Abstract
SNAP-25 and its ubiquitously expressed homologue, SNAP-23, are SNARE proteins that are essential for regulated exocytosis in diverse cell types. Recent work has shown that SNAP-25 and SNAP-23 are partly localized in sphingolipid/cholesterol-rich lipid raft domains of the plasma membrane and that the integrity of these domains is important for exocytosis. Here, we show that raft localization is mediated by a 36-amino-acid region of SNAP-25 that is also the minimal sequence required for membrane targeting; this domain contains 4 closely spaced cysteine residues that are sites for palmitoylation. Analysis of endogenous levels of SNAP-25 and SNAP-23 present in lipid rafts in PC12 cells revealed that SNAP-23 (54% raft-associated) was almost 3-fold more enriched in rafts when compared with SNAP-25 (20% raft-associated). We report that the increased raft association of SNAP-23 occurs due to the substitution of a highly conserved phenylalanine residue present in SNAP-25 with a cysteine residue. Intriguingly, although the extra cysteine in SNAP-23 enhances its raft association, the phenylalanine at the same position in SNAP-25 acts to repress the raft association of this protein. These different raft-targeting signals within SNAP-25 and SNAP-23 are likely important for fine-tuning the exocytic pathways in which these proteins operate.
The secretion of molecules from the cell and the transport of newly synthesized proteins and lipids to the plasma membrane are dependent upon the fusion of intracellular carrier vesicles with the plasma membrane; this fusion process is termed “exocytosis.” Exocytosis is mediated by a complex series of protein-protein and protein-lipid interactions that mediate the targeting of vesicles to the plasma membrane and the subsequent fusion of these two membranes (1, 2). Central to the process of exocytosis are SNARE1 proteins (3-5). The interaction of plasma membrane SNARE proteins with SNAREs present on exocytic vesicles draws the two membranes into close apposition and may initiate membrane fusion (6).
There has been much interest recently in the domain distribution of exocytic SNARE proteins at the plasma membrane. Exocytosis is mediated by the interaction of the vesicle SNARE protein, vesicle-associated membrane protein, with the plasma membrane SNAREs syntaxin and SNAP-25/SNAP-23. A number of recent studies have found that exocytic SNARE proteins are partly localized in cholesterol/sphingolipid-rich lipid raft domains (7-15). Furthermore, disruption of lipid rafts by cholesterol depletion affects the integrity of exocytosis, suggesting that these domains play a key role in this process. It is possible that rafts function in exocytosis by spatially coordinating proteins and protein complexes within the plasma membrane. In addition, the lipids enriched within lipid rafts may impact directly on membrane fusion (15).
The raft association of proteins can occur by several mechanisms, and protein acylation has been identified as an important raft-targeting signal (16). There are many data detailing the role of N-terminal dual acylation of proteins in raft targeting, the combination of one myristate and one palmitate group being sufficient to promote accumulation in lipid raft domains (17). In contrast, much less is known about the relationship between multiple palmitoylation (three or more palmitate groups) of proteins and raft association. This is particularly true for proteins that are multiply palmitoylated at a central cysteine-rich domain and for which palmitoylation is a prerequisite for membrane targeting.
One of the most interesting examples of a multiply palmitoylated raft-associated protein is SNAP-25. This protein has a central membrane-targeting domain containing 4 cysteines. Mutation of any one of these cysteines significantly reduces palmitate incorporation into the protein, suggesting that all 4 cysteines are sites for palmitoylation (18). Indeed, an earlier study demonstrated that 3– 4 moles of palmitate were present per mole of protein (19). SNAP-25 is most abundant in neuronal and neuroendocrine cells, whereas its homologue SNAP-23 is expressed fairly ubiquitously (20, 21). Perhaps the most intriguing and conspicuous difference between these protein homologues is the presence of an additional cysteine in the membrane-targeting domain of SNAP-23; the relevance of this additional cysteine is not known.
In this study, we have analyzed the sequence elements present within SNAP-25 and SNAP-23 that are important for raft association. We present novel data showing that the palmitoylation of SNAP-25 is required for raft association. Furthermore, we demonstrate that endogenous SNAP-23 displays an almost 3-fold enrichment in lipid rafts relative to SNAP-25. Mutational analysis of both SNAP-25 and SNAP-23 reveals that this difference in raft association is due to the additional cysteine residue in the membrane-targeting domain of SNAP-23. Interestingly, although this extra cysteine enhances the raft association of SNAP-23, a highly conserved phenylalanine at the same position in SNAP-25 acts to repress the raft association of this protein. These results demonstrate that the cysteine-rich membrane-targeting domains of SNAP-25 and SNAP-23 have different affinities for lipid raft domains as a result of a phenylalanine/cysteine switch. The different affinity of these SNARE proteins for raft domains may play an important role in fine-tuning the exocytosis machinery in diverse cell types.
