Abstract
BACKGROUND
The HOXA10 homeobox gene controls embryonic uterine development and adult endometrial receptivity. The three-amino-acid loop extension (TALE) family homeobox genes like myeloid ecotropic viral integration site 1 (MEIS) provide enhanced target gene activation and specificity in HOX-regulated cellular processes by acting as HOX cofactors.
METHODS AND RESULTS
Analysis of an Affymetrix data set in the public domain showed high expression of MEIS1 in human endometrium. MEIS1 expression was confirmed during the human menstrual cycle by RT–PCR and in situ hybridization and was increased during the secretory compared with proliferative phase of the cycle (P = 0.0001), the time of implantation. To assess the importance of maternal Meis1 expression in a mouse model, the uteri of Day 2 pregnant mice were injected with Meis1 over-expression or small interfering RNA (siRNA) constructs. Blocking Meis1 expression by siRNA before implantation significantly reduced average implantation rates (P = 0.00001). Increased or decreased Meis1 expression significantly increased or decreased the expression of integrin β3, a transcriptional target of HOXA10 and an important factor in early embryo-endometrium interactions (P = 0.006). Manipulating Meis1 expression before implantation also dramatically affected the number of pinopodes, uterine endometrial epithelial projections that develop at the time of endometrial receptivity.
CONCLUSIONS
The results suggest that in mouse, meis1 contributes to regulating endometrial development during the menstrual cycle and establishing the conditions necessary for implantation.
Keywords: embryonic implantation, endometrium, HOXA10, integrin β3, MEIS1
Introduction
The molecular mechanisms responsible for endometrial development and receptivity to embryonic implantation are poorly understood (Cross et al., 1994; Wang and Dey, 2006). In the last few years, therefore, a large number of high-throughput expression profiling studies have been performed on both human and animal model tissues to identify the molecular signaling involved in the endometrial development and embryo-endometrium interactions leading to successful embryonic implantation. These include DNA-microarray studies (reviewed in Horcajadas et al., 2007; see also Chen et al., 2006; Pan et al., 2006), as well as SAGE (serial analysis of gene expression) studies (Blomberg et al., 2005; Ma et al., 2006). In addition, two databases have been set up that contain expression data on genes thought to be involved in endometrium development and function: www.endometrialdatabase.com and endometrium.bcm.tmc.edu.
HOX-class homeobox genes are leading candidates for the regulation of endometrium differentiation in preparation for embryonic implantation (Taylor, 2000a; Eun Kwon and Taylor, 2004). HOX genes assign the correct identity to undifferentiated body segments along several developmental axes (Krumlauf, 1994; Carroll, 1995; Moens and Selleri, 2006; Wellik, 2007; Zakany and Duboule, 2007). They are, however, not only typically expressed during embryogenesis but also persistently active in both the mouse and human female reproductive tract (Taylor et al., 1997; Taylor, 2000b). The continued expression of these HOX genes allows the reproductive tract to maintain developmental plasticity and to differentiate appropriately during each menstrual cycle and throughout pregnancy.
The development of the para-mesonephric duct in the embryo is dictated in part by the abdominal-B group of HOX genes (Taylor et al., 1997, 1998; Taylor 2000b; Eun Kwon and Taylor, 2004). Specifically, HOXA10 is responsible for proper uterine development in both mouse and man (Taylor et al., 1997; Block et al., 2000). Female Hoxa10 null-mutant mice demonstrate uterine factor infertility (Satokata et al., 1995). These mice ovulate normally and produce normal embryos, but their uteri will not support the implantation of their own or transplanted wild-type embryos. In humans, HOXA10 expression is up-regulated in the mid-luteal phase at the time of implantation, and its expression is driven by estrogen and progesterone (Taylor et al., 1998). HOXA10 is a well-characterized marker of endometrial receptivity, which exerts pleiotropic effects on multiple aspects of adult endometrial development such as stromal decidualization, leukocyte infiltration and pinopode development (Bagot et al., 2001; Daftary and Taylor, 2004; Qian et al., 2005).
Pinopodes (also called uterodomes) are the best-characterized structural markers of endometrial receptivity in rodents and humans (Nikas, 2000; Nardo et al., 2002; Nikas and Makrigiannakis, 2003). Defective endometrial HOXA10 expression has been described in several conditions that result in diminished human embryo implantation such as endometriosis, polycystic ovarian syndrome, and hydrosalpinx fluid (Taylor et al., 1999; Daftary and Taylor 2002; Cermik et al., 2003). One of the known downstream target genes of Hoxa10 in endometrial cells is Itgb3 (Daftary et al., 2002).
HOX genes perform their function by acting as transcription factors. They bind to the regulatory regions of downstream target genes through their homeobox domain, and thereby activate or repress transcription. HOX proteins bind only weakly to DNA by themselves and/or exhibit a high degree of redundancy in binding site specificity (Pellerin et al., 1994). It has become increasingly evident that HOX proteins usually act as a part of hetero-dimeric (van Dijk et al., 1995; Kurant et al., 1998) or hetero-trimeric complexes (Jacobs et al., 1999; Shen et al., 1999; Schnabel et al., 2000). The TALE (three-amino-acid loop extension) family of homeodomain proteins represents the most important group of HOX cofactors. The unique TALE motif within their homeodomain allows TALE proteins to interact with other homeobox-containing proteins (Bürglin, 1997; Piper et al., 1999). Unlike most other homeobox gene families, TALE genes share sequence homology outside of the homeobox domain. This extensive homology appears linked to their function. Single TALE proteins have only low DNA-binding activity and specificity, but once complexed with a TALE protein from another subfamily and a HOX-class homeobox protein via these homologous domains, they show powerful and specific downstream target promoter regulation (reviewed in Geerts et al., 2003, 2005). Abd-B-like HOX transcription factors, like HOXA10, can form dimers or trimers with TALE proteins that thereby regulate the multiple HOX downstream target genes necessary for the implantation process.
The myeloid ecotropic viral integration site 1 (MEIS) genes belong to the TALE homeobox family. MEIS proteins are not just involved in TALE complex DNA binding and complex stabilization on the target DNA, but can also recruit other TALE partner proteins into the nucleus (Rieckhof et al., 1997). Meis1 was first identified as a major integration site for leukemogenic virus in a murine leukemia model (Moskow et al., 1995). Also in human leukemia, high expression of MEIS1 was found in bone marrow cells of acute myelogenous leukemia (AML) patients, where it was always co-expressed with HOXA9 (Smith et al., 1997; Lawrence et al., 1999). HOXA9 expression was found to cause transformation of normal hematopoietic cells to AML cells only when expressed together with MEIS1 (Kroon et al., 1998). Specifically, MEIS1 expression is vital for keeping HOXA9-immortalized myeloid cells in an undifferentiated, proliferating state, refractory to granulocyte-colony stimulating factor-induced terminal differentiation (Calvo et al., 2001).
Though co-expression and interaction of MEIS1 with Abd-B-like HOX transcription factors was convincingly demonstrated in leukemia, the data on MEIS1 gene expression and function in the endometrium were still scarce. In the current study, we performed data-mining on gene expression sets in the public domain to investigate the expression of the MEIS genes (MEIS1–3) in the human female reproductive tract. We next characterized the menstrual cycle-specific pattern of MEIS1 in human endometrium. To block or over-express maternal Meis1 expression with Meis1 small interfering RNA (siRNA) or over-expression constructs in a mouse model, we used liposome-mediated gene transfection in the murine uterus, to investigate whether maternal Meis1 expression in the endometrium contributes to implantation. We conclude from these experiments that manipulation of adult Meis1 expression significantly affected fertility in the mouse.
Materials and Methods
Meis gene expression in normal tissue
Affymetrix data for a set of 87 different normal human tissue types (the ‘human body index’ set) representing a total of 504 tissue samples were retrieved from public gene expression omnibus (GEO) data sets on the National Center for Biotechnology Information (NCBI) website (Barrett et al., 2005, 2007). CEL data from the Affymetrix GeneChip Human Genome U133 Plus 2.0 array data sets were downloaded, and intensity values and the accompanying P-values assigned with GeneChip® Operating Software (GCOS) using the MASS5.0 algorithm (Affymetrix, Santa Barbara, CA, USA). Annotations and clinical data for the tissue samples analyzed were available from http://www.ncbi.nlm.nih.gov/geo/query/ through its GEO ID: GSE7303.
Human tissue collection
Endometrial tissue was collected during routine endometrial biopsy from women with a normal menstrual cycle who were eligible for IVF and embryo transfer because of (partial) tubal blockage. Informed consent was obtained from all patients. The tissue collection met with the conditions described in the ‘Declaration of Helsinki for Medical Research involving Human Subjects’ as described in (http://www.wma.net/e/plicy/pdf/17c/pdf). Also, approval was obtained from the Tongji Hospital research and ethics committee. One half of the tissue sample was immediately frozen in liquid nitrogen and stored at −72°C. The other half was fixed in formalin, embedded in paraffin, sectioned and stained with hematoxylin and eosin. Menstrual cycle dating was determined by menstrual history and confirmed by histological examination using the Noyes criteria (Noyes et al., 1955). The early and late proliferative phase, and the early, mid and late secretory phase were defined as Days 5–9, 10–14, 15–19, 20–24 and 25–28 of the menstrual cycle, respectively.
