Abstract
Motivation: Order and Disorder prediction using Conditional Random Fields (OnD-CRF) is a new method for accurately predicting the transition between structured and mobile or disordered regions in proteins. OnD-CRF applies CRFs relying on features which are generated from the amino acids sequence and from secondary structure prediction. Benchmarking results based on CASP7 targets, and evaluation with respect to several CASP criteria, rank the OnD-CRF model highest among the fully automatic server group.
Availability: http://babel.ucmp.umu.se/ond-crf/
Contact: Uwe.Sauer@ucmp.umu.se
1 INTRODUCTION
Many proteins carry out important biological functions by means of intrinsically unstructured sequence intervals (Dunker et al., 2002; Romero et al., 1999). Identification of disordered regions in protein sequences can help to reduce bias in sequence similarity analyses and delineate boundaries of protein domains to guide structural and functional studies (Ferron et al., 2006).
Several state-of-the-art approaches have been proposed for prediction of ordered and disordered residues, such as neural networks (NNs) and support vector machines (SVMs). Similar to NNs and SVMs, the conditional random fields (CRFs) (Lafferty et al., 2001) are discriminative supervised machine learning methods. CRFs need training with labeled empirical data in order to learn the classification. However, compared to NNs and SVMs, CRFs are able to take into account inter-relation information between two labels of neighboring residues.
Here, we describe how to apply CRFs to build an order and disorder prediction model. We then compare the OnD-CRF method to prediction methods that successfully participated in CASP7 (Bordoli et al., 2007).
2 METHODS
The OnD-CRF training dataset is derived from high-resolution crystal structures. It contains 215 612 residues, of which 13 909 are defined as disordered (Cheng et al., 2005a) since they are part of a crystallized protein but lack a coordinate entry in the PDB file. The training dataset does not contain any of the CASP7 targets.
Performance is assessed with respect to the area under the ROC curve (AUC), the average of sensitivity and specificity (ACC) and a weighted score, Sw, that considers the rates of ordered and disordered residues in the dataset (Jin and Dunbrack, 2005). The AUC is a measure of the overall predictor quality, with a value of 1.0 for a perfect predictor and 0.5 for a random predictor. The weighted score Sw and the ACC, introduced in CASP6 and CASP7, are used to evaluate the overall prediction accuracy based on an imbalanced dataset.
Throughout, we use CRF++ 0.49 (http://crfpp.sourceforge.net/) to generate the OnD-CRF. The template file contains the rules for selecting the features that we use for training the OnD-CRF model. The features are extracted only from the amino acid sequence and, using SSpro (Cheng et al., 2005b), from the predicted secondary structure.
After 10-fold cross validation, we find that a sliding window size of nine amino acids yields the best template file. The set of parameters which give rise to an AUC value of 0.864 for the OnD-CRF build on the training dataset are: 1.018 for the hyper-parameter ‘C’, which trades the balance between overfitting and underfitting and 5 for the parameter ‘f’, which sets the cut-off threshold for the features. For all other parameters we use the default CRF++ 0.49 values. As a result of 10-fold cross validation, we find an optimal P-value cut-off of P < 0.05 for ordered and P ≥ 0.05 for disordered amino acids. Using this cut-off the OnD-CRF model achieves an ACC of 0.790 and a weighted score Sw of 0.580, based on the training dataset.
3 BENCHMARKING RESULTS
For benchmarking, we use all 96 targets available during CASP7 and compare the results obtained with OnD-CRF to the results of the 15 methods that predicted 93 or more targets. Evaluation is done with respect to the AUC, the sensitivity, Ssens, the specificity, Sspec, their product, Sprod, the ACC and Sw. The sensitivity and specificity can be interpreted as the fraction of correctly identified disordered and ordered residues, respectively.
The benchmarking results for all 16 disorder prediction methods, subdivided into the fully automated server group and the human expert group, are listed in Table 1. Within the automatic server group, the OnD-CRF method reaches the best overall performance with highest scores for AUC, Ssens, Sprod, ACC and Sw. The performance of OnD-CRF method is comparable to the best human expert methods such as ISTZORAN and fais. The results show, that OnD-CRF is an accurate and effective method for the fully automated prediction of disorder in proteins.
Table 1.
Comparing OnD-CRF with prediction methods that participated in CASP7
| Method | AUC | Ssens | Sspec | Sprod | ACC | Sw |
|---|---|---|---|---|---|---|
| CASP7 Automatic Server Group | ||||||
| OnD-CRFa | 0.839 | 0.688 | 0.813 | 0.560 | 0.750 | 0.501 |
| DISproa | 0.822 | 0.597 | 0.854 | 0.510 | 0.726 | 0.451 |
| GeneSilicoMetaServerd | 0.804 | 0.527 | 0.912 | 0.481 | 0.720 | 0.440 |
| BIME@NTU_serva | 0.798 | 0.591 | 0.839 | 0.496 | 0.715 | 0.430 |
| DISOPREDa | 0.837 | 0.425 | 0.953 | 0.405 | 0.689 | 0.378 |
| Distilla | 0.724 | 0.558 | 0.788 | 0.440 | 0.673 | 0.346 |
| MBI-NTU-serva | 0.796 | 0.327 | 0.971 | 0.318 | 0.649 | 0.298 |
| DRIPPREDb | 0.758 | 0.383 | 0.908 | 0.348 | 0.646 | 0.291 |
| CASP7 Human Expert Group | ||||||
| ISTZORANb | 0.860 | 0.725 | 0.837 | 0.607 | 0.781 | 0.562 |
| faisa | 0.844 | 0.556 | 0.924 | 0.514 | 0.740 | 0.481 |
| CBRC-DRa | 0.850 | 0.454 | 0.966 | 0.439 | 0.710 | 0.420 |
| BIME@NTUc | 0.804 | 0.536 | 0.883 | 0.473 | 0.710 | 0.419 |
| IUPredb | 0.777 | 0.396 | 0.947 | 0.375 | 0.672 | 0.343 |
| CBRC-DP_DRa | 0.704 | 0.338 | 0.971 | 0.328 | 0.655 | 0.309 |
| Okab | 0.609 | 0.280 | 0.937 | 0.262 | 0.609 | 0.218 |
| Softberrya | 0.704 | 0.201 | 0.971 | 0.195 | 0.586 | 0.172 |
The entries are sorted with respect to the weighted score Sw.
