Abstract
The early preimplantation mouse embryo is a unique system where it is possible to explore the foundations of totipotency and differentiation. Following fertilization, a single cell, the zygote, will give rise to all tissues of the organism. The first signs of differentiation in the embryo are evident at the blastocyst stage with the formation of the trophectoderm, a differentiated tissue that envelopes the inner cell mass. The question of when and how the cells start to be different from each other in the embryo is central to developmental biology: as cell fate decisions are undertaken, loss of totipotency comes about. Although the blastomeres of the preimplantation embryo are totipotent, as the embryo develops some differences appear to develop between them which are, at least partially, related to the epigenetic information of each of these cells. The hypothesis of epigenetic asymmetries acting as driver for lineage allocation is presented. Although there are now some indications that epigenetic mechanisms are involved in cell fate determination, much work is needed to discover how such mechanisms are set in play upon fertilization and how they are transmitted through cell division. These considerations are further discussed in the context of preimplantation genetic diagnosis: does it matter to the embryo which cell is used for genetic diagnosis? The exquisite complexity and richness of chromatin-regulated events in the early embryo will certainly be the subject of exciting research in the future.
Keywords: cell fate, epigenetics, histone methylation, mouse embryo, pluripotency
Embryonic development starts from a single cell, the zygote. In this cell, the two gametes convey and contribute information to start a new developmental programme. The formation of the newly fertilized zygote constitutes therefore the climax of totipotency because of the resulting zygote’s inherent ability to produce all cell types in a new organism.
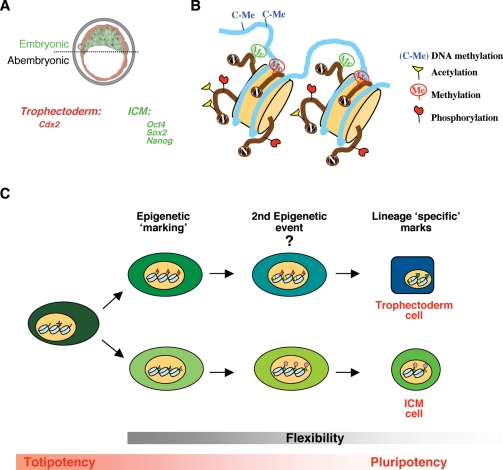
In the mammalian embryo, the first differentiative event occurs as inner cells form upon cell division at the 8-cell stage. As a result of this division, an ‘inner’ and an ‘outer’ population of cells can be distinguished in the 16-cell stage embryo, which will respectively occupy different positions in the morula (Johnson and Ziomek, 1981). Much work has been done in trying to understand how cell polarity develops in the inner and outer cells, but I will not deal with this topic here and instead will refer the reader to an excellent review published elsewhere (Johnson and McConnell, 2004). The inner cells will develop into the inner cell mass (ICM) and the outer layer of cells will differentiate into the trophectoderm (Tarkowski and Wroblewska, 1967; Ziomek and Johnson, 1982). Morphologically, however, the first overt signs of differentiation are evident only at the blastocyst stage, with the formation of the trophectoderm, which is the first differentiated tissue to form as an epithelial layer that envelops the ICM (Fig. 1A). The latter, in contrast to the trophectoderm, retains its pluripotent character and the ability to self renew. While the ICM will give rise to the embryo proper, the trophectoderm will give rise to the extraembryonic tissues that will support development of the embryo during gestation. The ICM will also give rise to yet another extraembryonic tissue, the primitive endoderm, which is first visible as a cuboidal layer of cells lining the blastocoelic cavity on the fourth day of development. The trophectoderm and the ICM each display molecular identity, which is reflected in part by the expression of specific genes that are, for the ICM, involved in its specification and the maintenance of pluripotency (such as Nanog and Oct4) or, for the trophectoderm, that are required for its differentiation (such as Cdx2) (Palmieri et al., 1994; Nichols et al., 1998; Chambers et al., 2003; Mitsui et al., 2003; Strumpf et al., 2005). The bifurcation of these two lineages is complete at the late blastocyst stage. At this stage, cells from the ICM do no longer have the potential to form trophectoderm derivates in vivo upon transplantation (Rossant and Lis, 1979), indicating that these cells have lost their totipotency and that lineage allocation has definitely occurred.
