Abstract
Cryptococcus neoformans (Cn) is the most common cause of fungal meningitis worldwide. In infected patients, growth of the fungus can occur within the phagolysosome of phagocytic cells, especially in non-activated macrophages of immunocompromised subjects. Since this environment is characteristically acidic, Cn must adapt to low pH to survive and efficiently cause disease. In the present work, we designed, tested, and experimentally validated a theoretical model of the sphingolipid biochemical pathway in Cn under acidic conditions. Simulations of metabolic fluxes and enzyme deletions or downregulation led to predictions that show good agreement with experimental results generated post hoc and reconcile intuitively puzzling results. This study demonstrates how biochemical modeling can yield testable predictions and aid our understanding of fungal pathogenesis through the design and computational simulation of hypothetical experiments.
Keywords: Biochemical Systems Theory, ceramide, Cryptococcus neoformans, S-system, sphingolipid
Introduction
Cryptococcus neoformans (Cn) is a fungal pathogen that infects humans via the respiratory tract. It is an environmental microorganism particularly present in pigeon droppings or associated with eucalyptus tree but it can be isolated from soil, water, milk, fruits, horse intestinal flora, bird nests, bats, burns, and cockroaches. Once inhaled in the lung, dissemination of the infection through the bloodstream leads to the development of a life-threatening meningoencephalitis, particularly in immunocompromised patients (Casadevall and Perfect, 1998; Perfect, 2005). An important characteristic that enables the fungus to cause disease is its ability to grow in alkaline, neutral, and acidic environments of the human body. Alkaline/neutral environments are found extracellularly, such as in alveolar spaces and in the bloodstream, whereas acidic environments are characteristically found intracellularly, within the phagolysosome of host phagocytic cells (Feldmesser et al, 2001). Indeed, Cn is a facultative intracellular pathogen and can move in and out without killing host cells (Alvarez and Casadevall, 2006; Ma et al, 2006). In doing so, it constantly needs to adapt to a new environment, for instance, by changing the organization of different cellular components, gene expression, protein activities, or arrangements of lipids within membranes.
It might be impossible to fully understand this adaptation using a purely reductionistic approach because of continual changes in the production and degradation of key cellular components and because of the complex interplay among these components during switches in the environment. Instead, the use of systems biological methods might offer an opportunity to complement reductionistic insights by explaining complex, systemic behaviors, such as cellular adaptation to environments, through the simultaneous investigation of stimuli and responses of several enzymes and metabolites in terms of space, time, and context. The spatial aspect is important because it accounts for compartmentalization and the topographic relationships among the components; the need to consider time is evident, given the dramatic dynamic changes in the molecular characteristics during adaptation; and addressing the cellular and environmental context at each time point accounts for the interdependencies between all components partaking in the adaptation process (Ahn et al, 2006a, 2006b). Thus, system biology has a chance of shedding light on the complexities that govern microbial metabolic pathways, and their regulation and adaptations may ultimately allow us to predict cellular or biological phenotypes without the need for large-scale wet experimentation. Specifically for the case of Cn, the application of systems biological concepts may provide significant insights into the mechanisms of fungal pathogenesis and into the complex interplay between the fungus and the host immunity in chronic fungal diseases.
In all eukaryotic cells, the main intracellular pH regulator is the plasma membrane H+ATPase (Pma1) pump, whose activation results in proton extrusion, maintaining the intracellular pH neutral (Serrano et al, 1986; Serrano, 1988) even if the extracellular pH is acidic (Perona et al, 1990; Portillo et al, 1991). In Saccharomyces cerevisiae (Sc) and in Cn, Pma1 is essential for cell viability (Soteropoulos et al, 2000), suggesting that Pma1 has a key role in regulating the intracellular pH. Many studies in Sc proposed a role for sphingolipids in the regulation of Pma1 function in the endoplasmic reticulum (ER) and at the plasma membrane (Patton et al, 1992; Lee et al, 2002; Wang and Chang, 2002; Gaigg et al, 2005, 2006; Toulmay and Schneiter, 2007).
The basic structural component of sphingolipids is a long-chain sphingoid base backbone (e.g., sphingosine or phytosphingosine (PHS)). The linkage of a fatty acid to the two-amino group of this backbone through an amide bond yields ceramide or phytoceramide. Complex sphingolipids are formed with the addition of a polar group to ceramide (or phytoceramide) via an ester bond at the C-1 position. The synthesis of sphingolipid occurs in all eukaryotic cells and, in addition to being common components of membranes, they have been recognized to function as signaling molecules in a variety of signaling pathways (reviewed in Futerman and Hannun, 2004). In mammalian cells, sphingolipids such as ceramide, ceramide-1-phosphate, and sphingosine-1-phosphate have key roles in the regulation of cellular proliferation, stress responses, cell cycle, apoptosis, inflammation, and immune response (reviewed in Luberto and Hannun, 1999; Hannun and Luberto, 2000; McQuiston et al, 2006).
In Cn, sphingolipids have emerged as important molecules required for growth and signaling in alkaline/neutral (Liu et al, 2005; Rittershaus et al, 2006; Saito et al, 2006) and acidic (Buede et al, 1991; Berne et al, 2005; Shea et al, 2006) environments. Thus, we hypothesize that when Cn shifts from a neutral/alkaline environment, such as extracellular alveolar spaces or the bloodstream, to an acidic environment, such as the phagolysosome of phagocytic cells, it should promote a biochemical response associated with its sphingolipid pathway, with a consequent metabolic adaptation to the new environment.