EXPERIMENTAL PROCEDURES
Materials
Rat HA antibody and Complete protease inhibitor tablets were purchased from Roche Applied Science. SNAP-23 and SNAP-25 antibodies were from Synaptic Systems (Göttingen, Germany). Anti-GFP was from Chemicon (Hampshire, UK). All media and sera were purchased from Invitrogen. Triton X-100 and all other reagents were of an analytical grade from Sigma.
Plasmids
Murine wild-type SNAP-23, C79F, and C83F mutants were generated by reverse transcription-PCR and cloned into pEGFPC2 (N-terminal GFP fusion). GFP-SNAP-25 and GFP-SNAP-25 (85–120) were kind gifts of Maurine Linder (Washington University School of Medicine, St. Louis, MO). HA-tagged (N terminus) SNAP-25B was provided by Bob Burgoyne (University of Liverpool, UK). The SNAP-23 (C79F), SNAP-23 (C83F), SNAP-23 (C79S), SNAP-23 (C83S), SNAP-25 (F84C), and SNAP-25 (F84S) mutations were introduced into the appropriate vector using QuikChange site-directed mutagenesis (Stratagene, La Jolla, CA).
Cell Culture and Transfection
PC12 cells were cultured in RPMI 1640 medium supplemented with 10% horse serum, 5% fetal calf serum in a humidified atmosphere containing 5% CO2. For electroporation, cells were trypsinized, and 10 μg of vector was added to 10 × 106 cells; cells were then pulsed three times at 250 V and 950 microfarads. Transfected cells were analyzed 2–3 days after transfection.
Fractionation of PC12 Cells
PC12 cells were homogenized with 20 strokes of a Dounce homogenizer in HES buffer (20 mm Hepes, 250 mm sucrose, 1 mm EDTA, pH 7.4) supplemented with protease inhibitors. Cytosol (supernatant) and membrane (pellet) fractions were separated by centrifugation at 196,000 × g for 30 min.
Detergent Solubilization and Sucrose Gradient Flotation
For analysis of endogenous proteins, detergent solubilization was performed on intact cells. In contrast, for analysis of transfected proteins, detergent solubilization was performed on purified membranes (see above). This protocol ensured that any differences in detergent insolubility of transfected proteins was not a consequence of their differential association with cytosol and membrane fractions. PC12 cells (∼25 × 106) or purified membranes were lysed in 0.5 ml of Mes-buffered saline (25 mm Mes, 150 mm NaCl, pH 6.5) containing 1% Triton-X-100 and supplemented with a protease inhibitor mix (Roche Applied Science). The samples were then incubated at 4 °C for 20 min with end-over-end rotation. The lysate was homogenized with 10 strokes of a Dounce homogenizer, and 0.4 ml of the homogenate was added to an equal volume of 80% (w/v) sucrose in Mes-buffered saline. The lysate (in 40% sucrose) was placed at the bottom of a centrifuge tube and overlaid successively with 2.2 ml of 30% sucrose and 1.4 ml of 5% sucrose. After centrifugation at 240,000 × g in a Beckman SW60 rotor for 18 h, 400-μl fractions were collected from the top of the gradient (designated fractions number 1 (top) through 11 (bottom)) and immediately supplemented with protease inhibitors. The pellet was resuspended by Dounce homogenization in 0.4 ml of Mes-buffered saline and designated fraction 12.
RESULTS
Palmitoylation of the 85–120 Membrane-targeting Domain Targets SNAP-25 to Lipid Rafts
SNAP-25 contains two α helical SNARE domains separated by a central cysteine-rich domain, which contains 4 cysteine residues. All of these cysteines are believed to be modified by the post-translational addition of palmitate (18, 19), and this palmitoylation is essential for membrane targeting of SNAP-25 (19, 20). This cysteine-rich cluster is contained within a 36-amino-acid domain of SNAP-25 (residues 85–120), which is sufficient to target GFP to the plasma membrane (22) (Fig. 1A).
Fig. 1. Amino acids 85–120 of SNAP-25 are sufficient to target GFP to lipid raft domains.
