Abstract
The ADP-ribosylating toxins (ADPRTs) produced by pathogenic bacteria modify intracellular protein and affect eukaryotic cell function. Actin-specific ADPRTs (including Clostridium perfringens ι-toxin and Clostridium botulinum C2 toxin) ADP-ribosylate G-actin at Arg-177, leading to disorganization of the cytoskeleton and cell death. Although the structures of many actin-specific ADPRTs are available, the mechanisms underlying actin recognition and selective ADP-ribosylation of Arg-177 remain unknown. Here we report the crystal structure of actin-Ia in complex with the nonhydrolyzable NAD analog βTAD at 2.8 Å resolution. The structure indicates that Ia recognizes actin via five loops around NAD: loop I (Tyr-60–Tyr-62 in the N domain), loop II (active-site loop), loop III, loop IV (PN loop), and loop V (ADP-ribosylating turn–turn loop). We used site-directed mutagenesis to confirm that loop I on the N domain and loop II are essential for the ADP-ribosyltransferase activity. Furthermore, we revealed that Glu-378 on the EXE loop is in close proximity to Arg-177 in actin, and we proposed that the ADP-ribosylation of Arg-177 proceeds by an SN1 reaction via first an oxocarbenium ion intermediate and second a cationic intermediate by alleviating the strained conformation of the first oxocarbenium ion. Our results suggest a common reaction mechanism for ADPRTs. Moreover, the structure might be of use in rational drug design to block toxin-substrate recognition.
Keywords: complex, crystal structure, protein recognition
ADP-ribosylating toxins (ADPRTs) facilitate scission of the N-glycosyl bond between nicotinamide and the N-ribose of NAD and transfer the ADP-ribose moiety to target proteins. ADPRTs are classified into four families based on their respective targets (1). Type I ADPRTs target heteromeric GTP-binding proteins. They include cholera toxin (CT) (2), pertussis toxin (PT) (3), and Escherichia coli heat-labile enterotoxin (LT) (4). Type II ADPRTs [diphtheria toxin (DT) (5) and Pseudomonas exotoxin A (ETA)] modify elongation factor 2 (EF2). Type III ADPRTs (Clostridium botulinum C3 exoenzyme) (6) ADP-ribosylate small GTP-binding proteins. Type IV ADPRTs ADP-ribosylate actin. These actin-specific ADPRTs include a family of binary toxins (7) comprising C. botulinum C2 toxin (8), C. perfringens ι-toxin (9), C. difficile toxin (10), C. spiroforme toxin (11), and Bacillus cereus vegetative insecticidal protein (VIP). The C2 toxin was the first reported actin-specific ADPRT; other clostridial actin-specific ADPRTs, such as ι-toxin, were later shown to share striking similarities, including the presence of an enzymatic component (C2I or Ia) and a cell-membrane-binding component (C2II or Ib). Both toxins ADP-ribosylate G-actin, but not F-actin, at Arg-177; this activity severely reduces the ability of actin to undergo polymerization, leading to disruption of the cytoskeletal architecture and cell death.
Recently, a nontypical binary-actin-specific ADPRT was discovered in Salmonella enterica. Salmonella spp. cause billions of human infections each year, resulting in >3 million annual human deaths due to typhoid fever alone (12). The spread of multidrug-resistant strains of Salmonella also has focused attention on this pathogen (13). During infection, Salmonella grows and replicates inside macrophages in a spatial membrane compartment known as the Salmonella-containing vacuole (14), after which it induces the death of infected macrophages. A number of Salmonella strains carry the spv virulence locus, encoding the ADP-ribosyl transferase SpvB protein, which depolymerizes the actin cytoskeleton during intracellular infection, leading to apoptosis. The SpvB effect might facilitate the cell-to-cell spread of the organism, thereby enhancing its virulence (14). The importance of type IV ADPRTs in virulence suggests that they might be attractive targets for antibacterial drug design.