Figure 1:
Cell lineages of the mammalian blastocyst and epigenetic marking
(A) Representation of the lineages in the mammalian blastocyst on the third day of gestation (E3.5). The blastocyst is composed of two distinct populations of cells: the trophectoderm (red) and the inner cell mass (ICM, green), which display molecular identity and epigenetic asymmetries. The embryonic-abembryonic regions (dotted line) of the blastocyst are determined by the position of the ICM, which lies within the embryonic region of the blastocyst. (B) Diagram illustrating some of the epigenetic marks. The DNA wrapped around the nucleosome (beige cylinders) is shown as light blue. The DNA is subject to DNA methylation, which constitutes one of the main epigenetic players. The core histones (beige) that form the nucleosome can be covalently modified (by acetylation, methylation, phosphorylation), particularly on their N-terminal tails. Each of these marks can have an effect on how the information contained in the DNA is read by modulating downstream events such as transcriptional activation or repression. For example, histone methylation (Me) can have a positive effect on transcription (green) or a repressive one (red). The marks can be present in different combinations and may change during the cell cycle. (C) Model for epigenetic marking and lineage allocation. In this model, an epigenetic mark would be laid down in a given cell during development. There could be other epigenetic event(s) that reinforce and/or are influenced by the first marking event. Cumulatively, this could result in determination of the fate of that cell towards a lineage in the blastocyst. The lineage specific marks could stabilize such cell identity and might be necessary for further differentiation. Alternatively, the acquisition of these epigenetic marks could be the result of cell fate determination. One should also consider that not only the nature of the mark would be important, but also the different regions of the chromatin that would be affected by such marks. Because the cells in the preimplantation embryo are totipotent and because the chromatin will still need to be dynamically remodelled during subsequent development, flexibility should be an important component of epigenetic mechanisms taking place during early development. As cell fate decisions are taken, a concomitant loss of totipotency takes place
Investigations during recent years have recreated an interest in whether the blastomeres of the mammalian embryo are truly alike throughout preimplantation development before the first differentiative division mentioned above. In other words, whether they acquire a ‘fate’ or whether they start to differ from each other prior to their spatial ‘inner/outer’ allocation within the embryo upon the formation of the morula. Of course, if this were to be the case, the big challenge would be to ascribe molecular mechanisms to these processes.
The question of when and how the cells start to be different from each other is not a trivial one. In particular, because as the first cell fate decisions are undertaken, a concomitant loss of totipotency occurs. The developmental time window when this first cell fate decision occurs comprises a number of epigenetic events (Morgan et al., 2005; Surani et al., 2007). These events include the reprogramming of the parental chromatin. Whether such epigenetic events are the cause or the consequence of reprogramming remains an exciting open question, but it is probably a combination of the two. Moreover, the two lineages of the blastocyst exhibit some epigenetic asymmetries.
This mini-review is divided in two parts, the first one will deal with the main epigenetic mechanisms that are known to occur during mammalian preimplantation development. The second one will give an overview on data obtained through experimental embryology manipulations and lineage tracing observations to study cell fate in the early mouse embryo.
Epigenetic mechanisms in the preimplantation embryo
In general terms, epigenetic mechanisms include DNA methylation, covalent modification of histones, chromatin remodelling and histone replacement through incorporation of the so-called histone variants (Fig. 1B). Histone marks have emerged as one of the main players involved in epigenetic mechanisms (Kouzarides, 2007). Histone modifications can be highly dynamic, or have a function in epigenetic memory. Although it is still unclear whether they are the actors of the epigenetic information or the epigenetic information itself, it is evident that covalent modifications of histones are essential components of the epigenome.
Histones can be modified by a number of enzymes that mediate methylation, acetylation, phosphorylation, ubiquitynation and ADP-ribosylation of specific amino acid residues (reviewed in Kouzarides, 2007). By and large, the highest density of modifications so far described occurs in histone H3, particularly on its tail. The effects of these modifications on the chromatin and on cellular processes are very diverse, and a modification of the same residue can even have opposite effects depending on the type of modification. For example, trimethylation of H3K9 is considered as a repressive mark, whereas acetylation of the same lysine has a positive effect on transcription (Bannister and Kouzarides, 2005). Likewise, methylation of arginine residues can have a positive effect on transcription (Chen et al., 1999) or a repressive effect (Pal et al., 2004), depending both on the targeted residue and on whether the methylation is symmetric or asymmetric. For some of the modified residues, there is a very clear view of the outcome of an eventual modification: H3K9me3 creates a specific docking site for the heterochromatin protein 1 (HP1), which subsequently recruits the H3K9 methyltransferase Su(var)3-9 and reinforces an autoregulatory loop for heterochromatin formation and maintenance (Bannister et al., 2001; Lachner et al., 2001; Nakayama et al., 2001).