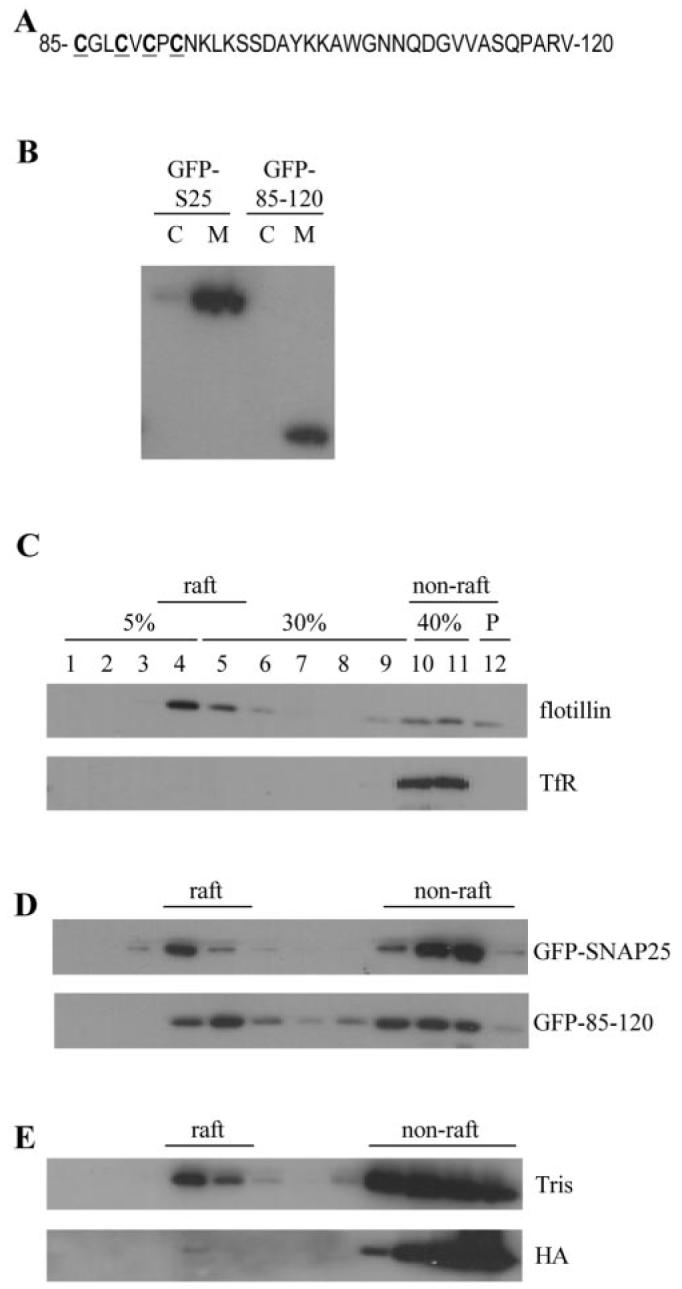
A, residues 85–120 of SNAP-25B, which is the minimal plasma membrane-targeting sequence of this protein. Cysteine residues are highlighted in bold and are underlined. B, transfected PC12 cells were fractionated into cytosol (C) and membrane (M) fractions. The distribution of GFP-SNAP-25 and GFP-85–120 between these fractions was determined by immunoblotting using a GFP-specific antibody. C, Triton X-100-solubilized PC12 cells were fractionated on a discontinuous sucrose gradient as detailed under “Experimental Procedures.” Gradient fractions were analyzed by immunoblotting using antibodies specific for flotillin and transferrin receptor (TfR). Fraction numbers and corresponding sucrose concentrations are indicated; P is the pellet fraction. D, PC12 cells were transfected with GFP-tagged constructs containing full-length SNAP-25B or the membrane-targeting domain (residues 85–120). Purified membranes from transfected cells were solubilized and fractionated on a discontinuous sucrose gradient. Collected fractions were analyzed by immunoblotting using a GFP-specific antibody. E, PC12 cell membranes were incubated in either 1 m hydroxylamine (HA), pH 7, or 1 M Tris, pH 7, at room temperature for 20 h. Membranes were washed, and rafts were purified as Triton-insoluble complexes. Fractions were probed with an antibody specific for SNAP-25.
To determine whether the raft targeting of SNAP-25 is mediated by the membrane-targeting domain or some other region of the protein, PC12 cells were transfected with plasmids encoding GFP-tagged SNAP-25 or GFP linked to the 85–120 membrane-targeting domain. Both constructs were associated with the membrane fraction of PC12 cells and were essentially absent from the cytosol (Fig. 1B). Lipid rafts were purified as detergent-resistant membranes based on Triton X-100 insolubility and sucrose gradient flotation (23). Fig. 1C confirms that this procedure effectively separates raft (flotillin) from non-raft (transferrin receptor) proteins. Analysis of the distribution of GFP constructs, fused to either full-length SNAP-25 or the 36-amino-acid (85–120) membrane-targeting domain, revealed that both constructs were present in raft fractions (Fig. 1D). Thus, the 85–120-amino-acid domain contains all the necessary information for SNAP-25 membrane targeting and lipid raft association. Interestingly, the 85–120 membrane-targeting domain displayed a consistently higher level of raft association than full-length SNAP-25 (17.5 ± 3.2% of GFP-SNAP-25 was raft-associated (n = 4), whereas 34.7 ± 3.7% of 85–120-GFP was raft-associated (n = 5)). As the palmitoylation of proteins is a well recognized raft targeting signal, an obvious possibility was that the palmitoylation of the 85–120 domain of SNAP-25 resulted in its accumulation in raft domains. To test this directly, membranes were isolated from PC12 cells and treated with 1 m hydroxylamine (pH 7), which cleaves thioester linkages (or 1 m Tris (pH 7) as a control). Hydroxylamine treatment has previously been shown to cause complete depalmitoylation of SNAP-25 but interestingly does not release the protein from membranes (24). Thus, palmitoylation is required for initial membrane targeting of SNAP-25, but once at the membrane, other interactions can preserve membrane association in the absence of palmitoylation (24). Hydroxylamine-treated membranes were solubilized in Triton, and rafts were isolated by flotation. These experiments showed that HA-induced depalmitoylation of SNAP-25 caused a marked loss of SNAP-25 immunoreactivity from raft domains (Fig. 1E), demonstrating that the palmitoylation of SNAP-25 is essential for its association with raft domains (quantification of three experiments of this type revealed that HA treatment caused an average 42% loss of SNAP-25 from raft fractions when compared with Tris treatment).