The structures of several enzymatic components from each type of ADPRT have been determined with or without NAD. Typical actin-specific ADPRTs possess two similar domains: the C domain, which is essential for catalytic activity; and the N domain, which is important for the interaction with the binding and translocation subunit. By contrast, SpvB from Salmonella and the type III ADPRT C3 have only one ADP-ribosyltransferase domain and lack the N-terminal adaptor domain. There is, however, a striking structural similarity among the catalytic domains of these ADPRTs. The structures and biochemical results of actin-specific ADPRTs have given us a detailed understanding of NAD binding and have suggested a possible catalytic mechanism (15–18). In all type IV ADPRTs, the EXE motif, including two key glutamate residues, is present at the catalytic center. The former glutamate of the EXE motif is thought to be a key residue for ADP-ribosyltransferase, which is deprotonated from Arg-177 in actin. The latter glutamate forms a hydrogen bond with the O′2 on N-ribose, which is thought to stabilize the oxocarbenium cation. Detailed knowledge of the molecular recognition and reaction mechanism, however, has remained elusive because of a lack of information about the complex structure of the ADPRT with the protein substrate. Recently, important new insights into the mechanism of toxin-protein recognition and ADP-ribosylation have been provided by the elucidation of the crystal structure of the ETA–EF2 complex (19). The nature of the structure suggests that the EF2 target residue, diphthamide (a modified histidine), is involved in triggering NAD cleavage and interacting with the proposed oxocarbenium ion intermediate during the nucleophilic substitution reaction, thus explaining the requirement of diphthamide for ADP-ribosylation. Although all ADPRTs share common features (20), their receptors and target residues differ. To understand the mechanisms of actin recognition and ADP-ribosylation of the Arg residue by type IV ADPRTs, a structural analysis of complexes between type IV ADPRTs and actin is needed. Moreover, structural information is essential to understand the mechanism of action of ADPRTs as virulence factors and to develop drugs that inhibit the ADPRT-target protein. To this end, we have determined the crystal structure of Ia in complex with actin and a nonhydrolyzable NAD analog, βTAD (21).
Results
Overall Structure of Actin–Ia–βTAD Complex.
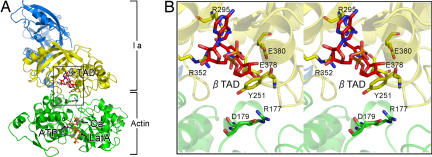
To obtain good diffraction-quality crystals of G-actin with Ia, we used ATP and latrunculin A, which is an inhibitor of actin polymerization. Moreover, to obtain stable complex crystals, we used the nonhydrolyzable NAD analog βTAD. Using these crystals, we collected diffraction data for the actin (with ATP and the polymerization-inhibiting drug latrunculin A)–Ia–βTAD complex at a resolution of 2.8 Å [supporting information (SI) Table S1]. The structure of the entire complex was revealed by molecular replacement using each Ia and actin coordinate (Fig. 1A). Ia consists of the N domain (residues 3–210) and the C domain (residues 211–413) with a similar structure. It is interesting to note that two actin residues, Arg-177 and Asp-179, are in close proximity to βTAD and Glu-378 of Ia (Fig. 1B). The electron densities of βTAD within the C domain cleft of Ia and the ATP and latrunculin A associated with actin are shown on the omit map (Fig. S1). The shape of bound βTAD is similar to that of NADH, which assumes an NMN ring-like conformation (17). Although we expected a structural change upon Ia binding (e.g., bending) at the surface of the actin, we found that the structure of actin within the complex was relatively unchanged from its monomeric form (Fig. S2). By contrast, actin binding induced a subtle shift of loops II, IV, and V of Ia (Fig. S2). These loops play important roles in actin recognition, as described below.
Fig. 1.
Whole structure of actin–Ia–β TAD complex. (A) Ribbon representation of actin–Ia–β TAD. Actin (green) and Ia (N domain, marine blue; C domain, yellow) molecules and cofactors [βTAD, ATP, latrunculin A (LatA), and calcium (Ca)] are labeled. (B) Detailed stereoview of the area around βTAD (boxed in A).
Molecular Recognition Between Actin and Ia.
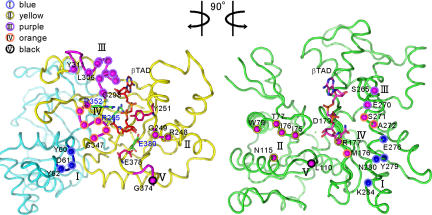
The structural components of Ia that mediate recognition of actin include five loops: loop I (Tyr-60–Tyr-62 in the N domain), loop II (the active-site loop connecting α7 and α8 helices), loop III (the loop between β9 and α10), loop IV [the PN loop situated 10 residues after the serine-threonine-serine (STS) motif that stabilize the NAD binding], and loop V [the ADP-ribosylating turn–turn (ARTT) loop containing the EXE motif] (Fig. 2 and Fig. S3). So far, to our knowledge, no direct structural analysis of the interactions of types III and IV ADPRTs with their protein substrates has been published, although a recent report has delineated the regions that are necessary for substrate binding of the C3-related ADPRTs, ExoS and ExoT, from Pseudomonas aeruginosa (22). These ADPRTs recognize their respective substrates by using three common regions: ARTT, PN, and the active-site loop. The present study directly reveals that Ia makes use of additional regions in substrate recognition. Sequence alignment of several type IV ADPRTs and C3, however, showed that these binding residues are poorly conserved (Fig. S4). To investigate the roles of interface residues in Ia, we replaced them with alanine (Table 1). The replacement of Tyr-62 on loop I and Arg-248 on loop II with alanine resulted in a drastic reduction of ADP-ribosyltransferase activity, but not NADase activity. By contrast, the replacements of Asp-61 on loop I and Tyr-306 and Tyr-311 on loop III with alanine had little effect on ADP-ribosylation activity and cytotoxicity. We carried out further analyses by using a deletion mutant of Ia that had the C domain (Ia 211–413), but not the N-terminal domain. The deletion of the N domain showed a drastic decrease in ADP-ribosyltransferase activity, whereas it showed ≈80% glycohydrolase activity (Table 1).