In this study, we thus developed a mathematical model of sphingolipid metabolism in the pathogenic fungus Cn and made reliable predictions on its biochemical sphingolipid adaptation to a shift from an alkaline to an acidic pH, mimicking the internalization of the fungus by phagocytic cells. The model was designed and analyzed within the framework of Biochemical System Theory (BST), which uses power-law representations for all enzymatic and transport processes (Savageau, 1969a, 1969b, 1976; Torres and Voit, 2002).
Results
The results of our studies fall into three categories. The first set describes experimental studies that identified inositol phosphorylceramide synthase (Ipc1), inositol phosphosphingolipid phospholipase C (Isc1), and phytoceramides of different lengths as important contributors to the response of Cn to H+ATPase pump (Pma1) and ATP-mediated shifts in pH. Especially the putative roles of the enzymes Ipc1 and Isc1 seem puzzling, because these enzymes catalyze opposite directions of the reversible reaction between phytoceramide and inositol phosphoryl ceramide (IPC). The second set of studies used a mathematical systems model to elucidate how the different contributors might lead to an appropriate stress response. This model in turn made predictions regarding Pma1 and the role of ATP, which we investigated experimentally in the third set of studies as model validation.
Experimental studies identifying Ipc1, Isc1, and phytoceramide as drivers of pH response
Effect of Ipc1 downregulation and Isc1 deletion on growth of Cn in acidic environments
Inositol phosphoryl ceramide synthase 1 (Ipc1) is a fungal enzyme localized in the Golgi apparatus in Sc (Levine et al, 2000) and Cn (M Del Poeta, unpublished data). It transfers inositol phosphate from phosphatidylinositol (PI) to phytoceramide, producing IPC and 1,2-sn-diacylglycerol (DAG) (Kuroda et al, 1999; Heidler and Radding, 2000). Isc1 is an enzyme localized in the ER and breaks down IPC, mannosyl-IPC (MIPC), and mannosyl diphosphoryl ceramide (M(IP)2C) to phytoceramide, inositol phosphate (MIP), and M(IP)2, respectively (Dickson and Lester, 1999). We tested a mutant in which Ipc1 is downregulated, GAL7:IPC1, grown on glucose medium, and found that it fails to adapt with sufficient speed to an acidic environment and, as a consequence, its growth is significantly retarded (Supplementary Figure 1A) (Luberto et al, 2001). Similarly, a mutant strain in which Isc1 was deleted, Δisc1, showed a delay in the adaptation to a low-pH environment (Supplementary Figure 1B) (Buede et al, 1991; Berne et al, 2005; Shea et al, 2006). Thus, two enzymes with ‘opposite' biochemical activities (Ipc1 uses phytoceramide as a substrate and produces IPC, whereas Isc1 uses IPC as a substrate and produces phytoceramide) have the same effect on viability in changing pH environments. This puzzling finding was quite intriguing and warranted further investigations at the systems level (see ‘Computational studies').
Measurement of phytoceramide subspecies in Cn wild type during growth at alkaline/neutral and acidic pH
Fungal cells produce variant species of phytoceramide that differ principally in the lengths of their acyl chains and their hydroxylation states (Vaena de Avalos et al, 2004, 2005). This variability led to the question of whether phytoceramide subspecies levels would change when cells are shifted from a neutral to an acidic environment. Targeted experiments indeed revealed that in Cn cells exposed to a low-pH environment, the total level of non-hydroxylated phytoceramide decreases to half the level in a neutral environment (from 3862.7±6.34 to 1588.7±6.32 pmol/pmol Pi; P<0.05), whereas the total level of hydroxylated phytoceramides increases significantly compared to that in neutral pH (from 1244.1±3.2 to 2147.2±3.5 pmol/pmol Pi) (Table I). Analysis of the different phytoceramide subspecies revealed that very long-chain phytoceramides (both C26 hydroxylated and non-hydroxylated forms) were significantly elevated at low compared to neutral pH, whereas short-chain non-hydroxylated phytoceramides (C14, C16, and C18) were significantly decreased at low compared to neutral pH (P<0.05). Phytoceramide measurements were also performed at alkaline pH (7.4). No differences were found in phytoceramide levels or phytoceramide subspecies between pH 7.0 and 7.4 (data not shown). These results suggest that Cn changes the metabolism of certain phytoceramide subspecies when shifted from a neutral/alkaline to an acidic environment.
Table 1.
Identification of ceramide species at 48 h of growth
| Ceramide species (pmol/pmol Pi) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phyto-C18 | Phyto-C18;1 | Phyto-C20 | Phyto-C24 | Phyto-C24:1 | Phyto-C26 | Phyto-C26:1 | α-OH-phyto-C18 | α-OH-phyto-C18:1 | α-OH-phyto-C24 | α-OH-phyto-C26 | α-OH-phyto-C26:1 | |
| pH 7 | 406.6 | 230 | 60 | 2260 | 640.1 | 170 | 96 | 3.1 | 1.04 | 570 | 360 | 310 |
| pH 4 | 215.3 | 180 | 4.5* | 396* | 400 | 222.5 | 170.4* | 4.2 | 3 | 810 | 710* | 620 |
*P<0.05 (pH 4 versus 7).