Endogenous SNAP-25 and SNAP-23 Display Different Levels of Raft Association in PC12 Cells
There are three SNAP-25 homologues; SNAP-25A and SNAP-25B are expressed predominantly in neuronal/neuroendocrine cells, whereas SNAP-23 has a more ubiquitous tissue distribution. Analysis of the membrane-targeting domains of these proteins reveals that SNAP-25A and SNAP-25B have a 4-cysteine motif, whereas SNAP-23 has 5 cysteines in this domain (Fig. 2A, and see also Ref. 25). As the palmitoylated membrane-targeting domain of SNAP-25 is sufficient for raft association (Fig. 1D), it was possible that the different cysteine clusters within the membrane-targeting domains of SNAP-25 and SNAP-23 have different affinities for rafts. To examine this possibility, the extent of the raft association of endogenous SNAP-25 and SNAP-23 in PC12 cells was determined. Intriguingly, SNAP-23 and SNAP-25 exhibited a marked difference in their levels of enrichment in lipid raft fractions (Fig. 2B). Quantification of several gradients revealed that SNAP-23 was present in raft domains to ∼2.65-fold higher levels than SNAP-25 (Fig. 2C); 20.3% ± 6.0 of SNAP-25 was raft-associated, 53.8% ± 5.1 of SNAP-23 was raft-associated (n = 5, p < 0.005). The observed differences in the association of SNAP-25 and SNAP-23 with detergent-insoluble rafts did not reflect different levels of the membrane association of these proteins, as both were essentially absent from the cytosol fraction of PC12 cells (Fig. 2D).
Fig. 2. Comparison of the association of endogenous SNAP-25 and SNAP-23 with lipid rafts in PC12 cells.
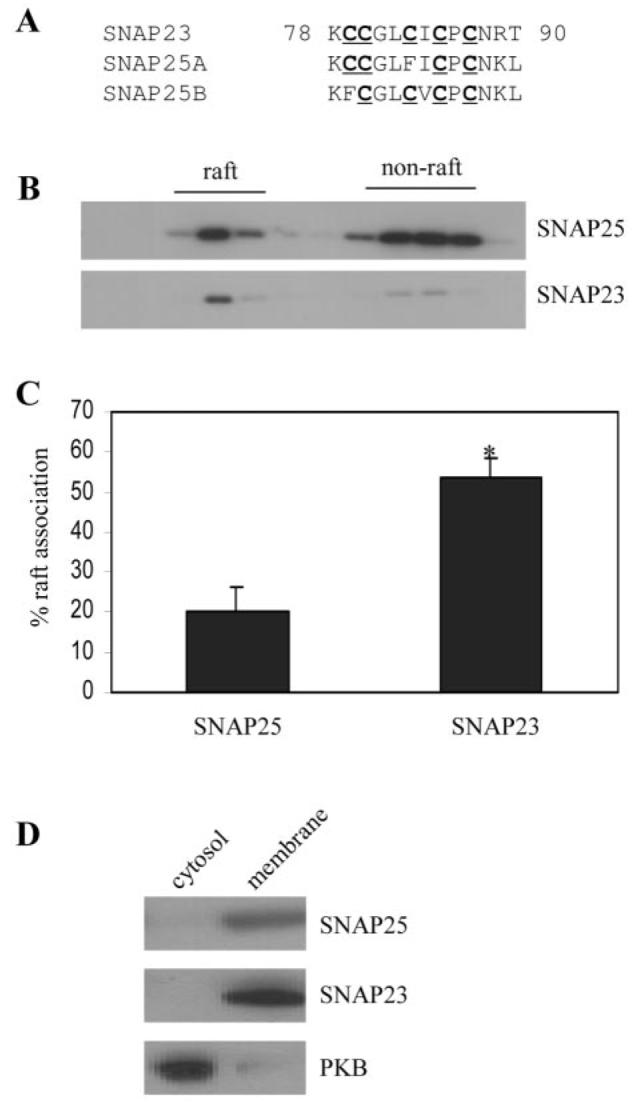
A, comparison of the cysteine-rich domains of SNAP-23, SNAP-25A, and SNAP-25B. Cells were solubilized in Triton X-100 and fractionated on a discontinuous sucrose gradient as described under “Experimental Procedures.” Recovered fractions were probed with antibodies specific to SNAP-25 and SNAP-23. B shows representative blots, whereas C is averaged data from five separate experiments. *, p < 0.005. D, distribution of SNAP-25, SNAP-23, and protein kinase B (PKB) between cytosolic and membrane fractions purified from PC12 cells.