RNA isolation and RT–PCR
Total RNA was extracted using the total RNA isolation reagent (TRI reagent, Molecular Research Center, Cincinnati, OH, USA) according to the manufacturer’s instructions. Four microgram total RNA from each sample was denatured at 70°C for 5 min and chilled rapidly on ice. The RNA was then reverse transcribed in a 20 µl reaction mixture containing 4 µl 5 × RT buffer, 2 µl 10 mM dNTP, 1 µl 0.5 µg/µl Oligo(dT15), 0.5 µl 50 U/µl ribonuclease inhibitor, 1 µl 200 U/µl Moloney murine leukemia virus transcriptase (Promega, Madison, WI, USA) with the following reaction condition: 42°C for 60 min; 95°C for 5 min in a Biometra T Gradient Thermocyler (Biometra, Göttingen, Germany). After first strand complementary DNA (cDNA) synthesis, 2 µl of cDNA was amplified in 50 µl of PCR mixture containing 5 µl 10 × PCR buffer, 4 µl 25 mM MgCl2, 1 µl 10 mM dNTP, 1.5 µl (1 U/µl) Taq polymerase (Fermentas, Foster City, CA, USA) and 1 µl (10 pmol/µl) of each primer pair. The primers used were (forward, reverse): for human MEIS1 (5′-TCCATAGCTCTTCACTTC-3′, 5′-GCTTGATGTGACAATTAG-3′), for mouse Meis1 (5′-ACGGCATCCACTCGTTCA-3′, 5′-TGGCTGTCCATCAGGGTT-3′), for mouse integrin β3 (Itgb3) (5′-GCCTTCGTGGACAAGCCTGTA-3′, 5′-GGACAATGCCTGCCAGTCTTC-3′), for human Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (5′-GGTCGGAGTCAACGGATTTGGTCG-3′, 5′-CTTCCGACGCCTGCTTCACCAC-3′) and for mouse β-actin (5′- TTCCAGCCTTCCTTCTTGGG-3′, 5′-TTGCGCTCAGGAGGAGCAAT-3′). The PCR reactions were performed as follows: 94°C for 5 min, followed by a number of cycles of 94°C for 40 s, annealing for 50 s at different annealing temperatures, and 72°C for 1 min, followed by incubation at 72°C for 10 min. Annealing temperatures for human MEIS1, mouse Meis1, mouse Itgb3, mouse β-actin and human GAPDH were 55°C, 50°C, 55°C, 50°C, and 55°C, respectively, and the numbers of cycles were 25, 30, 28, 30 and 25, respectively. The PCR products were separated on 2% agarose gels and visualized by ethidium bromide (EtBr) staining under UV light. The resulting PCR products were 411 bp for human MEIS1 [representing nucleotides (nt) 2261–2671 of RefSeq NM_002398), 405 bp for mouse Meis1 (nt 1106–1510 of NM_010789), 337 bp for mouse Itgb3 (nt 571–907 of NM_016780), 786 bp for human GAPDH (nt 117–902 of NM_002046) and 224 bp for mouse β-actin (nt 850–1073 of NM_007393). Mouse Meis1 primers used in the experiments described yielded a 405 bp PCR product containing endogenous mouse Meis1 as well as human MEIS1 (nt 1115–1519 of NM_002398), resulting from the pcDNA4-encoded transgene. Human MEIS1 or mouse Meis1 and Itgb3 mRNA levels were calculated as a ratio of the densitometric values for MEIS1, Meis1, or Itgb3 to their corresponding GAPDH or β-actin values. Care was taken to ensure a logarithmic signal for the PCR products measured. RT–PCR reactions performed for human MEIS1/GAPDH showed differential MEIS1 and equal GAPDH expression for all samples when RT–PCR was performed for 28, 25 or 22 cycles. For all three cycle numbers used, MEIS1 and GAPDH signal intensities were different between cycle numbers. Similar results were obtained for mouse Meis1/β-actin (35, 30 or 25 cycles) and mouse itgb3/β-actin (30, 28 or 25 cycles) (results not shown).
In situ hybridization
Diethyl pyrocarbonate-treated Milli-Q water was used in each step. Sections (4 µm thick) were immersed in 30% H2O2 solution for 30 min to eliminate endogenous peroxidase activity. After rinsing in 100 mM phosphate-buffered saline (PBS), sections were treated with pepsin [0.02% (w/v) in 3% citric acid] at 37°C for 10 min. Thereafter, sections were fixed in freshly prepared 4% (w/v) paraformaldehyde in PBS for 20 min at 4°C, rinsed in PBS and immersed in 0.25% acetic anhydride, 100 mM triethanolamine and 0.9% (w/v) NaCl consecutively, each for 10 min at room temperature. The probe for human MEIS1 was a 1:1 molar mixture of single stranded DNA oligonucleotides complementary to the 5′-CACTCGCATCAGTACCCGCACACAGCTCATACCAA-3′ (nt 593–627 of NM_002398) and 5′-ACACCTTATAATCCTGATGGACAGCCCATGGGAGG-3′ (nt 1493–1527 of NM_002398) MEIS1 sequences, labeled with digoxigenin. Sections were hybridized overnight at 40°C in hybridization buffer containing digoxigenin-labeled oligonucleotide probe at a final concentration of 400 mg/l. After hybridization, sections were washed in fresh 2 × SSC (saline sodium citrate) at 37°C for 10 min, 0.5 × SSC at 37°C for 15 min, and 0.2 × SSC at 37°C for 15 min, then incubated in 5% (w/v) bovine serum albumin for 30 min at room temperature, and in biotin-anti-digoxigenin at 37°C for 1 h, and finally washed and incubated in streptavidin–biotin complex (SABC) for 20 min at 37°C. Probe labeling, and digoxigenin, biotin-anti-digoxigenin and SABC incubations were performed using the MK2502 kit (Boster Bio-technology, Wuhan, Hubei, China). The brown precipitate generated at the site of digoxigenin-probe hybridization by reaction of the anti-digoxigenin antibody bound peroxidase with diaminobenzidine tetrahydrochloride (DAB) (AR1022, Boster Bio-technology) was scored as presence of MEIS1 mRNA. Hematoxylin was used for counterstaining. A total of 33 samples were examined. The staining of 100 consecutive cells in five non-adjacent microscopic fields was evaluated to quantify the expression of MEIS1.
Western blot analysis
Tissues from each mouse uterus were washed twice with PBS and lysed on ice in lysis buffer [50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 0.1% (w/v) sodium dodecyl sulfate (SDS), 0.5% (w/v) sodium deoxycholate, 1% (v/v) Nonidet-P40, 0.02% (w/v) sodium azide and freshly added protease inhibitors (phenylmethylsulphonyl fluoride to 10 µg/μl and aprotinin to 1 µg/ml)]. Solid cellular debris was removed by centrifugation at 1200g for 5 min. Protein concentration was measured by the Coomassie Brilliant Blue G-250 assay. Protein samples (45 µg each) were subjected to 10% SDS–polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane using a Bio-Rad electroblot apparatus (Bio-Rad, Beijing, China). Non-specific binding sites were blocked in 5% (w/v) non-fat dry milk in PBS containing 0.05% (v/v) Tween-20. The primary goat anti-MEIS1 polyclonal antibody (sc-10599, Santa Cruz, CA, USA) was used at 1:100 and incubated at 4°C overnight. This antibody was raised against a C-terminal epitope of human MEIS1, and recognizes both human MEIS1 and murine Meis1, since their protein sequences are identical in that region (also see below, section MEIS1 constructs). As secondary antibody rabbit peroxidase-conjugated anti-goat immunoglobulin (Ig)G (ZDR-5308, Zhongshan Biotechnology, Beijing, China) was used at 1:200, and incubated at 37°C for 1.5 h. Rabbit anti-actin antibody (sc-101616-R, Zhongshan Biotechnology, as a loading control) was used at 1:500 and incubated at 4°C overnight, and the secondary antibody goat anti-rabbit IgG (ZDR-5306, Zhongshan Biotechnology) was used at 1:500, and incubated at 37°C for 1.5 h. Protein bands were visualized by enhanced chemiluminescence (32109, Pierce Biotechnology, Rockford, IL, USA).
MEIS1 constructs
The complete human MEIS1 coding sequence was isolated by PCR using neuroblastoma tumor cDNA as a template with forward primer 5′-ccggaattccggATGGCGCAAAGGTACGAC-3′ and reverse primer 5′-ccgcgtcgacggCTACTGAGCATGAATGTCCAT-3′. The forward primer consisted of a non-coding flag containing an EcoRI recognition site (underlined) and the ATG initiation codon (in bold capitals) followed by codons 2–6 (in capitals), representing nt 458–475 of the human MEIS1 NCBI RefSeq NM_002398 coding sequence. The reverse primer consisted of a non-coding flag containing a SalI recognition site (underlined) and the TAG stopcodon (in bold capitals) preceded by the final codons 385–390 (in capitals), representing nt 1630–1610 of the NM_002398 coding sequence. This sequence encoded MEIS1B, the most widely expressed MEIS1 splice variant (see also Geerts et al., 2005 and the MEIS1 locus in TranscriptView at http://bioinfo.amc.uva.nl/human-genetics/transcriptviewb/). The derived human MEIS1B protein (NCBI sequence NP_002389, 390 residues) was identical to mouse Meis1B protein (NCBI sequence Q60 954), except for substitution of the Ile residue at position 201 in the human sequence by an analogous Val residue at position 201 in the mouse sequence. The 1196 bp MEIS1 PCR product was restricted with EcoRI and SalI, purified by agarose gel electrophoresis using Qiaex II gel extraction (20021, Qiagen, Venlo, The Netherlands) and ligated into EcoRI–SalI restricted pCI-neo vector (E1841, Promega Benelux, Leiden, The Netherlands) backbone. The MEIS1 sequence was excised from this construct using EcoRI and the 3′ polylinker NotI site and subcloned into the pcDNA4/TO/myc-HisA mammalian expression vector (V1030-20, Invitrogen, Breda, The Netherlands) restricted with EcoRI and NotI, resulting in pcDNA4/MEIS1. The construct was verified by sequencing, and expression of MEIS1 protein by the construct was demonstrated by western blot analysis of transiently transfected HEK293T cells and stably transfected human neuroblastoma cells (Revet et al., manuscript in preparation, see also Fig. 2 in Geerts et al., 2005).