Fig. 2.
Butterfly representation of recognition residues between Ia and actin. Roman numerals (I–V) show the five binding loops in Ia. The actin-recognition residues on five loops of Ia are shown as circles on the left. The catalytic residues of Ia around NAD (Tyr-251, Arg-295, Arg-352, Glu-378, and Glu-380) are shown as sticks. The Ia-recognition residues of actin are shown as circles on the right.
Table 1.
Cytotoxic and enzymatic activities of wild-type and variant Ia
| Region | Variant | Cytotoxic activity*, % | ARTase activity*, % | NADase activity*, % |
|---|---|---|---|---|
| Wild type | 100.0 | 100.0 | 100.0 | |
| Loop I | Y60A | ND | ND | 0.6 ± 0.2 |
| Loop I | D61A | 99.2 ± 4.1 | 77.6 ± 8.1 | 85.3 ± 7.3 |
| Loop I | Y62A | 1.2 ± 0.2 | 5.1 ± 0.5 | 60.3 ± 8.6 |
| Loop II | R248A | 1.3 ± 0.3 | 4.7 ± 0.6 | 55.1 ± 4.8 |
| Loop III | L306A | 67.2 ± 4.6 | 71.4 ± 0.5 | 100.5 ± 6.2 |
| Loop III | Y311A | 98.2 ± 6.8 | 92.3 ± 8.3 | 81.5 ± 7.4 |
| ΔN-dom | Ia 211–413 | — | 1.1 ± 0.4 | 81.1 ± 5.7 |
ND, not detected; —, not done; ΔN-dom, deletion of N domain.
*Activity shown as a percentage of wild-type activity.
Recognition Interface of Ia to Actin.
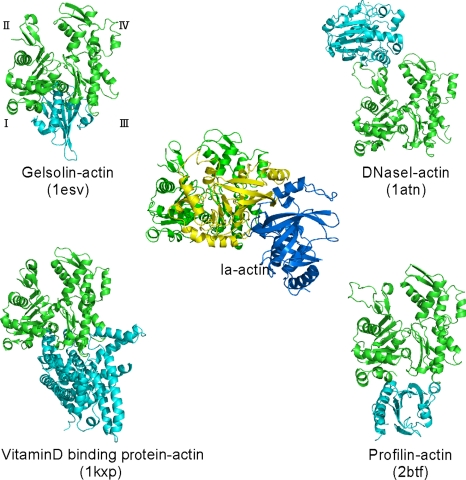
The structure of the actin–Ia–βTAD complex showed that the recognition interface of Ia differs from that of other known actin-binding proteins (Fig. 3). We compared the actin-binding interface of Ia with that of other actin-binding proteins, including gelsolin (23), vitamin D-binding protein (24), DNaseI (25), and profilin (26). These actin-binding proteins control the F-actin/G-actin ratio and the rate of actin assembly. DNaseI binds with high affinity to actin subdomains II and IV, whereas the other three proteins bind to actin via subdomains I and III. Ia mainly binds to actin through subdomains I, III, and IV, which cover its surface in the region that includes Arg-177.
Fig. 3.
Comparison of the molecular recognition interface of Ia and actin-binding proteins. Actin (green), Ia (N domain, marine blue; C domain, yellow), and representative actin-binding proteins (cyan) are shown. Roman numerals (I–IV) show the actin subdomains.
Actin-Specific ADP-Ribosylation Mechanism.