Mass spectrometric analysis of different phytoceramide and alpha hydroxyl phytoceramide species during late-log phase in a Cn H99 WT strain. Determinations are to neutral pH or acidic pH. The mass of each species was normalized to phosphorous levels of each sample. Results are the means of three separate experiments. The concentrations are reported as pmol/pmol Pi for phyto- (phytoceramide) and α-OH-phyto (alpha hydroxyl phytoceramide) with different length fatty acid chains.
Ebselen causes elevated inhibition in Isc1 mutant
As the plasma membrane proton pump H+ATPase (Pma1) is one of the major regulators of intracellular H+ (Serrano et al, 1986; Serrano, 1988), we wondered whether the absence of Isc1 or downregulation of Ipc1 would affect the susceptibility of yeast cells to Pma1 inhibitors. To address this question, we examined the minimal inhibitory concentration (MIC) and minimal fungicidal concentration (MFC) of ebselen, a well-established inhibitor of fungal Pma1 (Perlin et al, 1997). We found that loss of Isc1 or downregulation of Ipc1 significantly increases the susceptibility of yeast cells to ebselen (Table II), suggesting that both Isc1 and Ipc1 may potentially regulate Pma1 function.
Table 2.
Loss of Isc1 sensitizes Cn to the Pma1 inhibitor ebselena
| Strain | pH 7 (μM) | pH 4 (μM) |
|---|---|---|
| WT | 3.12 | 2.48 |
| Δisc1 | 3.12 | 0.78* |
| Δisc1REC | 3.12 | 3.12 |
| GAL7:IPC1-glucose | 3.12 | 0.39* |
*P<0.05 (pH 4 versus 7).
aEbselen is fungicidal to WT, Δisc1, Δisc1REC, and GAL7:IPC1 strains at an MFC of 3.12 μM at a neutral pH. The Δisc1 mutant and GAL7:IPC1 strain grown on glucose (Ipc1 downregulated) showed MFCs of 0.78 and 0.39 μM, respectively. Results are the means of three separate experiments.
Computational studies
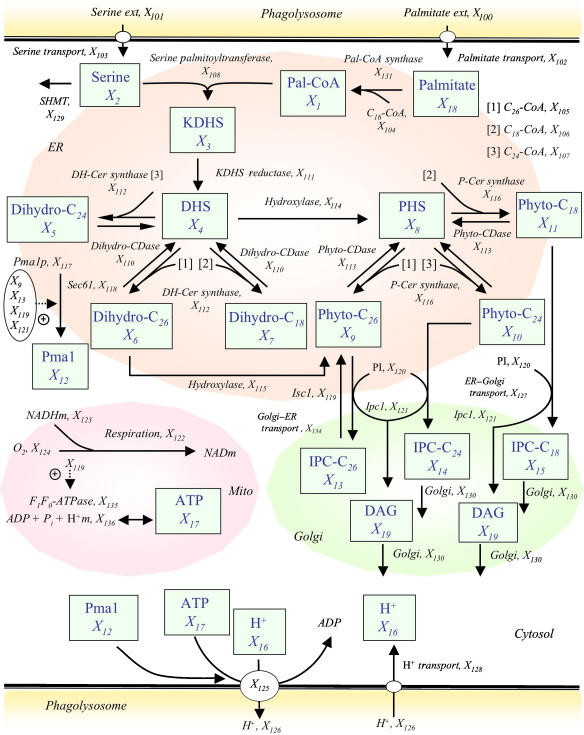
The diagram of all reaction steps that were deemed important in the sphingolipid pathway of Cn and on which the model is based is presented in Figure 1. Most analyses reflect the late-log phase of growth at acidic pH.
Figure 1.