The Meis1 siRNA expression constructs were prepared as follows: oligos T14F 5′-gatcccTCCAGAACTGGATAACTTGttcaagagaCAAGTTATCCAGTTCTGGAtttttggaaa-3′ and T14R 5′-agcttttccaaaaaTCCAGAACTGGATAACTTGtctcttgaaCAAGTTATCCAGTTCTGGAgg-3′, targeting the TCAAGAACTGGATAACTTG sequence in MEIS1 mRNA (nt 871–889 in the human MEIS1 RefSeq NM_002398, and 862–880 in the murine Meis1 RefSeq NM_010789) or 16F 5′-gatcccCTTGATGATTCAAGCCATAttcaagagaTATGGCTTGAATCATCAAGtttttggaaa-3′ and 16R 5′-agcttttccaaaaaCTTGATGATTCAAGCCATAtctcttgaaTATGGCTTGAATCATCAAGgg-3′, targeting the CTTGATGATTCAAGCCATA sequence in MEIS1 mRNA (nt 886–904 in NM_002398, and 877–895 in NM_010789) were annealed and ligated into pTER restricted with BglII and HindIII as described (van de Wetering et al., 2003), to create pTER/Meis1-T14 and pTER/Meis1-T16, respectively. A non-targeting pTER construct contained a similar sequence that had no homology to the human or murine genome, and was used as a negative control. The constructs were verified by sequencing and siRNA-mediated MEIS1 protein and mRNA knockdown by both constructs was demonstrated (Revet et al., manuscript in preparation, see also Fig. 2 in Geerts et al., 2005). The T14 and T16 constructs have complete homology to human MEIS1 as well as to murine Meis1, and target all annotated MEIS1 splice variants (i.e. isoforms A-D, see also Geerts et al., 2005).
DNA/liposome preparation
Plasmid DNA (1–2 µl) was mixed with 50 µl Opti-MEM reduced serum medium (31985, Invitrogen, Carlsbad, CA, USA), added to 4 µl liposome [a 3:1 (w/w) formulation of 2,3-dioleyloxy-N-[2(sperminecarboxamido)ethyl]-N-N-dimethyl-1- propanaminium trifluoroacetate (DOPSA) and dioleoylphosphatidyl ethanolamine (DOPE) (11668-027, Invitrogen)] in 50 µl OptiMEM and incubated for 20 min at room temperature. Final concentrations were 16 µg/ml DNA and 40 µg/ml liposome.
In vivo gene transfection
Nulliparous female and male Kunming mice of reproductive age were supplied by Center of Experimental Animals, Tongji Medical College (Wuhan, China). Female mice were mated and examined every 8 h until detection of a vaginal plug, which was designated Day 1 of the pregnancy. The mice were anesthetized 30–60 h following plug detection with 1% butaylone given by i.p. injection. A laparotomy was performed to expose the uterus and 25 µl of the DNA/liposome complex was injected into the base of each uterine horn using a 27-gage needle, as described (Bagot et al., 2000; Hsieh et al., 2002). The incision was closed in two layers (peritoneal and cutaneous) with 4–0 vicryl suture. The mice were euthanized by cervical vertebrae dislocation under anesthesia before excision of the uteri. For the experiments studying the effect of MEIS1 expression manipulation on MEIS1 expression, litter size and pinopode formation, the group sizes were five mice each transfected with pcDNA4/MEIS1, empty pcDNA4, pTER/Meis1 siRNA construct or non-targeting pTER, respectively (20 mice in total). For the experiments studying the effect of MEIS1 expression manipulation on Itgb3 expression, 25 mice each were transfected with pcDNA4/MEIS1, empty pcDNA4, pTER/Meis1 siRNA construct or non-targeting pTER, respectively (100 mice in total). The numbers of mice available for Meis1 expression analysis in Fig. 5 were 19, 17, 19 and 19, respectively (the remainder were not pregnant in spite of the detection of the vaginal plug or died of post-surgical complications). The presented work conformed to the ‘Guiding principles in the Care and Use of Animals’ as described in DHEW publication No. (NIH) 85-23, Revised 1985, Office of Science and Health Reports, DRR/NIH, Bethesda, MD 20982, USA, was approved by the ethical committee of Tongji Medical College and was also covered by Chinese animal husbandry legislation.
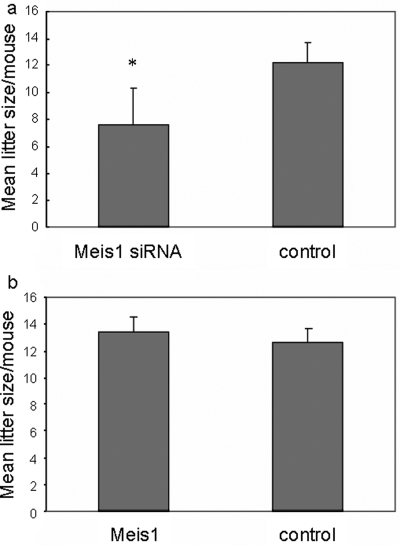
Figure 5:
Effect of Meis1 expression on mouse litter size.
Litter size, as determined by counting the number of implantation sites per uterus examined, at Day 9 of pregnancy. Mice were transfected 1 day after detection of a vaginal plug with pTER/Meis1, control pTER, pcDNA4/MEIS1 or empty pcDNA4. (a) Litter size was reduced by 40% in the group of mice transfected with pTER/Meis1 siRNA compared with pTER controls. This difference was statistically significant according to a two-tailed unpaired t-test (P = 0.00001). (b) Litter size at Day 9 of gestation was not affected by transfection with pcDNA4/MEIS1 compared with pcDNA4-transfected controls (P = 0.08). The experiment was repeated three times, with similar results
Scanning electron microscropy
Uteri were dissected from mice 90 h after vaginal plug detection. The specimens were dehydrated in an acetone series, dried in a critical point drier using carbon dioxide, mounted on the specimen holder, coated with gold and examined under a Stereoscan 520 Scanning Electron Microscope (Hitachi, Tokyo, Japan). From each biopsy, six to eight tissues fragments, measuring ∼2 mm in thickness and 10 mm in length, were evaluated. A minimum of an aggregated 4–5 mm2 of well-preserved epithelial surface was analyzed, to increase the likelihood that the observations were representative by negating the influence of local differences in endometrial morphology. The evaluations were performed with coded tissue and read blindly by an experienced investigator according to Nikas and Makrigiannakis (2003).
Immunohistochemistry
The expression of mouse Itgb3 protein in the uterus was evaluated by immunohistochemistry using a goat polyclonal antibody to Itgb3 (sc-6626, Santa Cruz). The full-thickness biopsies were obtained at the time of histological evaluation from each uterine horn. The specimens were embedded in paraffin and serial 5 µm sections obtained. The sections were deparaffinized in xylene and ethanol. Endogenous peroxidase was blocked with 3% H2O2 at room temperature for 10 min. After blocking for 45 min with 1.5% normal horse serum in PBS, the sections were incubated overnight at 4°C with primary Itgb3 antibody (1:200) in PBS. The sections were then incubated with biotinylated rabbit anti-goat IgG (ZDR-5308, Zhongshan Biotechnology) as a secondary antibody at 1:50 and incubated at 37°C for 30 min, subsequently with avidin and biotinylated peroxidase at room temperature for 45 min, and finally with DAB (400 mg/ml) at room temperature for 5 min. Hematoxylin was used for counterstaining. The staining of 100 consecutive cells in five non-adjacent microscopic fields was evaluated. Immunoreactivity of Itgb3 was scored as follows: 1+, <25% of cells stained; 2+, >25% but <50% of cells stained; 3+, >50% but <75% of cells stained; and 4+, >75% of cells stained. Overall intensity was scored as weak (1+), moderate (2+) or strong (3+). The sum of cell stained scores and intensity scores was calculated to quantify the expression of Itgb3.
Statistical analysis
All numerical results are expressed as the mean value ± SD, except for the Affymetrix expression values of MEIS1–3 mRNAs in normal human tissue, which are represented as mean value ± SEM. The data of MEIS1 mRNA in endometrium during the menstrual cycle were analyzed by analysis of variance. Comparisons between the expression of Meis1 in each group of murine tissue, Itgb3 in each group of murine tissue and litter size of each group of mice were performed using two-tailed unpaired t-tests. The differences of pinopode development between each group of murine tissue were analyzed using a chi-square test. P < 0.05 was considered significant in all tests. The Statistical Package for the Social Sciences software package for Windows (Version 13.0) was used for all statistical analyses.