The levels of regulation of epigenetic events in the preimplantation mouse embryo are multiple. They include the regulation of the subcellular localization of DNA methyltransferase activity, highlighted by the cytoplasmic retention of Dnmt1o (Carlson et al., 1992); the exclusion of a particular histone modification from the chromatin, which is exemplified by the lack of detection of H3K9me3 in the paternal pronucleus after fertilization resulting in an asymmetry of histone marks between the two pronuclei (Arney et al., 2002; Santos et al., 2005), the differential incorporation of chaperons and histone variants in the parental chromatin (van der Heijden et al., 2005; Torres-Padilla et al., 2006) and the acquisition of highly specific histone variants in the gametes (Clarke et al., 1992; Tanaka et al., 2001; Govin et al., 2007). Further, the maternal and paternal pronuclei exhibit different patterns of global DNA methylation: while the paternal pronucleus is rapidly demethylated—presumably through an active mechanism—right after fertilization, the maternal pronucleus is only passively demethylated through the subsequent rounds of replication and cell division that follow the first mitosis of the embryo (Mayer et al., 2000). Moreover, while the centromeric and pericentric paternal chromatin remain DNA methylated, the maternal DNA loses methylation in such regions (Rougier et al., 1998).
The changes in the levels of DNA methylation as development proceeds in the preimplantation embryo are dynamic. Global levels of DNA methylation have been analysed by immunofluorescence, bisulphate sequencing and restriction digestion (Rougier et al., 1998; Mayer et al., 2000; Santos et al., 2002; Aranyi and Paldi, 2006). Bisulphate sequencing and restriction digestion have also been used to analyse the methylation status of repeat sequences (such as L1 and IAP repeats) and some single-copy sequences (such as actin) (Howlett and Reik, 1991; Oswald et al., 2000). These studies have revealed that although global levels of DNA methylation decrease until the blastocyst stage, changes in DNA methylation do not occur to the same extent on all genes. Remarkably, imprinted genes (such as H19) and some repeat sequences (such as IAPs) do not undergo demethylation (Tremblay et al., 1997).
During early stages of development, decisions involved in cell fate determination and pluripotency have to be assumed. These processes require the chromatin to be dynamically remodelled to ensure its plasticity. This implies that the mechanisms involved in regulation of chromatin structure need to ensure stability across generations and cell division, but they also need to be flexible (Reik, 2007). The double nature of a covalent modification either on histones and/or on the DNA as dynamic (because in principle it can be added and removed) and at the same time its potential ability to propagate a memory, fits well with these aforementioned needs. Moreover, in keeping with the importance of epigenetic mechanisms during early development, the possibility for an epigenetic mark(s) underlying these phenomena appears very attractive.
Concerning the epigenetic asymmetries of the ICM and the trophectoderm, the ICM displays, in global terms, higher levels of DNA methylation compared with the trophectoderm (Dean et al., 2001; Santos et al., 2002). Specific histone marks such as trimethylation of lysines 9 and 27 of histone H3 (H3K9me3 and H3K27me3, respectively) are enriched in the ICM compared with the trophectoderm (Erhardt et al., 2003). Likewise, the trophectoderm retains an imprinted form of X inactivation, where the paternal X chromosome is silenced (Heard and Disteche, 2006). This is in contrast to the ICM, where there is reactivation of the inactive X chromosome and a subsequent round of inactivation occurs at random in which either the maternal or the paternal chromosome is inactivated (Mak et al., 2004; Okamoto et al., 2004).
The aforementioned epigenetic asymmetries of the two lineages of the blastocyst are evident once lineage allocation has taken place and might reinforce their molecular identity. However, epigenetic asymmetry could also act as a driver for lineage allocation, in which case, the former would precede the latter (Fig. 1C). This constitutes a fascinating current working hypothesis.
Development of cell identity in the mouse embryo
From experimental embryology, we have learnt from pioneer experiments performed in the 50’s that after mechanical separation of the blastomeres of a 2-cell stage embryo and transfer into foster mothers, each of these two cells gives rise to an adult mouse (Tarkowski, 1959). This indicates that mouse embryos are very flexible in what people have referred to as developmental potential. Derivation of twins from mouse blastomeres at later stages of development (e.g. 4-cell stage or later) has not been possible. Although this has been linked to the low number of cells present in the resulting embryos and hence their inability to form an ICM with a normal cell number, rather than to their developmental potential or identity (Tarkowski and Wroblewska, 1967; Rossant, 1976). Indeed, when random single 4- and 8-cell stage blastomeres are aggregated with ‘carrier’ blastomeres, their progeny is able to contribute to all the tissues of the embryo (Kelly, 1977; Tarkowski et al., 2005), and in this sense, the blastomeres were considered to be totipotent. So, what are these carrier cells providing? Is it a simple matter of cell number? and/or of an appropriate environment? It is also possible however that some subtle intrinsic differences of these blastomeres might be masked by the limitations of the outcome of transplantation procedures, given that the results of these studies are very often far from 100% and the transplantation efficiency is never absolute. Whether this is solely related to technical difficulties linked to these challenging manipulations or to an intrinsic property of specific blastomeres of the embryo is impossible to ascertain.