Model diagram of sphingolipid metabolism in Cn. Metabolites in boxes represent dependent variables that are defined through differential equations and are numbered from X1 to X19. Independent variables are numbered from X100 to X136. Solid arrows show flow of material. Plus signs associated with dotted arrows represent activation. The acylation state is coded as (1) C26-CoA, (2) C18-CoA, and (3) C24-CoA; these are substrates for the DH-Cer synthase reaction or for the enzyme P-Cer synthase (see main text and Supplementary information for details). Dependent variables: Pal-CoA (X1), palmitoyl-CoA; serine (X2); KDHS (X3), 3-ketodihydrosphingosine; DHS (X4), dihydrosphingosine; dihydro-C24 (X5), dihydroceramide C24; dihydro-C26 (X6), dihydroceramide C26; dihydro-C18 (X7), dihydroceramide C18; PHS (X8), phytosphingosine; phyto-C26 (X9), phytoceramide C26; phyto-C24 (X10), phytoceramide C24; phyto-C18 (X11), phytoceramide C18; Pma1 (X12), newly synthesized Pma1; IPC-C26 (X13), inositol phosphorylceramide C26; IPC-C24 (X14), inositol phosphorylceramide C24; IPC-C18 (X15), inositol phosphorylceramide C18; intracellular protons (X16); ATP (X17), adenosine-5′-triphosphate; palmitate (X18); DAG (X19), sn-1,2-diacylglycerol. Independent variables: palmitate ext (X100), palmitate external; serine ext (X101), serine external; palmitate transport (X102); serine transport (X103); Ac-CoA (X104), acetyl CoA; C26-CoA (X105), very long-chain fatty acid (C26-CoA); C18-CoA (X106), fatty acid (C18-CoA); C24-CoA (X107), fatty acid (C24-CoA); serine palmitoyltransferase (X108); ADP, adenosine biphosphate (X109); dihydro-CDase (X110), dihydroceramide ceramidase; KDHS reductase (X111), 3-ketodihydrosphingosine reductase; DH-Cer synthase (X112), dihydroceramide synthase; phyto-CDase (X113), phytoceramidase; hydroxylase (X114); hydroxylase (X115); P-Cer synthase (X116), phytoceramide synthase; Pma1p (X117), newly synthesized Pma1 in the ER; Sec61 (X118), Sec61 as probable ER insertion protein; Isc1 (X119), inositol phosphosphingolipid phospholipase C; PI (X120), phosphatidylinositol; Ipc1 (X121), inositol phosphorylceramide synthase; alternative respiration (X122); NADHm (X123), nicotinamide adenine dinucleotide; oxygen (X124); Pma1-H+ATPase (X125), synthesized plasma membrane H+-ATPase; H+ (X126), protons external; ER–Golgi transport (X127); H+ transport (X128), proton transport; SHMT (X129) serine hydroxymethyl transferase; Golgi membrane (X130); Pal-CoA synthase (X131), palmitoyl-CoA synthase; ATP total (X132); AMP (X133), adenosine monophosphate; Golgi–ER transport (X134); F0F1-ATPase (X135), F0F1-ATP synthase; H+ m (X136), mitochondrial protons.
We adapted an earlier sphingolipid model for yeast (Alvarez-Vasquez et al, 2004, 2005) to the peculiarities of Cn. Importantly, we incorporated several organelles that separate the physical synthesis of metabolites. Also the new model was developed during late-log phase and it contains several newly determined metabolite levels and enzyme activities in wild-type (WT) Cn grown at acidic pH. In addition to allowing for ceramides with different fatty acid chain lengths, the new model accounts for mechanisms of transporting protons between the cytosol and the phagolysosome, which are crucial for the survival of Cn. Since Pma1 trafficking (Gaigg et al, 2005) and function (Lee et al, 2002) are controlled by very long-chain phytoceramides, the mathematical model of Cn sphingolipid pathway proposed here pays particular attention to the dynamics of Pma1, as well as to short-chain and very long-chain phytoceramides and the enzymes that directly and indirectly regulate their production.
The model analyses fall into two categories. As it is standard in BST, we first diagnosed the model by thoroughly investigating its stability and robustness and evaluating sensitivities and gains, especially with respect to enzyme activities. Most of the results of these analyses are unremarkable and are therefore presented in Supplementary information. The gains provided us with some insights into which metabolites are most affected by changes in particular enzymes and transport steps. However, they did not reveal compelling explanations of how Cn coordinates responses to shifts in pH. By contrast, exploration of the consequences of alterations in Ipc1 and Isc1 suggested interesting mechanisms of handling protons. All analyses were executed with the free software PLAS© (Ferreira, 2000).
The cellular compartments involved in the model are as follows.
Cytoplasm
We include the transport of palmitate and serine, required for the first step of sphingolipid biosynthesis. In acidic conditions, as they exist inside macrophages, protons move into the cell. The inclusion of proton transport is important for the relationships with H+ATPase (Pma1) and cytoplasmic pH. Proton import into the fungal cytosol is modeled as a simple (first order) transport process, whereas the export mechanism depends highly on Pma1. A simple transport process implies direct proportionality between proton influx (inside the fungus) and the external proton concentration (outside the fungus but inside the phagolysosome). Our model considers the fungal ATP synthesis and degradation (Supplementary Table S7) and includes the F0F1-ATPase in the fungal mitochondria (equation 17). Our model assumes that the fungus is inside the phagolysosome (its physiological environment once internalized by host macrophages) with no direct contact with the host mitochondria. For instance, we refer to a simple transport process when the proton influx depends directly on the electrochemical gradient. By contrast, Pma1 (X125) utilizes energy obtained directly from ATP hydrolysis to transport protons against the electrochemical differences across the plasma membrane.
Endoplasmic reticulum
The first steps of the sphingolipid pathway take place in the ER. The condensation of serine and the acyl group transferred from palmitoyl-CoA form 3-ketodihydrosphingosine (KDHS). This reaction is catalyzed by serine palmitoyltransferase (Buede et al, 1991). KDHS is rapidly converted into dihydrosphingosine (DHS). In the model, DHS can be converted by a hydroxylase to PHS. At the same time, under the action of a specific ceramide synthase (sphingoid base N-acyl transferase), DHS and a long-chain fatty acid CoA may form dihydroceramides C18, C24, and C26. PHS may combine with long-chain fatty acid CoA to form phytoceramides C18, C24, and C26. Both dihydroceramides C18, C24, and C26 and phytoceramides C18, C24, and C26 may undergo hydrolytic reactions to form DHS and PHS, respectively, which are catalyzed by specific ceramidases (Mao and Obeid, 2000; Mao et al, 2000). In the model, dihydroceramide C26 may be hydroxylated directly to phytoceramide C26. Once produced, the transport of phytoceramides C18, C24, and C26 from ER to Golgi apparatus follows a first-order process.