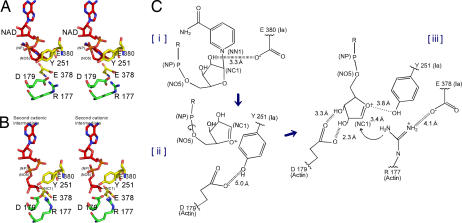
Two different mechanisms could be considered for the ADP-ribosylation of Arg-177. One is the SN2-like mechanism, in which Arg-177 plays an important role in a nucleophilic attack. The other is an SN1 mechanism suggested by studies of the Ia structure and its site-directed mutagenesis (17). The positively charged Arg-295 and Arg-352 interact electrostatically with NAD phosphate and contribute to the highly folded conformation of NMN. The specific conformation appears to induce the equilibrium shift toward the production of oxocarbenium cation. The present work showed that the distance between NC1 of N-ribose and the guanidyl nitrogen of Arg-177 is ≈8 Å (Fig. 4A), which might make a direct nucleophilic attack impossible by the SN2 reaction. Even if the reaction proceeds by the SN1 mechanism, it is unclear how to reduce the nucleophile–electrophile distance. To address this problem, we propose that our SN1 mechanism occurs via two intermediates: an oxocarbenium ion intermediate and a cationic intermediate (Fig. 4B) (27). After the oxocarbenium ion intermediate is produced, rotation occurs via NP-NO5 of ADP-ribose. This second cationic intermediate allows NC1 of N-ribose to approach the guanidyl nitrogen of Arg-177 to within a distance of 3.4 Å. (Fig. 4 B and C). In this model, Asp-179 of actin plays a stabilizing role by making contact with the N-ribose 2′ OH (Fig. 4 B and C). Finally, the nucleophile Arg-177 of actin, which associates with Glu-378 of Ia, attacks NC1 of the oxocarbenium ion, leading to α-selective ADP-ribosylation. It was previously suggested that Tyr-251 binds to actin (17); in fact, our structure shows this residue to be in close proximity to Asp-179 of actin. Instead of binding to actin, however, it appears that Tyr-251 guides the NP-NO5 rotation to position the N-ribose appropriately. Furthermore, Tyr-251 might play a role in catching the oxocarbenium ion before it is transferred to the guanidyl nitrogen of Arg-177. Interestingly, the residues around the N-ribose might create a negative environment to protect the oxocarbenium cation and to prevent an unfavorable reaction with water; these comprise Tyr-246, Tyr-251, Ser-338, Tyr-375, Glu-378, and Glu-380. Also, it is notable that there is an open space within the interface of the complex, suggesting that it does not prevent reactions with water. Instead, the space makes it possible to rotate N-ribose, thereby allowing NC1 of the oxocarbenium ion to come sufficiently close to react with the guanidyl nitrogen of Arg-177.
Fig. 4.
Mechanism of Sn1 ADP-ribosylation based on the strain-alleviation model. (A) Actin–Ia–NAD model. The structure is the same as that for actin–Ia–βTAD except that NAD is substituted for βTAD. (B) Second cationic intermediate. The N-ribose of ADP-ribose is rotated via NP-NO5 based on NAD. The torsion angles of three residues (Arg-177 and Asp-179) were manipulated to maintain appropriate bond lengths between atoms. (C) Schematic of the Sn1 mechanism of Ia: First, nicotinamide cleavage occurs via an Sn1 reaction induced by an NMN ring-like structure; second, the first oxocarbenium cation intermediate is formed with a strained conformation; third, the second cationic intermediate is induced by alleviation of the strained conformation by NP-NO5 rotation; fourth, actin Arg-177 of Ia nucleophilically attacks the NC1.
Discussion
Actin-Ia Recognition.
This article studied a complex structure comprising actin and a type IV ADPRT. Despite the limited primary sequence homology among different ADPRTs, the central cleft bearing the NAD-binding pocket formed by the two perpendicular β-sheet cores is remarkably well conserved between bacterial ADPRTs and eukaryotic mono-ADPRTs. In particular, types IV and III have a similar tertiary structure, with an α3 (α8 in Ia) helix on the enzymatic domain (28). Both have been suggested to have an ARTT motif that is important for the recognition of the substrates Rho and actin, respectively (28): The turn 1 aromatic residue plays a role in substrate recognition, whereas the turn 2 hydrogen-bonding side chain is involved in the recognition of Asn for Rho and Arg for Ia, respectively. Based on the complex structure, it was shown that Glu-378 on turn 2 is in close proximity to Arg-177, but that Tyr-375 on turn 1 does not interact with actin. Previous interaction studies of the substrate protein with ExoS and ExoT proposed that helix 1 (only ExoT), the active-site loop, the ARTT motif, and the PN loop might play roles in the recognition of target host proteins (22). Furthermore, Ras and Crk showed complementary electrostatic properties toward ExoS and ExoT, respectively. Because Ia and VIP2 share similar electrostatic potentials at the proposed enzyme–substrate interface and target the same actin (17), electrostatic potentials appear to be important for the substrate recognition of ADPRTs (22). The present crystallographic work showed directly that Ia binds to actin not only with the active-site loop, the ARTT motif, and the PN loop, but also with the other regions, loop I on the N domain, and loop III. The structure-based mutagenesis of Ia also suggested that Tyr-62 on loop I and Arg-248 on loop II are essential for the Ia–actin recognition. Furthermore, the deletion of the N domain showed a drastic decrease in ADP-ribosyltransferase activity. This result suggests that the role of the N domain of Ia is important for the binding to actin. This feature might be common for all type IV ADPRTs that consist of two domains. In summary, actin is recognized by five loops of Ia by using not only ionic interactions, but also van der Waals interactions based on shape complementarity.
