, Peyret P, Duffieux F, Méténier G, Gouy M, Michot B, Vivarès CP. Microsporidian Encephalitozoon cuniculi, a unicellular eukaryote with an unusual chromosomal dispersion of ribosomal genes and a LSU rRNA reduced to the universal core. Nucleic Acids Res. 1998;26:3513–3520. doi: 10.1093/nar/26.15.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katinka MD, Duprat S, Cornillot E, Méténier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, Delbac F, El Alaoui H, Peyret P, Saurin W, Gouy M, Weissenbach J, Vivarès CP. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- http://gmod.mbl.edu/perl/site/antonospora01?page=intro
- Slamovits CH, Fast NM, Law JS, Keeling PJ. Genome compaction and stability in microsporidian intracellular parasites. Curr Biol. 2004;14:891–896. doi: 10.1016/j.cub.2004.04.041. [DOI] [PubMed] [Google Scholar]
- Zhang J. Protein-length distributions for the three domains of life. Trends Genet. 2000;16:107–109. doi: 10.1016/S0168-9525(99)01922-8. [DOI] [PubMed] [Google Scholar]
- Polonais V, Prensier G, Méténier G, Vivarès CP, Delbac F. Microsporidian polar tube proteins: highly divergent but closely linked genes encode PTP1 and PTP2 in members of the evolutionarily distant Antonospora and Encephalitozoon groups. Fung Genet Biol. 2005;42:791–803. doi: 10.1016/j.fgb.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Burri L, Keeling PJ. Characterization of a divergent Sec61beta gene in microsporidia. J Mol Biol. 2006;359:1196–1202. doi: 10.1016/j.jmb.2006.04.028. [DOI] [PubMed] [Google Scholar]
- Corradi N, Akiyoshi DE, Morrison HG, Feng X, Weiss LM, Tzipori S, Keeling PJ. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS ONE. 2007;2:e1277. doi: 10.1371/journal.pone.0001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BA, Slamovits CH, Patron NJ, Fast NM, Keeling PJ. A high frequency of overlapping gene expression in compacted eukaryotic genomes. Proc Natl Acad Sci USA. 2005;102:10936–10941. doi: 10.1073/pnas.0501321102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradi N, Gangaeva A, Keeling PJ. Comparative profiling of overlapping transcription in the compacted genomes of microsporidia Antonospora locustae and Encephalitozoon cuniculi. Genomics. 2008 doi: 10.1016/j.ygeno.2007.12.006. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL. Mechanisms and rates of genome expansion and contraction in flowering plants. Genetica. 2002;115:29–36. doi: 10.1023/A:1016015913350. [DOI] [PubMed] [Google Scholar]
- Hinkle G, Morrison HG, Sogin ML. Genes coding for reverse transcriptase, DNA-directed RNA polymerase, and chitin synthase from the microsporidian Spraguea lophii. Biol Bull. 1997;193:250–251. doi: 10.1086/BBLv193n2p250. [DOI] [PubMed] [Google Scholar]
- Mittleider D, Green LC, Mann VH, Michael SF, Didier ES, Brindley PJ. Sequence survey of the genome of the opportunistic microsporidian pathogen, Vittaforma corneae. J Eukaryot Microbiol. 2002;49:393–401. doi: 10.1111/j.1550-7408.2002.tb00218.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Pan G, Fang L, Li J, Tian X, Li T, Zhou Z, Xiang Z. The varying microsporidian genome: existence of long-terminal repeat retrotransposon in domesticated silkworm parasite Nosema bombycis. Int J Parasitol. 2006;36:1049–1056. doi: 10.1016/j.ijpara.2006.04.010. [DOI] [PubMed] [Google Scholar]
- Dia N, Lavie L, Méténier G, Toguebaye BS, Vivarès CP, Cornillot E. InterB multigenic family, a gene repertoire associated with subterminal chromosome regions of Encephalitozoon cuniculi and conserved in several human-infecting microsporidian species. Curr Genet. 2007;51:171–186. doi: 10.1007/s00294-006-0114-x. [DOI] [PubMed] [Google Scholar]
- Franzen C, Nassonova ES, Scholmerich J, Issi IV. Transfer of the members of the genus Brachiola (microsporidia) to the genus Anncaliia based on ultrastructural and molecular data. J Eukaryot Microbiol. 2006;53:26–35. doi: 10.1111/j.1550-7408.2005.00066.x. [DOI] [PubMed] [Google Scholar]