Single Cysteine Mutations in SNAP-23 Reduce Its Raft Association to a Similar Level as Endogenous SNAP-25
Although there were a number of possible explanations as to why SNAP-23 was more abundant in rafts than SNAP-25, we were particularly intrigued by the additional cysteine residue in the membrane-targeting domain of SNAP-23. This cysteine residue is replaced by a phenylalanine in SNAP-25. To investigate the possibility that the increased raft association of SNAP-23 was due to this extra cysteine, we fused SNAP-23 to GFP and generated two mutants by site-directed mutagenesis; one mutant construct had the cysteine residue at amino acid position 79 replaced with a phenylalanine (C79F), and another mutant replaced cysteine 83 with a phenylalanine (C83F). Thus, these mutant proteins have membrane-targeting domains that have the same number and distribution of cysteine and phenylalanine residues as SNAP-25B (C79F) and SNAP-25A (C83F).
The GFP-tagged SNAP-23 constructs were transfected into PC12 cells, and their distribution was examined by immunofluorescence labeling. Fig. 3A shows that GFP-SNAP-23 was localized, as expected, to the plasma membrane. In addition, the C79F and C83F mutant proteins displayed an identical distribution to wild-type SNAP-23. Furthermore, the transfected proteins all associated efficiently with PC12 cell membranes (Fig. 3B).
Fig. 3. Analysis of the association of SNAP-23 cysteine mutants with lipid rafts.
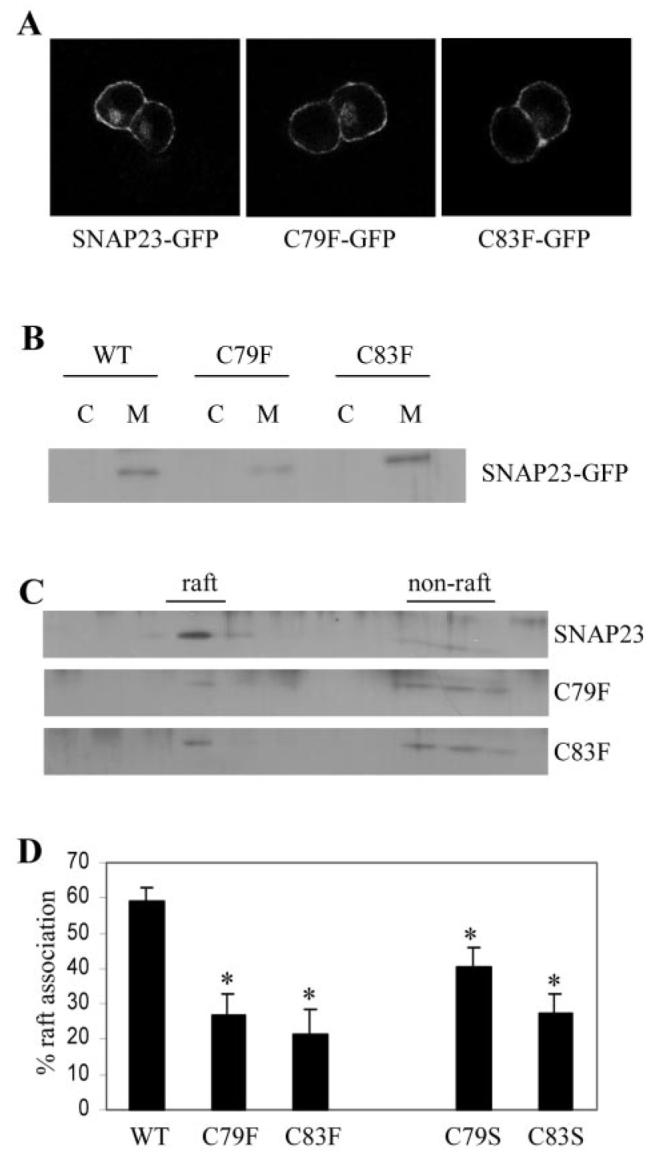
A, immunofluorescence analysis revealed that wild-type and mutant SNAP-23 proteins were strongly enriched at the cell surface. B, distribution of SNAP-23 and C79F and C83F mutants in cytosol (C) and membrane (M) fractions purified from PC12 cells. WT, wild type. C, membranes prepared from transfected PC12 cells were solubilized in Triton X-100 and fractionated on a discontinuous sucrose gradient as detailed under “Experimental Procedures.” Distribution of the GFP-tagged SNAP-23, C79F, and C83F proteins in the recovered gradient fractions was analyzed by immunoblotting using an antibody specific for GFP. D, averaged data from five separate experiments. *, p < 0.006 for C79F and C83F mutants when compared with wild-type SNAP-23. Also shown for comparison are normalized results for the raft association of C79S and C83S mutants, which displayed a marked reduction in raft association when compared with wild-type SNAP-23 (*, p < 0.02 for C79S and p < 0.0005 for C83S, n = 8)
Although SNAP-23 and the C79F and C83F mutant proteins were all localized correctly to the plasma membrane when expressed in PC12 cells, the cysteine mutants displayed a marked reduction in the level of raft association (Fig. 3, C and D). Averaged data from five experiments revealed that whereas 59.2% ± 3.6 of wild-type SNAP-23 was present in lipid raft fractions, the level of the raft association of C79F and C83F mutants was 26.6% ± 6.1 and 21.6% ± 6.6, respectively (Fig. 3C). These results demonstrate that the 5-cysteine motif present in SNAP-23 is responsible for the enhanced raft association of this protein when compared with SNAP-25.