Actin-specific ADPRTs have different substrate specificities: C2I ADP-ribosylates only nonmuscle actin, whereas Ia ADP-ribosylates both nonmuscle and skeletal muscle actin. Among the 25 amino acid exchanges between muscle and nonmuscle actin, only three residues were found on the interaction surface with actin (176 M/L, 272 A/C, and 279 Y/F). Although we analyzed the actin–Ia complex structure, it remained difficult to explain the substrate-specificity. Mutational studies of actin might provide a mechanism to account for this phenomenon.
Common Reaction Mechanisms Based on Structural Relations with Other ADPRTs.
In this article, we propose an SN1 mechanism for the reaction of Arg-177 ADP-ribosylation that involves the following three steps: the generation of the first oxocarbenium intermediate by the cleavage of nicotinamide, rotation to form the second cationic intermediate around the NP-NO5 bond by releasing the conformational strain, and the nucleophilic attack of Arg-177 that is activated by the interaction with Glu 378. This accounts for the ADP-ribosylation reaction in other type IV ADPRTs. Given the large extent of structural similarity, the structure of our complex also is likely to provide an insight into the reaction mechanisms of type III ADPRTs (28, 29) and mammalian ecto-ADP-ribosyltransferase (30). Furthermore, NAD structures also are similar in all ADPRTs, sharing a compact and energetically unfavorable conformation (31). In this regard, the reaction mechanism might be delineated in a similar way for all ADPRTs, including types I and II. Therefore, we compared the two complex structures of Ia–actin and ETA–EF2, which belong to different ADPRT families, and demonstrated the following similarities.
First, there was no structural change during the ADP-ribosylation reaction, meaning that there were few differences among the respective structures of ETA and EF2 within the apo complex, the putative Michaelis complex (EF2–ETA–βTAD), and the ADP-ribosylated (ADPR)–EF2–ETA postreaction complex. We did not determine the structure of apo actin–Ia or the postreaction ADPR–actin–Ia complex. However, actin assumes the same conformation in the monomeric ATP form (1IJJ) in our complex and in the ADPR–actin form (32). This finding suggests that no structural changes occur during the ADP-ribosylation reaction.
Second, the large gap between NC1 of NAD and the nitrogen atom of the substrate residue was consistent with the proposed SN1 mechanism for ADP-ribosylation by both ETA and type IV ADPRTs. The energetically unfavorable conformations of NMN have been seen in all ADPRTs, supporting the idea that an SN1 mechanism that cleaves the glycosyl bond could be common to all ADPRTs (31, 33). In Ia, the subtle shift of the loop around NAD upon actin binding might help to trigger the cleavage of nicotinamide, thereby increasing the strain of NAD further.
Third, there might be a common mechanism to reduce the large gap between NC1 of NAD and the nitrogen atom of the substrate residue. Jorgensen et al. (19) discussed several possible mechanisms by which the nucleophile–electrophile distance could be reduced, including the possibility that the oxocarbenium ion might migrate relative to ETA or the loop region containing diphthamide might undergo a transient conformational change during the reaction. Here we present a simple and plausible mechanism in which the NP-NO5 rotation of ADP-ribose permits N-ribose access to Arg-177. This finding could be the result of alleviating the strained conformation after the cleavage of the NC1–NN1 bonds of NAD.
Fourth and finally, there was a common residue in the substrate protein to fix the N-ribose. Asp-696 of ETA forms a hydrogen bond with N-ribose 2′ OH in ADP-ribosylated EF2. Similarly, within actin–Ia–βTAD, Asp-179 of Ia was located in an ideal position to fix the N-ribose 2′ OH of ADP-ribose.
In recent years, health problems related to Salmonella have greatly increased in terms of both the incidence and severity of cases. The World Health Organization reports that strains of Salmonella have emerged that are resistant to a range of antimicrobials, including first-choice agents for the treatment of humans, and are threatening to become a serious public health problem. With structural knowledge of the actin–Ia complex, we were able to model complexes between SpvB and actin (Fig. S5). Interestingly, SpvB differs from other type IV ADPRTs in that it lacks the N domain and contains a 30-amino acid insert as a helical subdomain of the C domain (Fig. S4). Although the function of the helical subdomain remains unknown, the structure of our actin–SpvB model clearly shows that it fits well with the DNaseI-binding loop of actin, suggesting that it plays an important role in substrate recognition. Instead of the loss of the N domain interaction, the helical subdomain interaction might be responsible for the binding. Such information might be of use in drug design against Salmonella infection. Because ADPRT is a key virulence factor in various infections, the model structure of actin–ADPRT might aid drug design for these conditions. Our representative structure of the complex between actin and type IV ADPRT is predicted to help in the development of small molecules and macromolecules that inhibit the molecular interaction between actin and ADPRTs, with the aim of targeting serious infectious diseases such as those caused by Salmonella.