Results
Expression of MEIS1, MEIS2 and MEIS3 genes in the normal human uterus
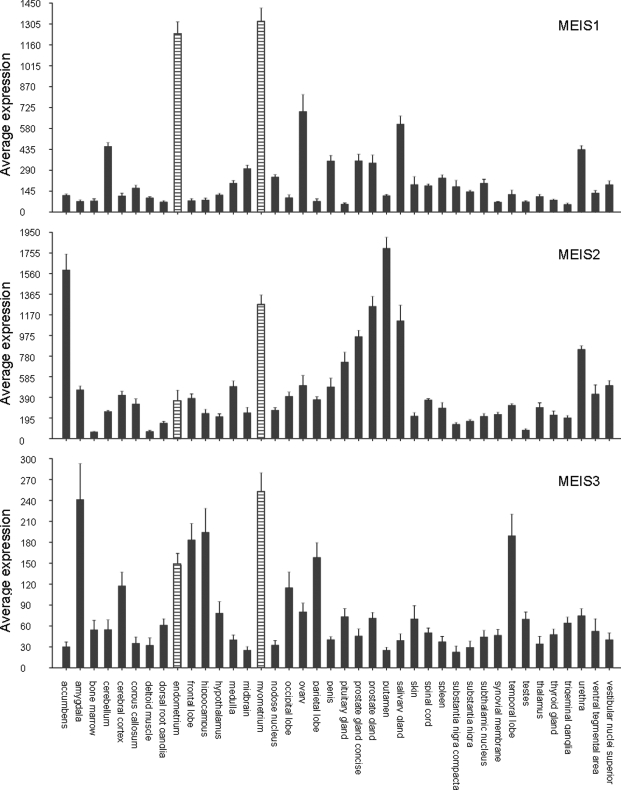
Affymetrix data for a set of 87 different normal human tissue types (the ‘human body index’ set) representing a total of 504 tissue samples were retrieved from the NCBI website. As shown in Fig. 1, MEIS1 was highly expressed in both uterine tissues present in the data set: myometrium and endometrium. In fact, the MEIS1 expression in these two tissues was the highest observed in all 87 tissue types investigated. In contrast, MEIS2 expression was high in myometrium, although several other tissues show even higher expression, but only moderate in endometrium. Finally, the MEIS3 gene showed overall much lower expression (5–7-fold) than the MEIS1 and MEIS2 genes. The high expression of MEIS1 compared with MEIS2 and MEIS3 in endometrium prompted us to investigate the expression and function of the MEIS1 gene in the reproductive cycle.
Figure 1:
Expression of MEIS1-3 mRNAs in normal human tissue.
Visual representation of MEIS1, MEIS2 and MEIS3 expression in the ‘human body index’ Affymetrix data set in the public domain: 504 samples representing 87 different normal human tissues. For reasons of presentation, only tissues consisting of five or more different samples were shown: 40 tissues representing 329 samples. MEIS1 expression in endometrium and myometrium was higher than in any of the non-shown sets. Average MEIS expression in all 87 tissues was: MEIS1 338.3, MEIS2 516.6 and MEIS3 77.1. For comparison: average expression of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and β-actin in this data set was 9240 and 11 890, respectively. Average MEIS gene expression in endometrium (18 samples) was: MEIS1 1241 ± 83, MEIS2 358 ± 99 and MEIS3 149 ± 16. Average MEIS expression in myometrium (27 samples) was: MEIS1 1330 ± 90, MEIS2 1273 ± 84 and MEIS3 254 ± 26. The average expression over the different samples per tissue is shown. The error bars represent the SEM
Expression of MEIS1 mRNA during the human reproductive cycle
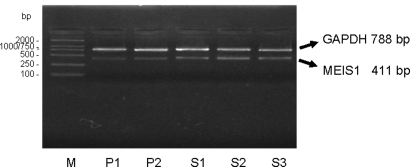
MEIS1 expression has been previously demonstrated in epithelial cells of the developing female reproductive tract (Dintilhac et al., 2005). To investigate the expression of MEIS1 in cyclic development of the endometrium, the menstrual cycle stage-specific expression was determined by RT–PCR. Representative EtBr stained agarose gel pictures of RT–PCR are shown in Fig. 2. MEIS1 mRNA expression was evident throughout the menstrual cycle.
Figure 2:
MEIS1 mRNA expression in human endometrium during the menstrual cycle.
RT–PCR analysis of MEIS1 expression. A representative agarose gel stained with ethidium bromide (EtBr) picture is shown. MEIS1 was expressed in all stages of the human menstrual cycle investigated; the first half of the proliferative phase (P1), the second half of the proliferative phase (P2) and the first third of the secretory phase (S1), the mid-secretory phase (S2) and the late secretory phase (S3). The sample numbers for the P1, P2, S1, S2 and S3 groups were 5, 7, 7, 8 and 6, respectively. M, size markers. No PCR product was observed in control samples that did not contain RT enzyme during RT synthesis or template DNA during PCR (not shown). RT–PCR analysis was repeated at least three times per sample. MEIS1 expression was analyzed by densitometry and normalized to GAPDH in 33 endometrial samples. The densitometric analysis was repeated three times, with similar results. The data were analyzed using analysis of variance. The average MEIS1/GAPDH ratio in each group was: P1 0.20 ± 0.02, P2 0.26 ± 0.03, S1 0.41 ± 0.07, S2 0.70 ± 0.10 and S3 0.44 ± 0.09. There was no statistical difference within the proliferative phase (P = 0.47), but the difference between the proliferative phase and the secretory phase was significant (P = 0.00001)
To assess potential regulation of MEIS1 expression during the reproductive cycle, densitometric analysis was performed on all samples, and the average abundance of MEIS1 during each stage of the menstrual cycle normalized to GAPDH, calculated. No difference was apparent between the levels of expression in the segments of the proliferative phase. In contrast, expression at the mid secretory phase was elevated relative to other stages (see legend to Fig. 2). These results were a first indication that MEIS1 expression varied during the menstrual cycle, markedly increasing at the mid secretory phase (S2), which corresponded to the time of implantation.
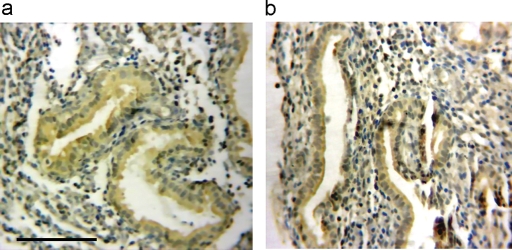
MEIS1 mRNA is expressed in the human endometrial stroma and glands
To investigate the site of MEIS1 expression in the human endometrium, and as a support of the MEIS1 expression detected with RT–PCR described above, in situ hybridization was performed. Fig. 3a and b shows high-power views of MEIS1 expression in proliferative and secretory endometrium. MEIS1 was expressed in both glandular and stromal cells. This supports the results of RT–PCR in Fig. 2, and further suggests that MEIS1 may play a role in embryonic implantation.
Figure 3:
MEIS1 mRNA expression in human endometrial tissue.
In situ hybridization analysis of MEIS1 expression in human endometrial samples. Views of two representative samples demonstrate MEIS1 expression in both the glandular and stromal cells of (a) proliferative phase and (b) secretory phase endometrium. MEIS1 mRNA is stained brown. No staining was observed when the MEIS1 probe was omitted (not shown). Scale bar = 100 µm. Each sample was analyzed at least in triplicate
Down-regulation of Meis1 expression affects murine embryo implantation rate
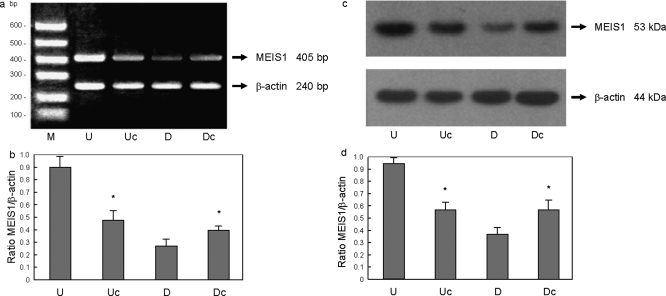
To assess whether an up-regulation of Meis1 expression during embryo implantation was essential to this process, the expression of maternal Meis1 was down-regulated in a mouse model by transfection in vivo, as described by (Bagot et al., 2000; Hsieh et al., 2002). A first indication of decreased Meis1 expression in the uterus was obtained by RT–PCR, and this was confirmed by western blot on Day 3 after transfection. Representative results for mRNA are shown in Fig. 4a and b. Densitometric analysis was performed on all samples, and the average abundance of Meis1 mRNA in each group normalized to β-actin mRNA was calculated. Levels of Meis1 mRNA and protein (Fig. 4b and c) were significantly decreased in endometrium transfected with pTER/Meis1 siRNA compared with the control.
Figure 4:
Meis1 expression manipulation in murine uterine tissue.