It was formally possible that the cysteine to phenylalanine mutants displayed a decreased affinity for rafts not due to the loss of a cysteine but due to the gain of a phenylalanine residue. In this respect, it should be noted that the phenylalanine residues that replace cysteines in SNAP-25A and B are highly conserved. Thus, we also constructed serine mutants and examined the raft association of C79S and C83S in PC12 cells. Similar to the phenylalanine mutants, the serine mutants displayed a marked reduction in raft association when compared with wild-type SNAP-23 (results with these mutants were normalized to allow comparison with the C79F and C83F mutants; the raft association of C79S was 40.6 ± 5.6% (n = 8), and C83S was 27.4 ± 5.4% (n = 8)). Although the raft association of the serine mutants was significantly reduced when compared with wild-type SNAP-23, these proteins appeared to display a greater level of raft association than the corresponding phenylalanine mutants (this was particularly true of substitutions made at amino acid position 79 in SNAP-23). Thus, although loss of cysteine residues in SNAP-23 has a direct and major effect on raft association, this also appears to be inhibited by the introduction of phenylalanine residues at these positions.
Introduction of an Extra Cysteine into the Membrane-targeting Domain of SNAP-25 Increases Its Level of Raft Association
The experiments detailed above demonstrate that SNAP-23 abundance in purified lipid raft fractions can be sharply modulated by mutation of specific cysteine residues within the membrane-targeting domain of this protein. Can the raft association of SNAP-25 be modulated by introducing an extra cysteine into the membrane-targeting domain (as present in SNAP-23)? Phenylalanine at amino acid position 84 in SNAP-25B was mutated to cysteine (F84C), making the number and distribution of cysteines the same as for SNAP-23. In addition, we generated a phenylalanine to serine (F84S) mutant to examine the role of Phe-84 in the raft association of SNAP-25. Immunofluorescence analysis of HA-tagged F84C and F84S revealed that these mutations had no effect on the targeting of SNAP-25 to the plasma membrane in PC12 cells (Fig. 4A). Similarly, all three proteins associated efficiently with the membrane fraction in PC12 cells (Fig. 4B). However, analysis of lipid raft association (as above) demonstrated that F84C had a significantly higher level of raft association than wild-type SNAP-25 (Fig. 4, C and D). In addition, the F84S mutant displayed a level of raft association intermediate between wild-type SNAP-25 and F84C. These results demonstrate that the Cys/Phe switch in SNAP-23/-25 has a dual effect on raft association; cysteine enhances and phenylalanine reduces raft association.
Fig. 4. Analysis of the association of SNAP-25 phenylalanine mutants with lipid rafts.
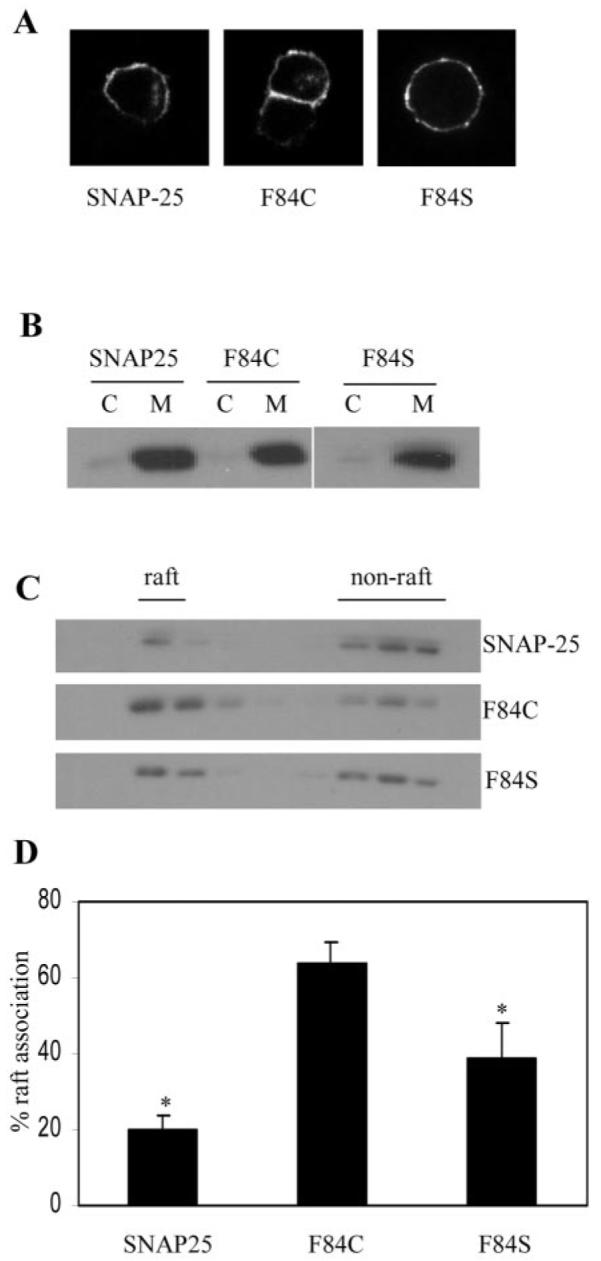
A, immunofluorescence analysis of the subcellular distribution of wild-type SNAP-25 and the F84C and F84S mutants. All three proteins show a strong enrichment at the plasma membrane. B, distribution of proteins between cytosol (C) and membrane (M) fractions purified from PC12 cells. C, membranes prepared from transfected PC12 cells were solubilized in Triton X-100 and fractionated on a discontinuous sucrose gradient as detailed under “Experimental Procedures.” The distribution of the HA-tagged proteins in the recovered gradient fractions was analyzed by immunoblotting using an antibody specific for the HA tag. D, averaged data from six separate experiments. *, F84C displayed an increased level of raft association when compared with wild-type SNAP-25 (p < 0.00005) and F84S (p < 0.035).