Despite a controversial viewpoint on whether there is any polarity in the early mouse embryo or not, most reports coincide with the interpretation that a blastomere at the 2-cell stage contributes to both the ICM and the trophectoderm (reviewed in Edwards and Beard, 1997; Zernicka-Goetz, 2006). The suggestion of a given blastomere at the 2-cell stage having a ‘preferential’ fate towards either of the blastocyst lineage is not resolved and some researchers have suggested that a slight, but distinct difference in the fate of 2-cell stage blastomeres might be disturbed by experimental manipulations (Alarcon and Marikawa, 2005; Hiiragi et al., 2006). The role of extrinsic factors to the embryo, such as the shape of the zona pellucida, in axis specification of the blastocyst is also a matter of controversy (Gardner, 2007; Kurotaki et al., 2007). However, there are some indications that a bias for a blastomere to contribute to a given region of the embryo in the blastocyst could already exist at the 4-cell stage (Fujimori et al., 2003; Piotrowska-Nitsche and Zernicka-Goetz, 2005). These conclusions are mostly based on lineage tracing experiments of labelled blastomeres, and their degree of invasiveness is debatable.
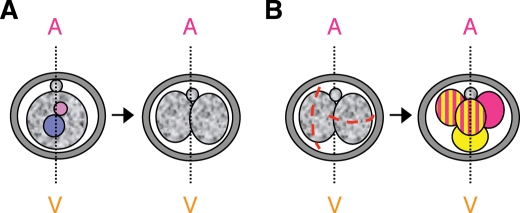
Some groups have used the plane of division in relation to the animal–vegetal axis of the embryo as a sort of guideline to distinguish and characterize blastomeres according to their cleavage plane. By convention, the animal pole is demarcated by the position of the second polar body (which is extruded after resumption of meiosis II upon fertilization) and hence the vegetal pole lies on the opposite side (Fig. 2A). The division from the 2- to the 4-cell stage would segregate for the first time the ‘animal’ and the ‘vegetal’ components of the zygote if it occurs equatorially, that is, perpendicular to the animal–vegetal axis of the conceptus (Gardner, 2002). Thus, whereas a cell that derives from a meridional division (parallel to the animal–vegetal axis) inherits both components, an equatorial division gives rise to an ‘animal’ and a ‘vegetal’ blastomere (Fig. 2B). By looking into the plane of division and the order at which this division occurs from the 2- to the 4-cell stage and subsequent lineage tracing, a subgroup of embryos was identified where it is possible, with a relatively high probability, to predict the future position of the blastomeres in the blastocyst (Piotrowska-Nitsche et al., 2005). This group of embryos are referred to as ME embryos (for Meridional–Equatorial, reflecting the type of cleavage plane and order of division that generated them (Fig. 2B) (Piotrowska-Nitsche et al., 2005; Piotrowska-Nitsche and Zernicka-Goetz, 2005). Although the ME type embryos constitute only a small part (20%) of a complete litter, they provide a very nice system where it is possible to explore the foundations of differentiation in the embryo. Indeed, the ‘vegetal’ blastomere would most often populate the abembryonic region of the blastocyst, which contains mainly mural trophectoderm.