(a) RT–PCR analysis of Meis1 expression in 20 murine endometria transfected with pcDNA4/MEIS1, Meis1 small interfering RNA (siRNA) constructs or respective controls (five animals in each group). Shown is a representative EtBr picture. (b) Meis1 mRNA expression was analyzed by densitometry and normalized to β-actin as described in Fig. 2, except that data were analyzed using a two-tailed unpaired t-test. No PCR product was observed in control samples that did not contain RT enzyme during RT synthesis or template DNA during PCR (not shown). RT–PCR and quantification was repeated at least three times, with similar results. Average Meis1/β-actin ratios in endometrium transfected with pcDNA4/MEIS1 (U) and control pcDNA4 DNA (Uc) were 0.90 ± 0.09 and 0.48 ± 0.07, respectively. There was a significant increase in Meis1 mRNA in the up-regulation group (U) compared with the control (Uc) (P = 0.0001). Average Meis1/β-actin level in endometrium transfected with Meis1 siRNA (D) and control pTER constructs (Dc) were 0.27 ± 0.05 and 0.40 ± 0.03, respectively. Levels of Meis1 mRNA were significantly decreased in Meis1 siRNA-transfected samples (D) compared with the control samples (Dc) (P = 0.002). (c) Western analysis performed on 20 murine endometria transfected with pcDNA4/MEIS1, Meis1 siRNA or respective controls (five animals in each group). A representative western blot image is shown. (d) Meis1 protein expression was analyzed by densitometry and normalized to β-actin values as described in Fig. 4b. No Meis1 band was detected on western blot in the absence of primary Meis1 antiserum (not shown). Western blot analysis and quantification was repeated at least three times, with similar results. The average Meis1/β-actin ratio in each group was: U 0.94 ± 0.05, Uc 0.57 ± 0.06, D 0.37 ± 0.05 and Dc 0.57 ± 0.08. There was a significant increase in Meis1 protein in endometria transfected with pcDNA4/MEIS1 (U) compared with pcDNA4-transfected controls (Uc) (P = 0.00001). Endometria transfected with pTER/Meis1 siRNA constructs (D) demonstrated a significant decrease in Meis1 protein compared with pTER controls (Dc) (P = 0.01)
At Day 9 of pregnancy, mice were killed and the uteri removed and examined for normal histology and for the number of implantation sites per uterus as a measure of litter size. The endometrium appeared histologically normal, and demonstrated adequate decidualization. Also the total number of pregnant mice did not vary significantly between Meis1 siRNA-treated mice and the control. Pups of mice transfected with Meis1 siRNA were born following a normal length gestation of 18–22 days, were of normal size and demonstrated no morphological abnormalities. However, the average number of implantation sites per uterus was 12.2 ± 1.5 in the mice transfected with the control pTER plasmid and 7.6 ± 2.7 in the mice transfected with a pTER/Meis1 siRNA construct (Fig. 5a). Decreased expression of Meis1 therefore resulted in a significant decrease in litter size (see legend to Fig. 5a). We concluded that the naturally occurring up-regulation of Meis1 in the reproductive cycle contributes to normal embryo implantation.
To evaluate the effect of up-regulation of Meis1 expression in the adult murine uterus, mice were transfected with the pcDNA4/MEIS1 expression construct, or with empty pcDNA4 plasmid (as a negative control). A first indication of increased Meis1 expression in the uterus was obtained by RT–PCR and this was confirmed by western blot. Levels of Meis1 mRNA and protein were significantly increased in endometrium transfected with the pcDNA4/MEIS1 expression construct compared with the control (Fig. 4c and d). As in the experiment described above, uterine histology was not affected by the transfection, and the number of mice pregnant and the weight and morphology of the pups did not differ between the groups. The average number of implantation sites was 13.4 ± 1.1 in the pcDNA4/MEIS1 treated mice and 12.6 ± 1.1 in the control mice (Fig. 5b). In contrast to Meis1 down-regulation, up-regulation of Meis1 expression by pcDNA4/MEIS1 treatment did not significantly alter litter size.
Effect of Meis1 on murine pinopode formation
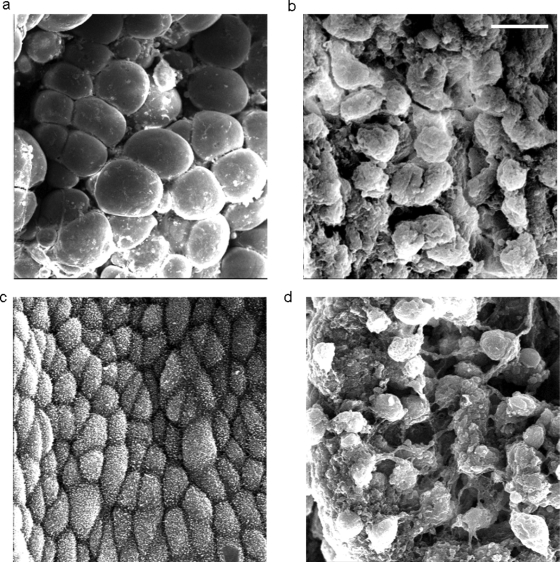
For further clues as to a possible function for Meis1 in murine implantation, uterine epithelial morphology was investigated. As described above, no uterine histological changes could be detected at the light microscopy level in mice transfected with Meis1 DNA constructs. To evaluate the effect of Meis1 on epithelial morphology, the uteri of these mice were examined with scanning electron microscopy for evidence of altered formation of pinopodes.
Mice which were transfected in vivo as described above were killed 4.5 days after vaginal plug detection, the uteri removed and prepared for electron microscopy analysis. ***Fig. 6a and b shows a typical section of endometrium from a pcDNA4/MEIS1-treated mouse, and an empty pcDNA4-treated control mouse; there was a significant increase in the development of pinopodes in the mouse transfected with MEIS1 over-expression plasmid. Fig. 6c and d shows a typical section of endometrium from pTER/Meis1 and control pTER-treated mouse uteri. None of the sections from Meis1 siRNA-treated mice contained normal pinopodes. The secretory cells were covered with dense microvilli, instead of with pinopodes, and were slightly bulging. In contrast, all sections from control animals contained a normal number of pinopodes. Apparently, decreasing Meis1 expression had a negative effect on efficient pinopode maturation. Absence of a normal amount of functioning pinopods in mice transfected with Meis1 siRNA could be a mechanism contributing to a less efficient embryonic implantation rate and diminished litter size in these animals.
Figure 6:
Effect of Meis1 expression on murine pinopode formation.
Pinopode development in transfected mouse uteri, examined with scanning electron microscopy. (a) Transfection with pcDNA4/MEIS1 demonstrated fully developed pinopodes on most (≥60%) secretory cells. (b) Transfection with empty pcDNA4 plasmid as a negative control showed a wide distribution of developing pinopodes, covering ∼50% of the endometrial luminal surface. However, there was a much lower occurrence (20%) of fully developed pinopodes in this group. (c) Transfection with Meis1 siRNA plasmid resulted in the majority of the endometrial surface expressing few, to no, pinopodes. Instead, the epithelial cells were covered with short and thick microvilli. (d) Transfection with non-targeting pTER plasmid as a control resulted in developing pinopodes on about half of the secretory cells. Scale bar = 10 µm. The experiment was repeated three times. The data were analyzed using a chi-square test
Effect of Meis1 on Itgb3 expression in the murine uterus
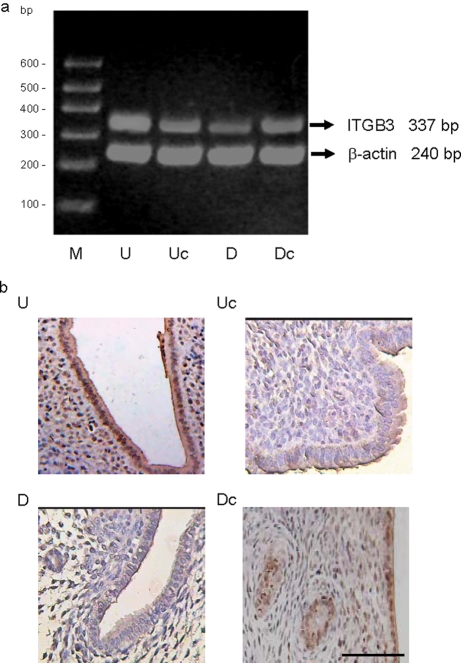
Maternal Hoxa10 has earlier been shown to be involved in pinopode formation in the mouse uterus (Bagot et al., 2001), in a manner resembling the effect of Meis1 on pinopode formation observed in this study. To investigate a possible connection between Meis1 and Hoxa10 function in murine uterine development and embryonic implantation, we evaluated the effect of Meis1 expression manipulation on Itgb3 expression. To obtain a first indication of differential Itgb3 mRNA expression, peri-implantation uteri from mice treated with pcDNA4/MEIS1 or empty pcDNA4 (as a negative control), or with pTER/Meis1 or non-targeting pTER (as a negative control) were analyzed by RT–PCR. Fig. 7a shows a representative EtBr gel picture of the RT–PCR. Densitometric analysis was performed on all samples, and the average abundance of Itgb3 in each group normalized to β-actin was calculated. Levels of Itgb3 mRNA appeared significantly decreased in endometrium transfected with pTER/Meis1 siRNA compared with the control. In contrast, levels of Itgb3 mRNA increased in endometrium transfected with the pcDNA4/MEIS1 expression construct compared with the control (see legend to Fig. 7).
Figure 7:
Effect of Meis1 expression manipulation on murine uterine integrin β3 (Itgb3) expression.