DISCUSSION
SNARE proteins have been localized to raft domains in diverse cell types, and this localization is thought to be important for efficient exocytosis. An important area of research is to understand the mechanisms of SNARE targeting to these lipid raft domains. The analysis reported here has revealed that the palmitoylation of the central cysteine-rich domain of SNAP-25 mediates raft targeting. Furthermore, the distinct cysteine-rich domains of SNAP-25 and SNAP-23 were shown to have a major effect on the level of the raft association of these proteins. We showed directly that a 5-cysteine domain has an almost 3-fold greater affinity for rafts than a 4-cysteine motif. This difference was accounted for by the inhibition of raft association by a phenylalanine within SNAP-25 and a positive influence of the substituted cysteine residue at this position within SNAP-23. The extra cysteine residue within SNAP-23 presumably has a positive effect on raft association by providing an extra palmitoylation site. In contrast, the phenylalanine (a bulky hydrophobic residue) may have a negative impact on raft association by perturbing the structure of the membrane-targeting domain of SNAP-25. This may prevent the tight association of palmitic acid residues with lipid raft domains or may impact directly on the palmitoylation of SNAP-25. The inhibitory effect of Phe-84 on the raft association of SNAP-25 can explain why we found that the GFP-85–120 construct was more abundant in rafts than full-length SNAP-25 (Fig. 1D); this construct does not contain Phe-84.
Two recent studies have suggested that the SNARE partner of SNAP-25, syntaxin 1A, does not have an intrinsic affinity for “raft” domains in model membranes (26, 27). We would emphasize that such model systems do not adequately reflect the lipid complexity and asymmetry of cellular membranes. Although simple mixtures of sphingolipid, cholesterol, and phospholipid can provide important information on the domain formation of these lipids, experiments examining protein association must be treated with caution. The data presented in this report clearly demonstrate that SNAP-25 has an intrinsic affinity for detergent-insoluble raft domains present in PC12 cells. Thus, even if syntaxin 1A does not have an intrinsic affinity for rafts, its interaction with SNAP-25 may be sufficient to result in the accumulation of syntaxin in rafts. Indeed, we have previously identified a syntaxin 1A-SNAP-25 complex in raft domains purified from PC12 cells.
The data presented in this study re-emphasize that the relationship between protein palmitoylation and raft targeting is not simple and certainly not easy to predict. For example, previous work from our group has shown that the cysteine string protein, which is palmitoylated on at least 11 cysteine residues, is completely excluded from raft domains in adipocytes, despite being localized exclusively to the plasma membrane in this cell type (10, 28).
Acknowledgments
We are grateful to Bob Burgoyne and Maurine Linder for the generous donation of plasmids.