Figure 2:
Blastomere division planes according to the animal-vegetal axis in the embryo
(A) The animal–vegetal (A–V) axis of the preimplantation embryo is demarcated, by convention, by the position of the second polar body, which marks the animal pole. The opposite side to the site of extrusion of the polar body is, by default, the vegetal pole of the embryo. (B) The division pattern from 2-to- 4-cell stage of a typical embryo that undergoes one meridional (M) and one equatorial (E) division (ME embryo) is represented. The cleavage plane is depicted by a red dashed line (embryo on the left). The blastomere that divides earlier is represented on the left. A Meridional division has a cleavage plane that is parallel to the A–V axis of the embryo and hence gives rise to two cells containing both ‘animal’ and ‘vegetal’ components (two cells with pink and yellow motifs on the embryo depicted on the right). In contrast, when a 2-cell stage blastomere divides equatorially, a segregation of the ‘animal’ and ‘vegetal’ cytoplasm occurs and follows derivation of an ‘animal’ (pink) and a ‘vegetal’ (yellow) blastomere
In looking for epigenetic marks that could be involved in an eventual cell fate decision of the blastomeres of these ME embryos, it was found that the ‘vegetal’ blastomere displays the lowest levels of dimethylated arginine 26 of H3 (H3R26me2). If H3R26me2 participates in lineage allocation, one might predict that modulating the levels of histone arginine methylation, would have an effect over cell fate. Overexpression of the histone methyltransferase that methylates this residue on H3, PRMT4/CARM1, into individual blastomeres not only induced upregulation of Nanog and Sox2, but also resulted in an almost complete allocation of these blastomeres into the ICM compartment (Torres-Padilla et al., 2007).
It is interesting to note that in the mouse, the differences described in histone H3 arginine methylation appear at the 4-cell stage (Torres-Padilla et al., 2007), that is. as early as one cell cycle after the major wave of embryonic genome activation occurs (Schultz, 2002; Hamatani et al., 2004), which suggests that these events might be, at least in part, linked to the transcriptional programme of the embryo. The developmental stage at which genome activation occurs in other mammalian species varies considerably: it takes place at the 1-to- 2-cell stage in mice, the 4-to- 8-cell stage in cows and humans, and the 8-to- 16-cell stage in sheep and rabbits (Schultz and Heyner, 1992). Would this anticipate a different timing for an eventual ‘cell fate path’ for other species? Normal fertile adults can be derived from single blastomeres from 2-, 4- and 8-cell stage embryos in the rabbit, sheep and cattle (Moore et al., 1968; Willadsen, 1981; Willadsen and Polge, 1981). Thus, these species indeed support blastomere isolation and further development at later stages than the mouse does.
Transplantation of isolated 4-cell blastomeres into morula stage embryos has demonstrated that the blastomeres at the 4-cell stage are totipotent (Kelly, 1977). In this context, it is important to note that aggregating the ‘vegetal’ cell from ME embryos to form chimeric embryos, showed that this cell is able to contribute to all tissues in the embryo. However, aggregating the same blastomere with other ‘vegetal’ blastomeres from ME embryos exclusively, results in a failure to proceed through development (Piotrowska-Nitsche et al., 2005). Thus, the environment where the blastomeres develop is crucial for the success of the embryo throughout development, and in a ‘normal’ situation, where the embryo has not been perturbed and a given cell develops in its niche, some differences appear to develop, which are, at least partially, related to the epigenetic information of each of them (Torres-Padilla et al., 2007). If epigenetic asymmetries of the early embryo are related to lineage allocation, it is still uncertain whether they are a cause or a consequence for lineage choice. Also, it remains unknown whether such epigenetic asymmetries would affect only particular regions of the genome. For example, whether genes involved in specification of the ICM such as Sox2 and Nanog would all be targeted by the same epigenetic marks in the same blastomere or whether such marks would vary among genes and/or among blastomeres. Further, are ‘inner’ and ‘outer’ cells at the 16-cell stage distinguishable in terms of their chromatin landscapes?
The experiments showing that blastomeres are able to respond to the overexpression of a histone modifier and change their fate, indicate that these cells have not yet acquired a ‘fix’ destiny, but that they can still be responsive to some kind of signals. These experiments have an important impact on showing that manipulating the epigenetic information can affect cell fate in the preimplantation embryo, in line with the importance of epigenetic mechanisms being crucial for early development. Moreover, these results do illustrate that such cells can still be flexible and accommodate themselves after a perturbing event (in this case, overexpression of a histone methyltransferase and the downstream effects on the information that is. imparted through specific histone modifications).