(a) RT–PCR analysis of Itgb3 expression in murine endometria transfected with pcDNA4/MEIS1 (U), empty pcDNA4 (Uc), pTER/Meis1 siRNA (D) or control pTER constructs (Dc). A representative agarose EtBr picture is shown. No PCR product was observed in control samples that did not contain RT enzyme during RT synthesis or template DNA during PCR (not shown). RT–PCR was repeated three times, with similar results. Itgb3 expression was analyzed by densitometry and normalized to β-actin as described in Fig. 4. Average Itgb3/β-actin ratio in each group was: U 0.83 ± 0.17, Uc 0.50 ± 0.07, D 0.28 ± 0.06 and Dc 0.44 ± 0.04. There was a significant increase in Itgb3 mRNA in endometrium transfected with pcDNA4/MEIS1 compared with endometrium transfected with empty pcDNA4 DNA (P = 0.02). Levels of Itgb3 mRNA in endometrium transfected with pTER/Meis1 siRNA were significantly decreased when compared with endometrium transfected with control pTER (P = 0.006). (b) Immunohistochemistry of Itgb3 protein expression in the endometrium of mice transfected with either pcDNA4/MEIS1 or pTER/Meis1 siRNA plasmid. Shown are representative images of each group: uterine sections from mice transfected with pcDNA4/MEIS1 (U), with empty pcDNA4 vector (Uc), with pTER/Meis1 siRNA (D) or with non-targeting pTER vector (Dc). No staining was observed in a negative control without Itgb3 antibody as the primary antiserum (not shown). Scale bar = 100 µm. Statistical analysis of Itgb3 immunoreactivity was performed as described in the ‘Materials and Methods’ section. The data were analyzed using a two-tailed unpaired t-test. The immunoreactivity analysis was repeated three times, with similar results. The average Itgb3 immunoreactivity scores in each group were: U 6.4 ± 0.5, Uc 4.0 ± 0.8, D 2.4 ± 0.9 and Dc 4.8 ± 0.7. Uterine sections from pcDNA4/MEIS1-transfected mice showed significantly increased glandular and stromal Itgb3 expression compared with control mice treated with empty pcDNA4 vector (P = 0.0015). Uterine sections from pTER/Meis1 siRNA-transfected mice showed significantly reduced glandular and stromal Itgb3 expression compared with control mice treated with non-targeting pTER vector (P = 0.03)
To support the results of the RT–PCR described above, the effect of Meis1 expression on Itgb3 protein levels was determined by immunohistochemistry on transfected uterine tissue of each group. Representative pictures are shown in Fig. 7b. Itgb3 expression levels appeared higher in mice transfected with pcDNA4/MEIS1 expression construct compared with the pcDNA4 control. In contrast, Itgb3 expression was lower in mice transfected with Meis1 siRNA expression constructs than in those transfected with the non-targeting siRNA construct. Statistic analysis of the Itgb3 immunoreactivity was performed on the samples, and the average abundance of Itgb3 in each group was calculated. Levels of Itgb3 expression were significantly decreased in endometrium transfected with pTER/Meis1 siRNA compared with the control. In contrast, Itgb3 protein levels were increased in endometrium transfected with the pcDNA4/MEIS1 expression construct compared with the control (see legend to Fig. 7). This was consistent with the results of the RT–PCR.
Together, these results showed that, in a murine uterine receptivity model, the expression of the known Hoxa10 target Itgb3 could be regulated by the manipulation of Meis1 expression, suggesting Meis1 action through co-operation, or possibly interaction, with Hoxa10.
Discussion
HOXA10 is essential for normal embryonic uterine development. In the developing mouse embryo, altered Hoxa10 expression as a result of targeted mutation (Satokata et al., 1995) or exposure to diethylstilbestrol leads to abnormal uterine development (Block et al., 2000). Hoxa10 expression persists in the adult uterus and is important for implantation of the embryo. Hoxa10 null-mutant mice or mice in which Hoxa10 has been blocked using antisense oligonucleotides demonstrate decreased litter size due to failure of implantation (Benson et al., 1996; Bagot et al., 2000).
HOX genes function as transcription factors and either activate or repress target genes. The pleiotropic effects of HOXA10 on endometrial differentiation and function are likely modulated through the regulation of multiple downstream targets that are necessary for the implantation process (Daftary et al., 2002; Troy et al., 2003). HOXA10 has been demonstrated to be involved in the regulation of pinopode development and expression of other genes involved in endometrial development such as ITGB3 and EMX2 (Bagot et al., 2001; Daftary et al., 2002; Troy et al., 2003).
HOX proteins require co-operation with other proteins to bind to their target DNA. MEIS1 in particular aids 5′ HOX proteins, like HOXA10, to gain this specificity. MEIS1 physically interacts with 5′ HOX proteins HOXA9–11 by forming heterodimeric binding complexes on a DNA target containing a MEIS1 site (TGACAG) and an Abd-B-like Hox site (TTTTACGAC) (Shen et al., 1997). Studies demonstrated that MEIS1 gene and HOXA9-13 genes are co-expressed throughout Müllerian duct differentiation (Dintilhac et al., 2005), which suggests that MEIS1 might play a role in embryonic female genital tract development. We have performed data-mining on Affymetrix gene expression database in the public domain showing that MEIS1 was highly expressed in human endometrium as well as in myometrium (Fig. 1), implying it may play a role in reproductive function in the adult human uterus.
We therefore investigated the expression pattern of the MEIS1 gene at different stages of the human menstrual cycle, demonstrating that MEIS1 appeared to be expressed in the endometrium at different levels, depending on the menstrual cycle stage. MEIS1 mRNA levels appeared significantly increased in the mid secretory phase, closely resembling the expression pattern of HOXA10. This observation pointed to the possibility that MEIS1 is an active cofactor of HOXA10 in the human endometrium.
On the basis of the results described above, we postulated that Meis1 and Hoxa10 might have closely related roles in vertebrate reproduction and in the regulation of endometrial cell proliferation or differentiation. Meis1 null-mutant mice die between embryonic days 11.5 and 14.5, because of internal hemorrhage, liver hypoplasia and anemia (Azcoitia et al., 2005). The embryo lethal phenotype has precluded investigation of the function of Meis1 in the adult reproductive tract of Meis1 mutant mice. Therefore, to investigate the role of Meis1 in adult reproductive function in a mouse model, we altered the expression of the Meis1 gene in the adult murine endometrium. When mouse endometrium was transfected with Meis1 siRNA at the time of implantation, the number of embryos that could implant in the uterus was reduced by 40%. This significant reduction in implantation sites demonstrated that, in normal embryonic development, optimal implantation benefits from maternal Meis1 expression.
We also investigate the effect of Meis1 on pinopodes. Pinopode formation is directly regulated by HOXA10 (Bagot et al., 2001). In this study, we examined whether the expression of pinopodes could be also affected by altering Meis1 expression levels. We demonstrated that blocking Meis1 expression reduced pinopode numbers and reduced their ability to differentiate into fully mature forms. In contrast, supernumerary pinopodes could be induced by over-expression of Meis1.
To elucidate potential mechanisms by which Meis1 might control endometrial function and receptivity to the implanting blastocyst, we tested the effect of altering Meis1 expression on Itgb3 expression. The ITGB3 gene has been identified as the first known target of HOXA10 gene regulation in the endometrium (Daftary et al., 2002). As a bridging molecule between the endometrium and trophoblast (Kiefer et al., 1989; Van Dijk et al., 1993; Sueoka et al., 1997), ITGB3 plays an important role in embryonic implantation. We found that a blockade of Meis1 expression with Meis1 siRNA was associated with a decrease in Itgb3 mRNA. Itgb3 mRNA levels significantly increased after up-regulation of Meis1 expression with MEIS1 cDNA. It appeared likely that Itgb3 was not regulated by Hoxa10 monomer, but by a heterodimeric or heterotrimeric transcription complex containing Meis1. The interaction of Abd-B-like HOX transcription factors and MEIS1 at the HOX site in the target gene promoter has been demonstrated in myeloid differentiation. The co-activation of HOXA9 and MEIS1 is a common event in AML (Lawrence et al., 1999; Camós et al., 2006), but co-regulation of MEIS1 and HOXA9, and also of HOXA10, might play a role in pathogenesis of acute lymphocytic and acute myeloid leukemias associated with ALL-1 fusions (Rozovskaia et al., 2001; Ferrando et al., 2003; Dik et al., 2005). In addition, Pineault et al. (2004) demonstrated that MEIS1 enhances the effect of a HOXA10 rearrangement releasing HOX expression necessary for leukemia induction. Finally, binding of MEIS1 to HOXA10 could be demonstrated in vitro (Williams et al., 2005).
CDKN1A (also known as P21 or CIP1) is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. CDKN1A reporter expression is enhanced after transfection with PBX1 and HOXA10, especially when MEIS1 was co-transfected (Bromleigh and Freedman, 2000). It is likely that a similar regulation occurs with HOXA10 and MEIS1 cofactor in the endometrium. Recently, Taylor et al. have confirmed that this potential protein–protein interaction does occur in endometrial cells. Ablation of the PBX cofactor half-site results in a loss of HOXA10–PBX2 binding to the EMX2 probe, suggesting the importance of the PBX2/HOXA10 protein–protein interaction in endometrium for high affinity HOXA10 binding to its target genes (Sarno et al., 2005).