The model considers that the biosynthesis of phytoceramide and Pma1 occurs in the ER. The Pma1 in the ER compartment has an activity in response to pH signals similar to the Pma1 inserted in the plasma membrane, as previously suggested (Portillo et al, 1989). The model predicts that Pma1 is stabilized by phytoceramide C26 produced by Isc1 (Lee et al, 2002). The model also considers Sec61, which was found to be upregulated during murine macrophage infection of Cn (Fan et al, 2005), a secretory factor that could mediate the insertion of Pma1 into vesicles. Pma1 travels from the ER to the Golgi apparatus and finally to the plasma membrane where it is inserted (Chang and Slayman, 1991).
Golgi apparatus
In the Golgi apparatus, phytoceramides C18, C24, and C26 are substrates for Ipc1. Ipc1 transfers inositol phosphate from PI to phytoceramide(s), producing IPC (IPC-C18, -C24, and -C26) and DAG. Ipc1 is localized in the Golgi apparatus, implying that phytoceramides are transported from the ER to the Golgi apparatus.
IPC(s) can also be mannosylated, forming MIPC, and MIPC can be transformed into M(IP)2C by the action of inositol phosphoryl transferase 1. For simplicity, MIPC, M(IP)2C, and It1 are not considered in the model. Also, we hypothesize that IPC-C26 is transported from the Golgi apparatus to the ER and used by the ER enzyme Isc1 to produce phytoceramide C26. The model does not differentiate among trans-, medial-, and cis-Golgi. The synthesis of complex sphingolipids (IPCs, MIPCs, and M(IP)2Cs) occurs in the Golgi apparatus and their different distribution in other membranes implies the existence of a sorting mechanism for complex sphingolipids. Indeed, the presence of inositol-containing sphingolipids in organelles of the secretory pathway has been suggested (Hechtberger et al, 1994). Synthesis and transport of sphingolipids in yeasts, especially complex sphingolipids, has been poorly studied. However, phytoceramide is indeed produced in the ER and transported to the Golgi apparatus, independent of vesicular traffic because IPC synthesis still continues when vesicular transport is blocked in sec mutants (Funato and Riezman, 2001).
Mitochondria
Mitochondria are the major producers of ATP. In yeast, the F0F1-ATP synthase is the protein that is responsible for the aerobic synthesis of ATP, and it is localized in the inner mitochondrial membrane (Fronzes et al, 2003). The F1F0-ATP synthase is important for the regulation of intracellular pH, implicating mitochondria as a major regulator of cytoplasmic pH (Matsuyama and Reed, 2000). Although in rat liver the cytosolic-mitochondrial delta pH can be between 0.06 and 0.31 pH units when the cytosol is acidified with permeant weak acids (Durand et al, 1999), there is no much literature on in vivo mitochondria-cytosol pH recordings.
In Cn, as in other eukaryotes, ATP is derived from mitochondria respiration. During respiration, the electron transfer chain with terminal cytochrome c oxidase (COX) pathway generates a proton gradient, which, among other functions, powers the synthesis of ATP through F1F0-ATP synthase. In addition to this pathway, Cn (and other fungi, higher plants, and some protozoa) possesses a unique cyanide-resistant electron transport chain in its mitochondria. This pathway is composed of a homodimeric protein identified as an alternative oxidase 1 (Aox1) (Akhter et al, 2003). As Aox1 is important for Cn to survive in acidic conditions, we included it in the model.
In Sc, Isc1 has been found in the mitochondria during the late-log phase (Vaena de Avalos et al, 2004). Thus, this enzyme can form phytoceramide in the mitochondria from IPC (Kitagaki et al, 2007). Interestingly, Sc Δisc1 mutant showed reduced levels of mitochondrial COX subunits Cox3 and Cox4 (Vaena de Avalos et al, 2005), suggesting that Isc1 play a role in the respiration by mitochondria.
Analytical model results
The model analyses fall into two categories. As it is standard in BST, we first diagnosed the model by analyzing its stability and evaluating its robustness through computation of sensitivities and (logarithmic) gains, especially with respect to enzyme activities. Most of the results of these analyses are not particularly interesting and were therefore moved to Supplementary information. The second type of analysis targets the dynamics of the system and is accomplished through simulation studies.
Eigenvalue analysis confirmed the stability of the steady state of the model. Furthermore, the system was quite robust, as measured by sensitivity and gain profiles with low magnitudes. A (logarithmic) gain quantifies the effect of a 1% change in some enzyme activity on some steady-state output quantity, such as a metabolite concentration or a flux. In the particular case of our model, no gains pointed to obvious problems or interesting control points. As in the earlier model for sphingolipid metabolism in yeast (Alvarez-Vasquez et al, 2004, 2005), the precursors serine, palmitate, and fatty acid-CoA, as well as the initial transport and kinetic steps, by and large have the highest gains, which is intuitively reasonable because they determine the entire input to the system, while most other reactions merely redistribute material within the system. Outside the ‘input mechanisms,' non-obvious positive, relatively high gains were found for dihydroceramide synthase (X112), dihydroceramide hydroxylase (X115), and Ipc1 (X121); increases in their activities result in changes in phytoceramide levels. High negative gains were seen for dihydroceramide ceramidase (X110), phytoceramidase (X113), hydroxylase (X114); increases in their activities result in decreases in phytoceramide levels. Of interest is that none of the gains are extraordinarily high and that gains with respect to Pma1 are largely unremarkable. A similar analysis of flux gains did not add further insight. The log gains are closely related to kinetic parameter sensitivities, which were therefore, not surprisingly, not remarkable either.