These studies have originated some interest from the part of the medical community, particularly, in the context of preimplantation genetic diagnosis (PGD) (Goldman, 2007). Does it matter to the embryo which cell is used for genetic diagnosis? Might the death of one of these cells have an effect on subsequent development? This question is equally valid on the impact of cell loss upon cryopreservation (Cohen et al., 2007). Although these are very delicate questions with very likely no easy answer, from the perspective of the mouse embryo, at least four things are to be considered. A tendency for a blastomere of some 4-cell stage embryos to contribute to a given region of the embryo has been documented. Second, the blastomeres in the 4-cell stage show clear differences not only in the levels of histone methylation, but also in their transcriptional activiy when they develop without being perturbed. However (third), the cells undergo a redirection of cell fate when a histone methyltransferase is overexpressed, indicating that they can readapt. Finally, the environment in which cells develop seems to be crucial for completing development and somehow the remaining cells in the embryo could compensate provided they are somehow different from each other. It is also important to note that a 4-cell stage mouse embryo might correspond to a very different developmental stage than a 4-cell embryo in other mammalian species, as illustrated by the differences in the onset of genome activation between them. Indeed, PGD is most often performed at the 8-cell stage and some reports document a better rate of development when 6-to- 9-cell stage embryos are diagnosed, as opposed to 3-to- 4-cell stage embryos (Wang et al., 2007). Moreover, the effects of in vitro fertilization procedures and culture on embryonic development are also extremely important, as they have been shown to alter epigenetic information in the mouse (Li et al., 2005).
As a final consideration, I would like to leave the reader with an open perspective of some ongoing questions in the field. Although there are strong indications that epigenetic mechanisms are involved in cell fate determination, we are still far from establishing a direct link between an epigenetic mark(s) and the derivation of a particular cell lineage in the embryo. Much work is still to be done to determine how these mechanisms are set in play upon fertilization and how they are transmitted during subsequent cleavage stages. Also, what other epigenetic marks contribute to the inheritability of cell fate decisions? How do these marks relate to and influence each other? Are different lineage-specific genes marked by a different combination(s) of epigenetic marks? Do the marking of these genes occur at different stages of development? It is also tempting to expand these notions into the stem cell field and question whether these mechanisms would also underlie the intrinsic self renewal ability of adult stem cells and their potential to differentiate into other cell types. The exquisite complexity and richness of chromatin-regulated events in the early embryo will certainly be the subject of exciting research in the future.
Funding
The author acknowledges support from the PNRRE/INSERM.
Acknowledgements
The author thanks Laszlo Tora and Stéphane Viville for comments on the manuscript.
References
- Alarcon VB, Marikawa Y. Unbiased contribution of the first two blastomeres to mouse blastocyst development. Mol Reprod Dev. 2005;72:354–361. doi: 10.1002/mrd.20353. [DOI] [PubMed] [Google Scholar]
- Aranyi T, Paldi A. The constant variation: DNA methylation changes during preimplantation development. FEBS Lett. 2006;580:6521–6526. doi: 10.1016/j.febslet.2006.11.018. [DOI] [PubMed] [Google Scholar]
- Arney KL, Bao S, Bannister AJ, Kouzarides T, Surani MA. Histone methylation defines epigenetic asymmetry in the mouse zygote. Int J Dev Biol. 2002;46:317–320. [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T. Reversing histone methylation. Nature. 2005;436:1103–1106. doi: 10.1038/nature04048. [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- Carlson LL, Page AW, Bestor TH. Properties and localization of DNA methyltransferase in preimplantation mouse embryos: implications for genomic imprinting. Genes Dev. 1992;6:2536–2541. doi: 10.1101/gad.6.12b.2536. [DOI] [PubMed] [Google Scholar]
- Chambers I, Colby D, Robertson M, Nichols J, Lee S, Tweedie S, Smith A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell. 2003;113:643–655. doi: 10.1016/s0092-8674(03)00392-1. [DOI] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Clarke HJ, Oblin C, Bustin M. Developmental regulation of chromatin composition during mouse embryogenesis: somatic histone H1 is first detectable at the 4-cell stage. Development. 1992;115:791–799. doi: 10.1242/dev.115.3.791. [DOI] [PubMed] [Google Scholar]