Materials and Methods
Crystallization and Structure Determination.
Expression and purification of Ia was carried out as described previously (34). Rabbit skeletal actin (Sigma) was mixed with latrunculin A in G buffer [2 mM Tris (pH 8.0), 0.2 mM ATP, 0.5 mM DTT, 0.2 mM CaCl2, and 0.02% NaN3] to final concentrations of 15 mg/ml actin and 0.34 mM latrunculin A (35). The sample was then mixed with 17–20 mg/ml Ia, after which βTAD was added to a final concentration of 0.5 mM. Crystallization was carried out by using hanging drop vapor diffusion against a reservoir solution containing 100 mM Mes (pH 6.5) and 20% PEG1000 at 4°C. The thin plate-like crystals, which grew to 0.1 mm × 0.1 mm × 0.05 mm, were transferred to cryoprotectant oil and flash frozen in liquid N2. We have so far been unable to crystallize sufficient amounts of the apo (no βTAD) and ADP-ribosylated (with NAD instead of βTAD) forms to enable analysis, although twin crystals were obtained for the former. Data collection was performed at PF-AR NW12 by using a wavelength of 1.0 Å. Diffraction was good despite the small size of the crystals, and the best dataset was collected at 2.8 Å. The crystal space group was determined to be P212121 (a = 57.0, b = 126.3, c = 147.1 Å) and to contain one actin and one Ia in an asymmetric unit. Crystals are highly nonisomorphous, and their structure was determined by molecular replacement by using MOLREP (36). Using the structure of Ia (1GIQ), we identified and fixed the Ia position, after which we searched for actin by using the structure of actin (1IJJ). The βTAD, ATP, and latrunculin A densities were clear, so these cofactors were built in. The model was then iteratively rebuilt and refined at a 2.8-Å resolution by using REFMAC (37). The final model was then refined to Rwork = 22.3% (Rfree = 29.6%) and consisted of Ia, actin, βTAD, ATP, latrunculin A, calcium, and 79 water molecules (Table S1). The figures were prepared by using PYMOL (38). There were no residues in the disallowed region of the Ramachandran plot.
Molecular Modeling of Actin–Ia–NAD, Pretransition State Model, and SpvB–Actin.
The actin–Ia–NAD complex was modeled by using NAD rather than βTAD after superimposing 1GIQ (NAD-bound Ia) onto Ia within the actin–Ia complex (rmsd = 0.7 Å). The pretransition state complex was modeled by using only the NO5–NC5 rotation of ADP-ribose in the actin–Ia–NAD model. To model the SpvB–actin complex, SpvB was superimposed on Ia within the actin–Ia–βTAD complex (rmsd = 2.9 Å).
Site-Directed Mutagenesis, Purification, and Assay of Biological Activities.
Site-directed mutagenesis was carried out by the unique restriction site-elimination technique by using the Transformer mutagenesis kit (Clontech) with synthetic appropriate oligonucleotide primers (34). All of the variants obtained were identified by DNA sequencing. The expression and purification from Ia and variant of Ia and the deletion of N domain were performed as described previously (34). The ADP-ribosyltransferase activity, NADase activity, and cytotoxic activity of wild-type Ia and variant Ia were determined as described previously (34).
Supplementary Material
Acknowledgments.
We thank E. Yamauchi for the discussion of the crystallization. This study was supported in part by the National Project on Protein Structural and Functional Analysis funded by the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan; a Grant-in-Aid for the Academic Frontier Promotion Program of MEXT, Japan; and the Intramural Research Program of the National Institutes of Health, Center for Cancer Research, National Cancer Institute–Frederick (V.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of the actin–Ia–βTAD complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3BUZ).
This article contains supporting information online at www.pnas.org/cgi/content/full/0801215105/DCSupplemental.