The gains provided us with modest insights into which metabolites are most affected by changes in particular enzymes and transport steps. However, they did not reveal compelling explanations of how Cn coordinates responses to shifts in pH. Therefore, we used simulations to explore mechanisms of proton handling and the role of alterations in Ipc1 and Isc1 in these mechanisms. All analyses were executed with the free software PLAS© (Ferreira, 2000). The Isc1 simulation was made with a 95% decrease in activity and Ipc1 perturbations were implemented with 85% decreases. These alterations were initiated in the simulations at time 1 min. We selected for our discussion of results the simulations most relevant for the analysis of regulation of Pma1 by phytoceramide with respect to internal proton concentrations.
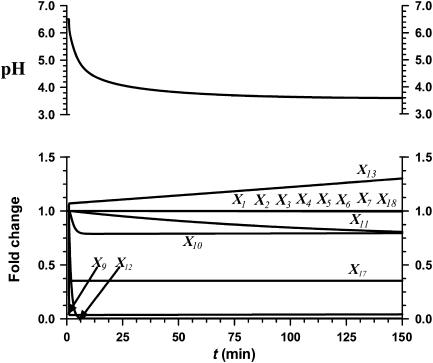
Effect of decreased Isc1 (X119)
The ‘static' gain analysis was not able to rationalize the means by which Cn responds to shifts in pH. Because our biological experiments pointed to Isc1 and Ipc1 as potential contributors to the acidity response, we simulated changes in their activities, which in reality would probably be achieved through alterations in gene expression, following sensing and signal transduction.
Figure 2 shows the responses to a 95% decrease in Isc1 activity. The transients in this figure are not as important as the final state of the system, because we are modeling an ‘inborn' mutation, as opposed to something like a sudden change in substrate. It is evident that phytoceramide C26 (X9) and ATP (X17) levels are dramatically and permanently decreased in this situation. Other key sphingolipids (X1–X7, X4–X15, and X18) are not significantly affected by this perturbation. By contrast, loss of Isc1 activity severely diminishes Pma1 activity (X12), and the cytoplasmic pH decreases from 6.5 to 3.6 and does not return to its nominal state, in which Isc1 shows normal activity.
Figure 2.
Simulation result of a 95% decrease in Isc1 (X119) activity. Phyto-C26 (X9), Pma1 (X12), and ATP (X17) decrease. The remaining metabolites stay close to their initial values, except for IPC-C26 (X13), which increases. The intracellular pH decreases from 6.5 to 3.6.
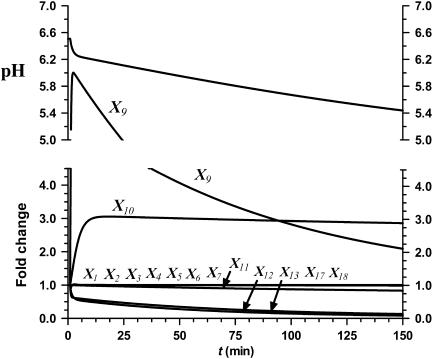
Effect of decreased Ipc1 (X121) activity
Decreases in the activity of Ipc1 (X121) similarly affect Pma1 function (see Figure 3 for the consequences of a 85% decrease). In this simulation of a low-pH environment, IPC-C26 (X13), phytoceramide C18 (X11), and Pma1 (X12) decrease, whereas phytoceramide C26 (X9) increases. The intracellular pH decreases from 6.5 to 5.4 and does not return to its normal state.
Figure 3.
Simulation result of an 85% decrease in Ipc1 (X121) activity. Phyto-C18 (X11), IPC-C26 (X13), and Pma1 (X12) decrease, whereas phyto-C26 (X9) and phyto-C24 (X10) increase. The remaining metabolites stay close to their initial values. The intracellular pH decreases from 6.5 to 5.4.
Validation of the model
The biochemical conditions considered in the model support the simplified scheme (Figure 1) of sphingolipid metabolism in Cn in acid and neutral pH environments and allow us to predict how changes are transduced to Pma1 activity. As the model predictions are very specific, it is not difficult in this particular case to test some of them in the laboratory. In other words, the model effectively narrowed the range of most promising experimental targets from potentially very many to just a few. Table III shows comparisons between model deductions and observations.
Table 3.
Comparison between model predictions and experimental data for acidic pH at 48 h of growth
| Metabolite | WT | Experimental data (pmol/pmol Pi) |
Model predictions (pmol/pmol Pi) |
||
|---|---|---|---|---|---|
| GAL7:IPC1 | Δisc1 | GAL7:IPC1 | Δisc1 | ||
| Phytoceramide C26 (X9) | 222.5 | 523.3 | 10 | 465.08 | 9.37 |
| Phytoceramide C18 (X11) | 215.3 | 114.6 | 235 | 179.65 | 173.80 |
| ATP (X17)a | 82.48 | 80.36 | 16.92* | 82.48 | 29.17 |
apmol/μg protein.
*P<0.05, Δisc1 versus WT.