- Cohen J, Wells D, Munne S. Removal of 2 cells from cleavage stage embryos is likely to reduce the efficacy of chromosomal tests that are used to enhance implantation rates. Fertil Steril. 2007;87:496–503. doi: 10.1016/j.fertnstert.2006.07.1516. [DOI] [PubMed] [Google Scholar]
- Dean W, Santos F, Stojkovic M, Zakhartchenko V, Walter J, Wolf E, Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc Natl Acad Sci USA. 2001;98:13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RG, Beard HK. Oocyte polarity and cell determination in early mammalian embryos. Mol Hum Reprod. 1997;3:863–905. doi: 10.1093/molehr/3.10.863. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development. 2003;130:5113–5122. doi: 10.1242/dev.00725. [DOI] [PubMed] [Google Scholar]
- Gardner RL. Experimental analysis of second cleavage in the mouse. Hum Reprod. 2002;17:3178–3189. doi: 10.1093/humrep/17.12.3178. [DOI] [PubMed] [Google Scholar]
- Gardner RL. The axis of polarity of the mouse blastocyst is specified before blastulation and independently of the zona pellucida. Hum Reprod. 2007;22:798–806. doi: 10.1093/humrep/del424. [DOI] [PubMed] [Google Scholar]
- Goldman B. The first cut. Nature. 2007;445:479–480. doi: 10.1038/445479a. [DOI] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol. 2007;176:283–294. doi: 10.1083/jcb.200604141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6:117–131. doi: 10.1016/s1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- Heard E, Disteche CM. Dosage compensation in mammals: fine-tuning the expression of the X chromosome. Genes Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hiiragi T, Alarcon VB, Fujimori T, Louvet-Vallee S, Maleszewski M, Marikawa Y, Maro B, Solter D. Where do we stand now? Mouse early embryo patterning meeting in Freiburg, Germany 2005. Int J Dev Biol. 2006;50:581–586. doi: 10.1387/ijdb.062181th. discussion 586-7. [DOI] [PubMed] [Google Scholar]
- Howlett SK, Reik W. Methylation levels of maternal and paternal genomes during preimplantation development. Development. 1991;113:119–127. doi: 10.1242/dev.113.1.119. [DOI] [PubMed] [Google Scholar]
- Johnson MH, McConnell JM. Lineage allocation and cell polarity during mouse embryogenesis. Semin Cell Dev Biol. 2004;15:583–597. doi: 10.1016/j.semcdb.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Johnson MH, Ziomek CA. The foundation of two distinct cell lineages within the mouse morula. Cell. 1981;24:71–80. doi: 10.1016/0092-8674(81)90502-x. [DOI] [PubMed] [Google Scholar]
- Kelly SJ. Studies of the developmental potential of 4- and 8-cell stage mouse blastomeres. J Exp Zool. 1977;200:365–376. doi: 10.1002/jez.1402000307. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kurotaki Y, Hatta K, Nakao K, Nabeshima Y, Fujimori T. Blastocyst axis is specified independently of early cell lineage but aligns with the ZP shape. Science. 2007;316:719–723. doi: 10.1126/science.1138591. [DOI] [PubMed] [Google Scholar]
- Li T, Vu TH, Ulaner GA, Littman E, Ling JQ, Chen HL, Hu JF, Behr B, Giudice L, Hoffman AR. IVF results in de novo DNA methylation and histone methylation at an Igf2-H19 imprinting epigenetic switch. Mol Hum Reprod. 2005;11:631–640. doi: 10.1093/molehr/gah230. [DOI] [PubMed] [Google Scholar]
- Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Mak W, Nesterova TB, de Napoles M, Appanah R, Yamanaka S, Otte AP, Brockdorff N. Reactivation of the paternal X chromosome in early mouse embryos. Science. 2004;303:666–669. doi: 10.1126/science.1092674. [DOI] [PubMed] [Google Scholar]
- Mayer W, Smith A, Fundele R, Haaf T. Spatial separation of parental genomes in preimplantation mouse embryos. J Cell Biol. 2000;148:629–634. doi: 10.1083/jcb.148.4.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui K, Tokuzawa Y, Itoh H, Segawa K, Murakami M, Takahashi K, Maruyama M, Maeda M, Yamanaka S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- Moore NW, Adams CE, Rowson LE. Developmental potential of single blastomeres of the rabbit egg. J Reprod Fertil. 1968;17:527–531. doi: 10.1530/jrf.0.0170527. [DOI] [PubMed] [Google Scholar]
- Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum Mol Genet. 2005;14 Spec No 1:R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Rice JC, Strahl BD, Allis CD, Grewal SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–113. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998;95:379–391. doi: 10.1016/s0092-8674(00)81769-9. [DOI] [PubMed] [Google Scholar]