References
- 1.Holbourn KP, Shone CC, Acharya KR. A family of killer toxins. Exploring the mechanism of ADP-ribosylating toxins. FEBS J. 2006;273:4579–4593. doi: 10.1111/j.1742-4658.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 2.Gill DM, Meren R. ADP-ribosylation of membrane proteins catalyzed by cholera toxin: basis of the activation of adenylate cyclase. Proc Natl Acad Sci USA. 1978;75:3050–3054. doi: 10.1073/pnas.75.7.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katada T, Ui M. Direct modification of the membrane adenylate cyclase system by islet-activating protein due to ADP-ribosylation of a membrane protein. Proc Natl Acad Sci USA. 1982;79:3129–3133. doi: 10.1073/pnas.79.10.3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss J, Garrison S, Oppenheimer NJ, Richardson SH. NAD-dependent ADP-ribosylation of arginine and proteins by Escherichia coli heat-labile enterotoxin. J Biol Chem. 1979;254:6270–6272. [PubMed] [Google Scholar]
- 5.Van Ness BG, Howard JB, Bodley JW. ADP-ribosylation of elongation factor 2 by diphtheria toxin. Isolation and properties of the novel ribosyl-amino acid and its hydrolysis products. J Biol Chem. 1980;255:10717–10720. [PubMed] [Google Scholar]
- 6.Aktories K, Weller U, Chhatwal GS. Clostridium botulinum type C produces a novel ADP-ribosyltransferase distinct from botulinum C2 toxin. FEBS Lett. 1987;212:109–113. doi: 10.1016/0014-5793(87)81566-1. [DOI] [PubMed] [Google Scholar]
- 7.Barth H, Aktories K, Popoff MR, Stiles BG. Binary bacterial toxins: Biochemistry, biology, and applications of common Clostridium and Bacillus proteins. Microbiol Mol Biol Rev. 2004;68:373–402. doi: 10.1128/MMBR.68.3.373-402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aktories K, et al. Botulinum C2 toxin ADP-ribosylates actin. Nature. 1986;322:390–392. doi: 10.1038/322390a0. [DOI] [PubMed] [Google Scholar]
- 9.Vandekerckhove J, Schering B, Barmann M, Aktories K. Clostridium perfringens iota toxin ADP-ribosylates skeletal muscle actin in Arg-177. FEBS Lett. 1987;225:48–52. doi: 10.1016/0014-5793(87)81129-8. [DOI] [PubMed] [Google Scholar]
- 10.Popoff MR, Rubin EJ, Gill DM, Boquet P. Actin-specific ADP-ribosyltransferase produced by a Clostridium difficile strain. Infect Immun. 1988;56:2299–2306. doi: 10.1128/iai.56.9.2299-2306.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popoff MR, Boquet P. Clostridium spiroforme toxin is a binary toxin which ADP-ribosylates cellular actin. Biochem Biophys Res Commun. 1988;152:1361–1368. doi: 10.1016/s0006-291x(88)80435-2. [DOI] [PubMed] [Google Scholar]
- 12.Pang T, Bhutta ZA, Finlay BB, Altwegg M. Typhoid fever and other salmonellosis: A continuing challenge. Trends Microbiol. 1995;3:253–255. doi: 10.1016/s0966-842x(00)88937-4. [DOI] [PubMed] [Google Scholar]
- 13.Torok TJ, et al. A large community outbreak of salmonellosis caused by intentional contamination of restaurant salad bars. J Am Med Assoc. 1997;278:389–395. doi: 10.1001/jama.1997.03550050051033. [DOI] [PubMed] [Google Scholar]
- 14.Guiney DG, Lesnick M. Targeting of the actin cytoskeleton during infection by Salmonella strains. Clin Immunol. 2005;114:248–255. doi: 10.1016/j.clim.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 15.Han S, Craig JA, Putnam CD, Carozzi NB, Tainer JA. Evolution and mechanism from structures of an ADP-ribosylating toxin and NAD complex. Nat Struct Biol. 1999;6:932–936. doi: 10.1038/13300. [DOI] [PubMed] [Google Scholar]
- 16.Barth H, Preiss JC, Hofmann F, Aktories K. Characterization of the catalytic site of the ADP-ribosyltransferase Clostridium botulinum C2 toxin by site-directed mutagenesis. J Biol Chem. 1998;273:29506–29511. doi: 10.1074/jbc.273.45.29506. [DOI] [PubMed] [Google Scholar]
- 17.Tsuge H, et al. Crystal structure and site-directed mutagenesis of enzymatic components from Clostridium perfringens iota-toxin. J Mol Biol. 2003;325:471–483. doi: 10.1016/s0022-2836(02)01247-0. [DOI] [PubMed] [Google Scholar]
- 18.Sakurai J, Nagahama M, Hisatsune J, Katunuma N, Tsuge H. Clostridium perfringens iota-toxin, ADP-ribosyltransferase: Structure and mechanism of action. Adv Enzyme Regul. 2003;43:361–377. doi: 10.1016/s0065-2571(02)00044-4. [DOI] [PubMed] [Google Scholar]