Phytoceramide C26 level is increased when Ipc1 is downregulated in the GAL7:IPC1-glucose strain compared to the control WT strains at pH 4. Values are concentrations in pmol phytoceramide C18 or C26/pmol phosphate (Pi). Results shown are the means of three independent experiments. ATP is significantly lower in the Δisc1 mutant compared to the control strain (WT) at pH 4.
According to mass spectrometric measurements, Cn WT produces more phytoceramide C26 in acidic compared to neutral conditions (Table I), but the same amounts of DAG (data not shown). Our early experiments (see ‘Experimental studies identifying Ipc1, Isc1, and phytoceramide as drivers of pH response') had suggested that both Ipc1 and Isc1 regulate Pma1 function. Moreover, it seems that the level of very long-chain phytoceramides is highly regulated by Isc1 but not by Ipc1 (Table IV). These experimental findings were tested against model predictions. For technical reasons, these tests were most reliably executed by comparing model results with experimental data for ATP and lipid measurements.
Table 4.
Experimental measurements of phytoceramides C18 and C26 in Cn WT, GAL7:IPC1, and Δisc1 mutant strains during in vitro growth at neutral and acidic pH
| Parameter | 12 h |
24 h |
48 h |
||||||
|---|---|---|---|---|---|---|---|---|---|
| WT | GAL7:IPC1 | Δisc1 | WT | GAL7:IPC1 | Δisc1 | WT | GAL7:IPC1 | Δisc1 | |
| pH 7 | |||||||||
| Phyto-C18 | 482 | 1333 | 430 | 517 | 1090 | 530 | 406.6 | 900 | 980 |
| Phyto-C26 | 225 | 244 | 75* | 280 | 263 | 40* | 170 | 245 | 150 |
| PH 4 | |||||||||
| Phyto-C18 | 207 | 45.2 | 180 | 220 | 96.6 | 240 | 215.3 | 114.6 | 235 |
| Phyto-C26 | 340 | 320 | 20* | 443 | 470 | 10* | 222.5 | 523.3 | 10* |
Ipc1 is downregulated in the GAL7:IPC1-glucose strain and Isc1 is deleted in the Δisc1 strain. Downregulation of Ipc1 shows an increase of phytoceramide C18 (phyto-C18) at neutral pH. In contrast, phytoceramide C26 (phyto-C26) is slightly increased in GAL7:IPC1 compared to WT strain at acidic pH but only at 48 h of growth.
The level of phytoceramide C26 is significantly reduced in the Δisc1 mutant compared to the WT strain (*P<0.05). Measurements of phytoceramides C18 and C26 were also performed in the Δisc1-reconstituted strain, and no significant differences were found in comparison with the WT strain (data not shown). Values are concentrations in pmol phytoceramide C18 or C26/pmol phosphate (Pi). Results shown are the means of three independent experiments.
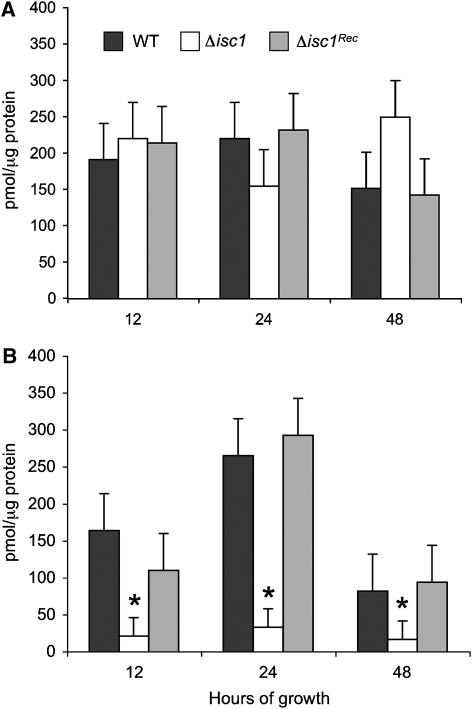
As ATP is required for Pma1 activity, we wondered whether deletion of Isc1 or downregulation of Ipc1 would affect the intracellular level of ATP. We found that the ATP level is dramatically reduced when Isc1 is deleted, compared to WT strain, but only in acidic and not neutral pH at 12, 24, and 48 h of growth (P<0.05) (Figure 4). Interestingly, downregulation of Ipc1 does not affect ATP levels at neutral (data not shown) or acidic conditions (Table III). The model predicts a 65% decrease in the ATP level in the Isc1 mutant, and the experimental data show an 80% decrease. At the same time, the model predicts no change in the ATP level upon Ipc1 downregulation, and the experimental results show no changes in ATP when Ipc1 activity is downregulated (Table III and Figure 4).
Figure 4.
Measurement of intracellular ATP in Δisc1 mutant and control strains during growth at neutral and acidic pH. (A) Production of ATP is not impaired in the Δisc1 strain when exposed to a neutral pH environment. (B) Production of ATP is significantly impaired in the Δisc1 strain when exposed to a low-pH environment. Values are reported as pmol/μg protein. Results are means±s.d. of three separate experiments. *P<0.05, Δisc1 versus WT.