In conclusion, this study shows for the first time that MEIS1 expression contributes to endometrial receptivity in mouse. MEIS1 can function in cancer as an inhibitor of differentiation, and have a positive influence on cellular proliferation (see ‘Introduction’). In addition, the MEIS genes are potent enhancers of early cell proliferation during development (reviewed in Geerts et al., 2003, 2005). MEIS1 could have a similar proliferative function during endometrial development. The expression pattern of MEIS1, its contribution to pinopode formation in mouse and its effect on Itgb3 expression suggest that the effects of MEIS1 on endometrial function occur through affecting the affinity and specificity of HOXA10–DNA interactions, leading to the regulation of downstream targets of HOXA10. The identification of MEIS1 as a potential HOXA10 co-operating gene could lead to elucidation of a partial HOXA10 network in endometrial development, but could also establish MEIS1 in a signaling network in its own right.
Author’s Role
B.X.: conception and design of experiments, acquisition of data, analysis and interpretation of data; writing and revising of manuscript; final approval of manuscript to be published.
D.G.: design of experiments, acquisition, analysis, and interpretation of data; writing and revising of the manuscript; final approval of manuscript to be published.
K.Q.: conception and design of experiments; revision of manuscript; final approval of manuscript to be published.
H.Z.: conception and design of experiments; final approval of manuscript to be published.
G.Z.: interpretation of data; revision of manuscript; final approval of manuscript to be published.
Funding
Dutch Cancer Society (‘KWF Kankerbestrijding’) (UVA2003–2849, UVA2005–3665 to D.G.); The ‘973’ Program of China (No. 2007CB948104).
Acknowledgements
The authors would like to thank Dr J. Koster (Department of Human Genetics, Academic Medical Center, University of Amsterdam, The Netherlands) for the GEO Affymetrix data mining. pTER was a generous gift from Prof. Dr H. Clevers (Hubrecht Laboratory, The Netherlands Institute for Developmental Biology, Utrecht, The Netherlands).
References
- Azcoitia V, Aracil M, Martínez AC, Torres M. The homeodomain protein Meis1 is essential for definitive hematopoiesis and vascular patterning in the mouse embryo. Dev Biol. 2005;280:307–320. doi: 10.1016/j.ydbio.2005.01.004. [DOI] [PubMed] [Google Scholar]
- Bagot CN, Troy PJ, Taylor HS. Alteration of maternal Hoxa10 expression by in vivo gene transfection affects implantation. Gene Ther. 2000;7:1378–1384. doi: 10.1038/sj.gt.3301245. [DOI] [PubMed] [Google Scholar]
- Bagot CN, Kliman HJ, Taylor HS. Maternal Hoxa10 is required for pinopod formation in the development of mouse uterine receptivity to embryo implantation. Dev Dyn. 2001;222:538–544. doi: 10.1002/dvdy.1209. [DOI] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles–database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson GV, Lim H, Paria BC, Satokata I, Dey SK, Maas RL. Mechanisms of reduced fertility in Hoxa-10 mutant mice: uterine homeosis and loss of maternal Hoxa-10 expression. Development. 1996;122:2687–2696. doi: 10.1242/dev.122.9.2687. [DOI] [PubMed] [Google Scholar]
- Block K, Kardana A, Igarashi P, Taylor HS. In utero diethylstilbestrol (DES) exposure alters Hox gene expression in the developing müllerian system. FASEB J. 2000;14:1101–1108. doi: 10.1096/fasebj.14.9.1101. [DOI] [PubMed] [Google Scholar]
- Blomberg LA, Long EL, Sonstegard TS, Van Tassell CP, Dobrinsky JR, Zuelke KA. Serial analysis of gene expression during elongation of the peri-implantation porcine trophectoderm (conceptus) Physiol Genom. 2005;20:188–194. doi: 10.1152/physiolgenomics.00157.2004. [DOI] [PubMed] [Google Scholar]
- Bromleigh VC, Freedman LP. p21 is a transcriptional target of HOXA10 in differentiating myelomonocytic cells. Genes Dev. 2000;14:2581–2586. doi: 10.1101/gad.817100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bürglin TR. Analysis of TALE superclass homeobox genes (MEIS, PBC, KNOX, Iroquois, TGIF) reveals a novel domain conserved between plants and animals. Nucleic Acids Res. 1997;25:4173–4180. doi: 10.1093/nar/25.21.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camós M, Esteve J, Jares P, Colomer D, Rozman M, Villamor N, Costa D, Carrió A, Nomdedéu J, Montserrat E, et al. Gene expression profiling of acute myeloid leukemia with translocation t(8;16)(p11;p13) and MYST3-CREBBP rearrangement reveals a distinctive signature with a specific pattern of HOX gene expression. Cancer Res. 2006;66:6947–6954. doi: 10.1158/0008-5472.CAN-05-4601. [DOI] [PubMed] [Google Scholar]
- Calvo KR, Knoepfler PS, Sykes DB, Pasillas MP, Kamps MP. MEIS1a suppresses differentiation by G-CSF and promotes proliferation by SCF: potential mechanisms of cooperation with HoxA9 in myeloid leukaemia. Proc Natl Acad Sci USA. 2001;98:13120–13125. doi: 10.1073/pnas.231115398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. doi: 10.1038/376479a0. [DOI] [PubMed] [Google Scholar]
- Cermik D, Selam B, Taylor HS. Regulation of HOXA-10 expression by testosterone in vitro and in the endometrium of patients with polycystic ovary syndrome. J Clin Endocrinol Metab. 2003;88:238–243. doi: 10.1210/jc.2002-021072. [DOI] [PubMed] [Google Scholar]
- Chen Y, Ni H, Ma XH, Hu SJ, Luan LM, Ren G, Zhao YC, Li SJ, Diao HL, Xu X, et al. Global analysis of differential luminal epithelial gene expression at mouse implantation sites. J Mol Endocrinol. 2006;37:147–161. doi: 10.1677/jme.1.02009. [DOI] [PubMed] [Google Scholar]
- Cross JC, Werb Z, Fisher SJ. Implantation and the placenta: key pieces of the developmental puzzle. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Hydrosalpinx fluid diminishes endometrial cell HOXA10 expression. Fertil Steril. 2002;78:577–580. doi: 10.1016/s0015-0282(02)03306-x. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Taylor HS. Pleiotrophic effects of Hoxa10 on the functional development of peri-implantation endometrium. Mol Reprod Dev. 2004;67:8–14. doi: 10.1002/mrd.20013. [DOI] [PubMed] [Google Scholar]
- Daftary GS, Troy PJ, Bagot CN, Young SL, Taylor HS. Direct regulation of beta3-integrin subunit gene expression by HOXA10 in endometrial cells. Mol Endocrinol. 2002;16:571–579. doi: 10.1210/mend.16.3.0792. [DOI] [PubMed] [Google Scholar]
- Dik WA, Brahim W, Braun C, Asnafi V, Dastugue N, Bernard OA, van Dongen JJ, Langerak AW, Macintyre EA, Delabesse E. CALM-AF10+ T-ALL expression profiles are characterized by overexpression of HOXA and BMI1 oncogenes. Leukemia. 2005;19:1948–1957. doi: 10.1038/sj.leu.2403891. [DOI] [PubMed] [Google Scholar]
- Dintilhac A, Bihan R, Guerrier D, Deschamps S, Bougerie H, Watrin T, Bonnec G, Pellerin I. PBX1 intracellular localization is independent of MEIS1 in epithelial cells of the developing female genital tract. Int J Dev Biol. 2005;49:851–858. doi: 10.1387/ijdb.052013ad. [DOI] [PubMed] [Google Scholar]
- Eun Kwon H, Taylor HS. The role of HOX genes in human implantation. Ann N Y Acad Sci. 2004;1034:1–18. doi: 10.1196/annals.1335.001. [DOI] [PubMed] [Google Scholar]
- Ferrando AA, Armstrong SA, Neuberg DS, Sallan SE, Silverman LB, Korsmeyer SJ, Look AT. Gene expression signatures in MLL-rearranged T-lineage and B-precursor acute leukemias: dominance of HOX dysregulation. Blood. 2003;102:262–268. doi: 10.1182/blood-2002-10-3221. [DOI] [PubMed] [Google Scholar]
- Geerts D, Schilderink N, Jorritsma G, Versteeg R. The role of the MEIS homeobox genes in neuroblastoma. Cancer Lett. 2003;197:87–92. doi: 10.1016/s0304-3835(03)00087-9. [DOI] [PubMed] [Google Scholar]
- Geerts D, Revet I, Jorritsma G, Schilderink N, Versteeg R. MEIS homeobox genes in neuroblastoma. Cancer Lett. 2005;228:43–50. doi: 10.1016/j.canlet.2005.01.047. [DOI] [PubMed] [Google Scholar]
- Horcajadas JA, Pellicer A, Simon C. Wide genomic analysis of human endometrial receptivity: new times, new opportunities. Hum Reprod Update. 2007;13:77–86. doi: 10.1093/humupd/dml046. [DOI] [PubMed] [Google Scholar]