- Becnel JJ, White SE, Shapiro AM. Review of microsporidia-mosquito relationships: from the simple to the complex. Folia Parasitol (Praha) 2005;52:41–50. [PubMed] [Google Scholar]
- Biderre C, Pagès M, Méténier G, David D, Bata J, Prensier G, Vivarès CP. On small genomes in eukaryotic organisms: molecular karyotypes of two microsporidian species (Protozoa) parasites of vertebrates. C R Acad Sci III. 1994;317:399–404. [PubMed] [Google Scholar]
- Malone LA, McIvor CA. DNA probes for two Microsporidia, Nosema bombycis and Nosema costelytrae. J Invertebr Pathol. 1995;65:269–273. doi: 10.1006/jipa.1995.1041. [DOI] [PubMed] [Google Scholar]
- Vossbrinck CR, Debrunner-Vossbrinck BA. Molecular phylogeny of the Microsporidia: ecological, ultrastructural and taxonomic considerations. Folia Parasitol (Praha) 2005;52:131–142. doi: 10.14411/fp.2005.017. [DOI] [PubMed] [Google Scholar]
- Muller A, Trammer T, Chioralia G, Seitz HM, Diehl V, Franzen C. Ribosomal RNA of Nosema algerae and phylogenetic relationship to other microsporidia. Parasitol Res. 2000;86:18–23. doi: 10.1007/PL00008501. [DOI] [PubMed] [Google Scholar]
- Slamovits CH, Williams BA, Keeling PJ. Transfer of Nosema locustae (Microsporidia) to Antonospora locustae n. comb. based on molecular and ultrastructural data. J Eukaryot Microbiol. 2004;51:207–213. doi: 10.1111/j.1550-7408.2004.tb00547.x. [DOI] [PubMed] [Google Scholar]
- Gilson PR, Su V, Slamovits CH, Reith ME, Keeling PJ, McFadden GI. Complete nucleotide sequence of the chlorarachniophyte nucleomorph: nature's smallest nucleus. Proc Natl Acad Sci USA. 2006;103:9566–9571. doi: 10.1073/pnas.0600707103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas S, Zauner S, Fraunholz M, Beaton M, Penny S, Deng LT, Wu X, Reith M, Cavalier-Smith T, Maier UG. The highly reduced genome of an enslaved algal nucleus. Nature. 2001;410:1091–1016. doi: 10.1038/35074092. [DOI] [PubMed] [Google Scholar]
- Warringer J, Blomberg A. Evolutionary constraints on yeast protein size. BMC Evol Biol. 2006;6:61. doi: 10.1186/1471-2148-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouze P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynie S, Cooke R, Saeys Y, Wuyts J, Jabbari K, Bowler C, Panaud O, Piegu B, Ball SG, Ral JP, Bouget FY, Piganeau G, De Baets B, Picard A, Delseny M, Demaille J, Van de Peer Y, Moreau H. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci USA. 2006;103:11647–11652. doi: 10.1073/pnas.0604795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. The Evolution of Genome Size. , John Wiley and Sons, Chichester; 1985. [Google Scholar]
- Méténier G, Vivarès CP. Genomics of microbial parasites: The microsporidial paradigm. In: Hirt RP and Horner DS, editor. Organelles, Genomes and Eukaryote Phylogeny. London, CRC Press; 2004. pp. 207–236. [Google Scholar]
- Streett DA. Analysis of Nosema locustae (Microsporidia: Nosematidae) chromosomal DNA with pulsed-field gel electrophoresis. J Invert Pathol. 1994;63:301–303. doi: 10.1006/jipa.1994.1055. [DOI] [Google Scholar]
- Chioralia G, Trammer T, Maier WA, Seitz HM. Morphologic changes in Nosema algerae (Microspora) during extrusion. Parasitol Res. 1998;84:123–131. doi: 10.1007/s004360050368. [DOI] [PubMed] [Google Scholar]
- Sokolova YY, Lange CE. An ultrastructural study of Nosema locustae Canning (Microsporidia) from three species of Acrididae (Orthoptera) Acta Protozool. 2002;41:229–237. [Google Scholar]
- Terry RS, Smith JE, Sharpe RG, Rigaud T, Littlewood DT, Ironside JE, Rollinson D, Bouchon D, MacNeil C, Dick JT, Dunn AM. Widespread vertical transmission and associated host sex-ratio distortion within the eukaryotic phylum Microspora. Proc Biol Sci. 2004;271:1783–1789. doi: 10.1098/rspb.2004.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]