Footnotes
This work was funded by the Biotechnology and Biological Sciences Research Council (Grant 17/C16435), the Wellcome Trust, and the Diabetes Research and Wellness Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked "advertisement" in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The abbreviations used are: SNARE, soluble NSF attachment protein receptors; SNAP, soluble NSF attachment protein; NSF, N-ethylmaleimide-sensitive factor; HA, hemagglutinin; GFP, green fluorescent protein; Mes, 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Jahn R, Südhof TC. Annu. Rev. Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- 2.Burgoyne RD, Morgan A. Physiol. Rev. 2003;83:581–632. doi: 10.1152/physrev.00031.2002. [DOI] [PubMed] [Google Scholar]
- 3.Söllner TH, Whiteheart SW, Brunner M, Erdjument-Bromage H, Geromanos S, Tempst P, Rothman JE. Nature. 1993;362:318–324. doi: 10.1038/362318a0. [DOI] [PubMed] [Google Scholar]
- 4.Söllner TH, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. Cell. 1993;75:409–418. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 5.Chen YA, Scheller RH. Nat. Rev. Mol. Cell. Biol. 2001;2:98–106. doi: 10.1038/35052017. [DOI] [PubMed] [Google Scholar]
- 6.Weber T, Zemelman BV, McNew JA, Westermann B, Gmachl M, Parlati F, Söllner TH, Rothman JE. Cell. 1998;92:759–772. doi: 10.1016/s0092-8674(00)81404-x. [DOI] [PubMed] [Google Scholar]
- 7.Lafont F, Verkade P, Galli T, Wimmer C, Louvard D, Simons K. Proc. Natl. Acad. Sci. U. S. A. 1999;96:3734–3738. doi: 10.1073/pnas.96.7.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chamberlain LH, Burgoyne RD, Gould GW. Proc. Natl. Acad. Sci. U. S. A. 2001;98:5619–5624. doi: 10.1073/pnas.091502398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang T, Bruns D, Wenzel D, Riedel D, Holroyd P, Thiele C, Jahn R. EMBO J. 2001;20:2202–2213. doi: 10.1093/emboj/20.9.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chamberlain LH, Gould GW. J. Biol. Chem. 2002;277:49750–49754. doi: 10.1074/jbc.M206936200. [DOI] [PubMed] [Google Scholar]
- 11.Pombo I, Rivera J, Blank U. FEBS Lett. 2003;550:144–148. doi: 10.1016/s0014-5793(03)00864-0. [DOI] [PubMed] [Google Scholar]
- 12.Foster LJ, de Hoog CL, Mann M. Proc. Natl. Acad. Sci. U. S. A. 2003;100:5813–5818. doi: 10.1073/pnas.0631608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taverna E, Saba E, Rowe J, Francolini M, Clementi F, Rosa P. J. Biol. Chem. 2004;279:5127–5134. doi: 10.1074/jbc.M308798200. [DOI] [PubMed] [Google Scholar]
- 14.Xia F, Gao X, Kwan E, Lam PPL, Chan L, Sy K, Sheu L, Wheeler MB, Gaisano HY, Tsushima RG. J. Biol. Chem. 2004;279:24685–24691. doi: 10.1074/jbc.M314314200. [DOI] [PubMed] [Google Scholar]
- 15.Salaün C, James DJ, Chamberlain LH. Traffic. 2004;5:255–264. doi: 10.1111/j.1600-0854.2004.0162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melkonian KA, Ostermeyer AG, Chen JZ, Roth MG, Brown DA. J. Biol. Chem. 1999;274:3910–3917. doi: 10.1074/jbc.274.6.3910. [DOI] [PubMed] [Google Scholar]
- 17.Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM. J. Cell Biol. 1994;126:353–363. doi: 10.1083/jcb.126.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lane SR, Liu Y. J. Neurochem. 1997;69:1864–1869. doi: 10.1046/j.1471-4159.1997.69051864.x. [DOI] [PubMed] [Google Scholar]
- 19.Veit M, Söllner TH, Rothman JE. FEBS Lett. 1996;385:119–123. doi: 10.1016/0014-5793(96)00362-6. [DOI] [PubMed] [Google Scholar]
- 20.Ravichandran V, Chawla A, Roche PA. J. Biol. Chem. 1996;268:24408–24414. doi: 10.1074/jbc.271.23.13300. [DOI] [PubMed] [Google Scholar]
- 21.Wang G, Witkin JW, Hao G, Bankaitis VA, Scherer PE, Baldini G. J. Cell Sci. 1997;110:505–513. doi: 10.1242/jcs.110.4.505. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalo S, Greentree WK, Linder ME. J. Biol. Chem. 1999;274:21313–21318. doi: 10.1074/jbc.274.30.21313. [DOI] [PubMed] [Google Scholar]
- 23.Chamberlain LH. FEBS Lett. 2004;559:1–5. doi: 10.1016/s0014-5793(04)00050-x. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo S, Linder ME. Mol. Biol. Cell. 1998;9:585–597. doi: 10.1091/mbc.9.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koticha DK, Huddleston SJ, Witkin JW, Baldini G. J. Biol. Chem. 1999;274:9053–9060. doi: 10.1074/jbc.274.13.9053. [DOI] [PubMed] [Google Scholar]
- 26.Saslowsky DE, Lawerence JC, Henderson RM, Edwardson JM. J. Membr. Biol. 2003;194:153–164. doi: 10.1007/s00232-003-2035-7. [DOI] [PubMed] [Google Scholar]
- 27.Bacia K, Schuette CG, Kahya N, Jahn R, Schwille P. J. Biol. Chem. 2004;279:37951–37955. doi: 10.1074/jbc.M407020200. [DOI] [PubMed] [Google Scholar]
- 28.Chamberlain LH, Graham ME, Kane S, Jackson JL, Maier VH, Burgoyne RD, Gould GW. J. Cell Sci. 2001;114:445–455. doi: 10.1242/jcs.114.2.445. [DOI] [PubMed] [Google Scholar]