- Hsieh YY, Lin CS, Sun YL, Chang CC, Tsai HD, Wu JC. In vivo gene transfer of leukemia inhibitory factor (LIF) into mouse endometrium. J Assist Reprod Genet. 2002;19:79–83. doi: 10.1023/A:1014495614792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Y, Schnabel CA, Cleary ML. Trimeric association of Hox and TALE homeodomain proteins mediates Hoxb2 hindbrain enhancer activity. Mol Cell Biol. 1999;19:5134–5142. doi: 10.1128/mcb.19.7.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer MC, Bauer DM, Barr PJ. The cDNA and derived amino acid sequence for human osteopontin. Nucleic Acids Res. 1989;17:3306. doi: 10.1093/nar/17.8.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. HoxA9 transforms primary bone marrow cells through specific collaboration with MEIS1A but not Pbx1b. Eur Mol Biol Org J. 1998;17:3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Kurant E, Pai CY, Sharf R, Halachmi N, Sun YH, Salzberg A. Dorsotonals/homothorax, the Drosophila homologue of MEIS1, interacts with extradenticle in patterning of the embryonic PNS. Development. 1998;125:1037–1048. doi: 10.1242/dev.125.6.1037. [DOI] [PubMed] [Google Scholar]
- Lawrence HJ, Rozenfeld S, Cruz C, Matsukuma K, Kwong A, Kömüves L, Buchberg AM, Largman C. Frequent co-expression of the HOXA9 and MEIS1 homeobox genes in human myeloid leukemias. Leukemia. 1999;13:1993–1999. doi: 10.1038/sj.leu.2401578. [DOI] [PubMed] [Google Scholar]
- Ma XH, Hu SJ, Ni H, Zhao YC, Tian Z, Liu JL, Ren G, Liang XH, Yu H, Wan P, et al. Serial analysis of gene expression in mouse uterus at the implantation site. J Biol Chem. 2006;281:9351–9360. doi: 10.1074/jbc.M511512200. [DOI] [PubMed] [Google Scholar]
- Moens CB, Selleri L. Hox cofactors in vertebrate development. Dev Biol. 2006;291:193–206. doi: 10.1016/j.ydbio.2005.10.032. [DOI] [PubMed] [Google Scholar]
- Moskow JJ, Bullrich F, Huebner K, Daar IO, Buchberg AM. Meis1, a PBX1-related homeobox gene involved in myeloid leukemia in BXH-2 mice. Mol Cell Biol. 1995;15:5434–5443. doi: 10.1128/mcb.15.10.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardo LG, Sabatini L, Rai R, Nardo F. Pinopode expression during human implantation. Eur J Obstet Gynecol Reprod Biol. 2002;101:104–108. doi: 10.1016/s0301-2115(01)00523-1. [DOI] [PubMed] [Google Scholar]
- Nikas G. Endometrial receptivity: changes in cell-surface morphology. Semin Reprod Med. 2000;18:229–235. doi: 10.1055/s-2000-12561. [DOI] [PubMed] [Google Scholar]
- Nikas G, Makrigiannakis A. Endometrial pinopodes and uterine receptivity. Ann N Y Acad Sci. 2003;997:120–123. doi: 10.1196/annals.1290.042. [DOI] [PubMed] [Google Scholar]
- Noyes RW, Hertig AT, Rock J. Dating the endometrial biopsy. Fertil Steril. 1955;1:3–25. doi: 10.1016/j.fertnstert.2019.08.079. [DOI] [PubMed] [Google Scholar]
- Pan H, Zhu L, Deng Y, Pollard JW. Microarray analysis of uterine epithelial gene expression during the implantation window in the mouse. Endocrinology. 2006;147:4904–4916. doi: 10.1210/en.2006-0140. [DOI] [PubMed] [Google Scholar]
- Pellerin I, Schnabel C, Catron KM, Abate-Shen C. Hox proteins have different affinities for a consensus DNA site that correlate with the positions of their genes on the hox cluster. Mol Cell Biol. 1994;14:4532–4545. doi: 10.1128/mcb.14.7.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineault N, Abramovich C, Ohta H, Humphries RK. Differential and common leukemogenic potentials of multiple NUP98-Hox fusion proteins alone or with Meis1. Mol Cell Biol. 2004;24:1907–1917. doi: 10.1128/MCB.24.5.1907-1917.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- Qian K, Chen H, Wei Y, Hu J, Zhu G. Differentiation of endometrial stromal cells in vitro: down-regulation of suppression of the cell cycle inhibitor p57 by HOXA10? Mol Hum Reprod. 2005;11:245–251. doi: 10.1093/molehr/gah147. [DOI] [PubMed] [Google Scholar]
- Rieckhof GE, Casares F, Ryoo HD, Abu-Shaar M, Mann RS. Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. Cell. 1997;91:171–183. doi: 10.1016/s0092-8674(00)80400-6. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T, Feinstein E, Mor O, Foa R, Blechman J, Nakamura T, Croce CM, Cimino G, Canaani E. Upregulation of Meis1 and HoxA9 in acute lymphocytic leukemias with the t(4:11) abnormality. Oncogene. 2001;20:874–878. doi: 10.1038/sj.onc.1204174. [DOI] [PubMed] [Google Scholar]
- Sarno JL, Kliman HJ, Taylor HS. HOXA10, Pbx2, and Meis1 protein expression in the human endometrium: formation of multimeric complexes on HOXA10 target genes. J Clin Endocrinol Metab. 2005;90:522–528. doi: 10.1210/jc.2004-0817. [DOI] [PubMed] [Google Scholar]
- Satokata I, Benson G, Maas R. Sexually dimorphic sterility phenotypes in Hoxa10-deficient mice. Nature. 1995;374:460–463. doi: 10.1038/374460a0. [DOI] [PubMed] [Google Scholar]
- Schnabel CA, Jacobs Y, Cleary ML. HoxA9-mediated immortalization of myeloid progenitors requires functional interactions with TALE cofactors Pbx and Meis. Oncogene. 2000;19:608–616. doi: 10.1038/sj.onc.1203371. [DOI] [PubMed] [Google Scholar]
- Shen WF, Montgomery JC, Rozenfeld S, Moskow JJ, Lawrence HJ, Buchberg AM, Largman C. Abd-B-like Hox proteins stabilize DNA binding by the Meis1 homeodomain proteins. Mol Cell Biol. 1997;17:6448–6558. doi: 10.1128/mcb.17.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen WF, Rozenfeld S, Kwong A, Kömüves LG, Lawrence HJ, Largman C. HOXA9 forms triple complexes with PBX2 and MEIS1 in myeloid cells. Mol Cell Biol. 1999;19:3051–3061. doi: 10.1128/mcb.19.4.3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JE, Jr, Bollekens JA, Inghirami G, Takeshita K. Cloning and mapping of the MEIS1 gene, the human homolog of a murine leukemogenic gene. Genomics. 1997;43:99–103. doi: 10.1006/geno.1997.4766. [DOI] [PubMed] [Google Scholar]
- Sueoka K, Shiokawa S, Miyazaki T, Kuji N, Tanaka M, Yoshimura Y. Integrins and reproductive physiology: expression and modulation in fertilization, embryogenesis, and implantation. Fertil Steril. 1997;67:799–811. doi: 10.1016/s0015-0282(97)81388-x. [DOI] [PubMed] [Google Scholar]
- Taylor HS. The role of HOX genes in human implantation. Hum Reprod Update. 2000;a 6:75–79. doi: 10.1093/humupd/6.1.75. [DOI] [PubMed] [Google Scholar]
- Taylor HS. The role of HOX genes in the development and function of the female reproductive tract. Semin Reprod Med. 2000;b 18:81–89. doi: 10.1055/s-2000-13478. [DOI] [PubMed] [Google Scholar]
- Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved Hox axis in the mouse and human reproductive system: late establishment and persistent expression of the Hoxa cluster genes. Biol Reprod. 1997;57:1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- Taylor HS, Arici A, Olive D, Igarashi P. HOXA10 is expressed in response to sex steroids at the time of implantation in the human endometrium. J Clin Invest. 1998;101:1379–1384. doi: 10.1172/JCI1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor HS, Bagot C, Kardana A, Olive D, Arici A. HOX gene expression is altered in the endometrium of women with endometriosis. Hum Reprod. 1999;14:1328–1331. doi: 10.1093/humrep/14.5.1328. [DOI] [PubMed] [Google Scholar]
- Troy PJ, Daftary GS, Bagot CN, Taylor HS. Transcriptional repression of peri-implantation EMX2 expression in mammalian reproduction by HOXA10. Mol Cell Biol. 2003;23:1–13. doi: 10.1128/MCB.23.1.1-13.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 2003;4:609–615. doi: 10.1038/sj.embor.embor865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dijk S, D’Errico JA, Somerman MJ, Farach-Carson MC, Butler WT. Evidence that a non-RGD domain in rat osteopontin is involved in cell attachment. J Bone Miner Res. 1993;8:1499–1506. doi: 10.1002/jbmr.5650081213. [DOI] [PubMed] [Google Scholar]
- van Dijk MA, Peltenburg LT, Murre C. Hox gene products modulate the DNA binding activity of PBX1 and Pbx2. Mech Dev. 1995;52:99–108. doi: 10.1016/0925-4773(95)00394-g. [DOI] [PubMed] [Google Scholar]
- Wang H, Dey S. Roadmap to embryo implantation: clues from mouse models. Nat Rev Genet. 2006;7:185–199. doi: 10.1038/nrg1808. [DOI] [PubMed] [Google Scholar]
- Wellik DM. Hox patterning of the vertebrate axial skeleton. Dev Dyn. 2007;236:2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- Williams TM, Williams ME, Innis JW. Range of HOX/TALE superclass associations and protein domain requirements for HOXA13:MEIS interaction. Dev Biol. 2005;277:457–471. doi: 10.1016/j.ydbio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17:359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]