Number of predicted targets: a96; b95; c94; d93; AUC: Area Under ROC Curve (Bordoli et al., 2007); Sens = TP/(TP + FN); Sspec = TN/(TN + FP); Sprod = Ssens × Sspec; ACC = (Ssens + Sspec)/2; Sw = (WdisorderNTP – WorderNFP +WorderNTN – WdisorderNFN)/(WdisorderNdisorder + WorderNorder).
4 OND-CRF PREDICTION EXAMPLE
We demonstrate the power of the OnD-CRF method on a particular example. The structure of the human cancer-related signaling adaptor protein CRK was recently determined by NMR. The protein harbors one SH2 and two SH3 domains, SH2-nSH3-cSH3 (pdb codes 2EYZ, 2EYV, 2EYW, 2EYX) (Kobashigawa et al., 2007).
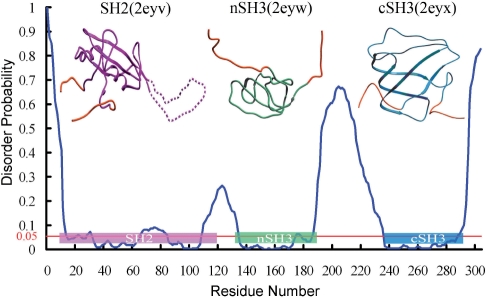
We compare the OnD-CRF prediction, blue curve, with the experimentally determined domains of CRK (Fig. 1) and superimpose their boundaries in the form of colored boxes onto the OnD-CRF curve. The OnD-CRF prediction of the ordered and disordered regions of CRK is in close agreement with the solution NMR structure of this molecule. This is remarkable, since the training dataset includes only high-resolution crystal structures. As shown in Figure 1, the SH2 domain (residues 10–120), the nSH3 domain (134–191) and the cSH3 domain (238–293) are located in the regions of the OnD-CRF plot with highest probability for ordered residues. Interestingly, the amino-acid interval with a high probability for disorder, located roughly in the middle of the CRK-SH2 domain, corresponds precisely to the highly dynamic loop (residues 65–85) connecting the βD and βE strands. The DE-loop can change its conformation to provide a crucial inter-domain contact surface when binding to the Abl SH3 domain (Donaldson et al., 2002). Besides the prediction of disordered sequence intervals, we suggest that the accuracy of the OnD-CRF predictions can be used to determine domain boundaries for 3D structure analysis.
Fig. 1.
OnD-CRF Prediction analysis for CRK. The blue curve represents the predicted disorder probability at each amino acid position. The horizontal red line at 0.05 probability, represents the boundary between order and disorder. The NMR structures of the three CRK domains are shown above the graph. Their boundaries are marked as magenta, green and blue bars, respectively, and overlap with the mostly ordered regions of the OnD-CRF prediction. Note the accurately predicted flexible ‘DE loop’ in the SH2 domain between residues 65–85 (dashed line).
ACKNOWLEDGEMENT
Funding: This work is supported by the Swedish Knowledge Foundation through the Industrial PhD program in Medical Bioinformatics at Karolinska Institute, Strategy and Development Office as well as by BIOVITRUM AB and Umetrics AB.
Conflict of Interest: none declared.
REFERENCES
- Bordoli L, et al. Assessment of disorder predictions in CASP7. Proteins. 2007;69(Suppl. 8):129–136. doi: 10.1002/prot.21671. [DOI] [PubMed] [Google Scholar]
- Cheng J, et al. Accurate prediction of protein disordered regions by mining protein structure data. Data Min. Knowl. Dis. 2005a;11:213–222. [Google Scholar]
- Cheng J, et al. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 2005b;33:W72–W76. doi: 10.1093/nar/gki396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson LW, et al. Structure of a regulatory complex involving the Abl SH3 domain, the Crk SH2 domain, and a Crk-derived phosphopeptide. Proc. Natl Acad. Sci. USA. 2002;99:14053–14058. doi: 10.1073/pnas.212518799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker AK, et al. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- Ferron F, et al. A practical overview of protein disorder prediction methods. Proteins. 2006;65:1–14. doi: 10.1002/prot.21075. [DOI] [PubMed] [Google Scholar]
- Jin Y, Dunbrack RL., Jr Assessment of disorder predictions in CASP6. Proteins. 2005;61(Suppl. 7):167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- Kobashigawa Y, et al. Structural basis for the transforming activity of human cancer-related signaling adaptor protein CRK. Nat. Struct. Mol. Biol. 2007;14:503–510. doi: 10.1038/nsmb1241. [DOI] [PubMed] [Google Scholar]
- Lafferty J, et al. Conditional Random Fields: Probabilistic Models for Segmenting and Labeling Sequence Data. 18th International Conference on Machine Learning (ICML); 2001. pp. 282–289. [Google Scholar]
- Romero P, et al. Folding minimal sequences: the lower bound for sequence complexity of globular proteins. FEBS Lett. 1999;462:363–367. doi: 10.1016/s0014-5793(99)01557-4. [DOI] [PubMed] [Google Scholar]