- 19.Jorgensen R, et al. Exotoxin A-eEF2 complex structure indicates ADP ribosylation by ribosome mimicry. Nature. 2005;436:979–984. doi: 10.1038/nature03871. [DOI] [PubMed] [Google Scholar]
- 20.Okazaki IJ, Moss J. Common structure of the catalytic sites of mammalian and bacterial toxin ADP-ribosyltransferases. Mol Cell Biochem. 1994;138:177–181. doi: 10.1007/BF00928460. [DOI] [PubMed] [Google Scholar]
- 21.Marquez VE, et al. Thiazole-4-carboxamide adenine dinucleotide (TAD). Analogues stable to phosphodiesterase hydrolysis. J Med Chem. 1986;29:1726–1731. doi: 10.1021/jm00159a027. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Maresso AW, Kim JJ, Barbieri JT. How bacterial ADP-ribosylating toxins recognize substrates. Nat Struct Mol Biol. 2004;11:868–876. doi: 10.1038/nsmb818. [DOI] [PubMed] [Google Scholar]
- 23.McLaughlin PJ, Gooch JT, Mannherz HG, Weeds AG. Structure of gelsolin segment 1-actin complex and the mechanism of filament severing. Nature. 1993;364:685–692. doi: 10.1038/364685a0. [DOI] [PubMed] [Google Scholar]
- 24.Otterbein LR, Cosio C, Graceffa P, Dominguez R. Crystal structures of the vitamin D-binding protein and its complex with actin: Structural basis of the actin-scavenger system. Proc Natl Acad Sci USA. 2002;99:8003–8008. doi: 10.1073/pnas.122126299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabsch W, Mannherz HG, Suck D, Pai EF, Holmes KC. Atomic structure of the actin:DNase I complex. Nature. 1990;347:37–44. doi: 10.1038/347037a0. [DOI] [PubMed] [Google Scholar]
- 26.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 27.Schmidt RR, Behrendt M, Toepfer A. Nitriles as solvents in glycosylation reactions: Highly selective β-glycosidase synthesis. Synlett. 1990:694–696. [Google Scholar]
- 28.Han S, Arvai AS, Clancy SB, Tainer JA. Crystal structure and novel recognition motif of rho ADP-ribosylating C3 exoenzyme from Clostridium botulinum: Structural insights for recognition specificity and catalysis. J Mol Biol. 2001;305:95–107. doi: 10.1006/jmbi.2000.4292. [DOI] [PubMed] [Google Scholar]
- 29.Evans HR, et al. The crystal structure of C3stau2 from Staphylococcus aureus and its complex with NAD. J Biol Chem. 2003;278:45924–45930. doi: 10.1074/jbc.M307719200. [DOI] [PubMed] [Google Scholar]
- 30.Mueller-Dieckmann C, Ritter H, Haag F, Koch-Nolte F, Schulz GE. Structure of the ecto-ADP-ribosyl transferase ART2.2 from rat. J Mol Biol. 2002;322:687–696. doi: 10.1016/s0022-2836(02)00818-5. [DOI] [PubMed] [Google Scholar]
- 31.O'Neal CJ, Jobling MG, Holmes RK, Hol WG. Structural basis for the activation of cholera toxin by human ARF6-GTP. Science. 2005;309:1093–1096. doi: 10.1126/science.1113398. [DOI] [PubMed] [Google Scholar]
- 32.Margarit SM, Davidson W, Frego L, Stebbins CE. A steric antagonism of actin polymerization by a salmonella virulence protein. Structure (London) 2006;14:1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 33.Bell CE, Yeates TO, Eisenberg D. Unusual conformation of nicotinamide adenine dinucleotide (NAD) bound to diphtheria toxin: A comparison with NAD bound to the oxidoreductase enzymes. Protein Sci. 1997;6:2084–2096. doi: 10.1002/pro.5560061004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nagahama M, Sakaguchi Y, Kobayashi K, Ochi S, Sakurai J. Characterization of the enzymatic component of Clostridium perfringens iota-toxin. J Bacteriol. 2000;182:2096–2103. doi: 10.1128/jb.182.8.2096-2103.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bubb MR, et al. Polylysine induces an antiparallel actin dimer that nucleates filament assembly: Crystal structure at 3.5-A resolution. J Biol Chem. 2002;277:20999–21006. doi: 10.1074/jbc.M201371200. [DOI] [PubMed] [Google Scholar]
- 36.Vagin A, Teplyakov A. An approach to multi-copy search in molecular replacement. Acta Crystallogr D. 2000;56:1622–1624. doi: 10.1107/s0907444900013780. [DOI] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.DeLano WL. The PyMOL Users Manual. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.