- Okamoto I, Otte AP, Allis CD, Reinberg D, Heard E. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science. 2004;303:644–649. doi: 10.1126/science.1092727. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmieri SL, Peter W, Hess H, Scholer HR. Oct-4 transcription factor is differentially expressed in the mouse embryo during establishment of the first two extraembryonic cell lineages involved in implantation. Dev Biol. 1994;166:259–267. doi: 10.1006/dbio.1994.1312. [DOI] [PubMed] [Google Scholar]
- Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–490. doi: 10.1242/dev.01602. [DOI] [PubMed] [Google Scholar]
- Piotrowska-Nitsche K, Zernicka-Goetz M. Spatial arrangement of individual 4-cell stage blastomeres and the order in which they are generated correlate with blastocyst pattern in the mouse embryo. Mech Dev. 2005;122:487–500. doi: 10.1016/j.mod.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Rossant J. Postimplantation development of blastomeres isolated from 4- and 8-cell mouse eggs. J Embryol Exp Morphol. 1976;36:283–290. [PubMed] [Google Scholar]
- Rossant J, Lis WT. Potential of isolated mouse inner cell masses to form trophectoderm derivatives in vivo. Dev Biol. 1979;70:255–261. doi: 10.1016/0012-1606(79)90022-8. [DOI] [PubMed] [Google Scholar]
- Rougier N, Bourc’his D, Gomes DM, Niveleau A, Plachot M, Paldi A, Viegas-Pequignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12:2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- Santos F, Peters AH, Otte AP, Reik W, Dean W. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev Biol. 2005;280:225–236. doi: 10.1016/j.ydbio.2005.01.025. [DOI] [PubMed] [Google Scholar]
- Schultz GA, Heyner S. Gene expression in pre-implantation mammalian embryos. Mutat Res. 1992;296:17–31. doi: 10.1016/0165-1110(92)90029-9. [DOI] [PubMed] [Google Scholar]
- Schultz RM. The molecular foundations of the maternal to zygotic transition in the preimplantation embryo. Hum Reprod Update. 2002;8:323–331. doi: 10.1093/humupd/8.4.323. [DOI] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 2005;132:2093–2102. doi: 10.1242/dev.01801. [DOI] [PubMed] [Google Scholar]
- Surani MA, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128:747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Hennebold JD, Macfarlane J, Adashi EY. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog. Development. 2001;128:655–664. doi: 10.1242/dev.128.5.655. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK. Experiments on the development of isolated blastomers of mouse eggs. Nature. 1959;184:1286–1287. doi: 10.1038/1841286a0. [DOI] [PubMed] [Google Scholar]
- Tarkowski AK, Wroblewska J. Development of blastomeres of mouse eggs isolated at the 4- and 8-cell stage. J Embryol Exp Morphol. 1967;18:155–180. [PubMed] [Google Scholar]
- Tarkowski AK, Ozdzenski W, Czolowska R. Identical triplets and twins developed from isolated blastomeres of 8- and 16-cell mouse embryos supported with tetraploid blastomeres. Int J Dev Biol. 2005;49:825–832. doi: 10.1387/ijdb.052018at. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Bannister AJ, Hurd PJ, Kouzarides T, Zernicka-Goetz M. Dynamic distribution of the replacement histone variant H3.3 in the mouse oocyte and preimplantation embryos. Int J Dev Biol. 2006;50:455–461. doi: 10.1387/ijdb.052073mt. [DOI] [PubMed] [Google Scholar]
- Torres-Padilla ME, Parfitt DE, Kouzarides T, Zernicka-Goetz M. Histone arginine methylation regulates pluripotency in the early mouse embryo. Nature. 2007;445:214–218. doi: 10.1038/nature05458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay KD, Duran KL, Bartolomei MS. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol Cell Biol. 1997;17:4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Dieker JW, Derijck AA, Muller S, Berden JH, Braat DD, van der Vlag J, de Boer P. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech Dev. 2005;122:1008–1022. doi: 10.1016/j.mod.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Wang WH, Kaskar K, Gill J, Desplinter T. A simplified technique for embryo biopsy for preimplantation genetic diagnosis. Fertil Steril. 2007;17 doi: 10.1016/j.fertnstert.2007.06.093. [Epub ahead of print] PMID: 17880956. [DOI] [PubMed] [Google Scholar]
- Willadsen SM. The development capacity of blastomeres from 4- and 8-cell sheep embryos. J Embryol Exp Morphol. 1981;65:165–172. [PubMed] [Google Scholar]
- Willadsen SM, Polge C. Attempts to produce monozygotic quadruplets in cattle by blastomere separation. Vet Rec. 1981;108:211–213. doi: 10.1136/vr.108.10.211. [DOI] [PubMed] [Google Scholar]
- Zernicka-Goetz M. The first cell-fate decisions in the mouse embryo: destiny is a matter of both chance and choice. Curr Opin Genet Dev. 2006;16:406–412. doi: 10.1016/j.gde.2006.06.011. [DOI] [PubMed] [Google Scholar]
- Ziomek CA, Johnson MH. The roles of phenotype and position in guiding the fate of 16-cell mouse blastomeres. Dev Biol. 1982;91:440–447. doi: 10.1016/0012-1606(82)90050-1. [DOI] [PubMed] [Google Scholar]