Next, we performed lipid measurements in conditions where Ipc1 was downregulated or Isc1 deleted. Loss of Isc1 leads to a specific depletion of phytoceramide C26, especially at pH 4, whereas no changes are observed with phytoceramide C18 (Table IV) or other subspecies (data not shown). Interestingly, loss of Isc1 leads to an increase of C16:1/18:1-DAG in neutral but not acidic pH (data not shown). Downregulation of Ipc1 increases phytoceramide C26 compared to the WT strain at acidic pH but only at 48 h of growth (Table IV). At neutral pH, phytoceramide C26 does not change when Ipc1 is downregulated compared with WT (Table IV). The total DAG level decreases when Ipc1 is downregulated (Heung et al, 2004, 2005), especially in the form of the Di-C16 DAG subspecies (data not shown). These results suggest that Isc1 specifically produces phytoceramide C26, which implies that it prefers IPC-C26 and/or C26 MIPC and/or C26-MIP2C compared to other complex sphingolipid substrates. By contrast, Ipc1 seems to use mainly phytoceramide C18 as a substrate and it appears that a rearrangement of phytoceramide species will occur at low pH (Table IV).
Finally, we measured the activity of Pma1 in Cn WT and mutant strains grown at pH 7.0 or 4.0. Pma1 activity was measured in extracted cell membranes by using an established protocol previously described for Cn (Soteropoulos et al, 2000). We then calculated Vmax and KM of Pma1 using the Lineweaver–Burk method, which linearizes the Michaelis–Menten equation by taking the reciprocal of both sides of the equation 1/v0=1/Vmax+(KM/Vmax)(1/[S])). We found that the KM of Pma1 increased by ∼9- and ∼7-fold in Δisc1 and GAL7:IPC1 mutants grown at pH 4.0, respectively, compared to WT or Δisc1-Rec, whereas no significant changes were observed in Vmax (Table V). A slightly increased value of KM in the mutants was also present when strains were grown at pH 7.0, but the difference was not significant (data not shown). These results suggest that the affinity of Pma1 for ATP is dramatically reduced when Isc1 is deleted or Ipc1 downregulated.
Table 5.
Experimental measurement of KM and Vmax of Pma1 in Cn WT, Δisc1, Δisc1-reconstituted (Δisc1-Rec), and GAL7:IPC1 strains in cell membranes extracted from cells grown at pH 4.0
| pH 4.0 |
||
|---|---|---|
| Vmax (nmol Pi/μg/min) | KM (mM) | |
| WT | 3.58 | 0.65 |
| Δisc1 | 4.58 | 5.96 |
| Δisc1-Rec | 3.18 | 0.84 |
| GAL7:IPC1 | 4.64 | 6.21 |
KM of Pma1 in Δisc1 and GAL7:IPC1 strains increased by ∼9- and 7-fold respectively compared to the KM of Pma1 in WT or Δisc1-Rec strains. No significant changes were observed in Vmax. Also, no significant changes in Vmax and KM were observed when strains were grown at pH 7.0 (data not shown).
Perhaps the oligomerization of Pma1 is impaired in the mutants compared to control strains and this effect is particularly present at pH 4.0. These results strongly validate our model in which very long-chain phytoceramides, such as the product of Isc1 phytoceramide C26, are predicted to regulate Pma1 oligomerization. They also suggest, however, that very long-chain phytoceramides are not the only regulators of Pma1 because Pma1 activity is also impaired in the GAL7:IPC1 strain in which phytoceramide C26 level is normal.
Discussion
This article presents a mathematical model that provides rationale for some expected and some counterintuitive changes in the sphingolipid pathway of Cn under neutral and acidic environments. The model focuses in particular on the metabolism of phytoceramides and predicts specifically that certain phytoceramide subspecies (C26) play a key role in the regulation of adaptation of Cn to low pH. To our knowledge, this is the first mathematical model proposed for Cn pathogenesis. While no model can be proved to be ‘correct,' our results indicate that the model is quite robust and accurate because all model predictions tested so far were validated successfully.
We experimentally confirmed that the concentration of phytoceramides is affected when cells are shifted from a neutral to an acidic environment. In acidic pH, the phytoceramide C26 subspecies increase compared to other subspecies and the production of phytoceramide C26 at low pH appears to be dependent on Isc1 activity. In conditions in which most Isc1 activity is lost, phytoceramide C26 cannot be formed and the intracellular ATP concentration is low. As a consequence, Pma1 activity is impaired by decreasing the affinity to the substrate. Under these conditions, cells exposed to an acidic environment do not die but, interestingly, need additional time to adapt before they can grow.
The transformation of complex sphingolipids into phytoceramide by the action of Isc1 is a process that mainly occurs in the ER but can also occur in other organelles. For instance, it was suggested that, in addition to ER localization, Isc1 in Sc may also be localized in the mitochondria (Vaena de Avalos et al, 2004). This localization hypothesis is supported by mammalian studies in which sphingomyelinase, the counterpart of Isc1 in mammalian cells, is localized in the mitochondria in addition to the ER (Birbes et al, 2001, 2005). In addition, recent studies showed that Isc1 produces phytoceramides in mitochondria, in particular α-hydroxylated phytoceramide C26 but also other species such as non-hydroxylated phytoceramides C18, C24, and C26 (Kitagaki et al, 2007). Production of these sphingolipids by Isc1 implies that IPCs formed in the Golgi apparatus are transported to the ER. Currently, it is not known whether complex sphingolipid transport occurs, but since the transport of phospholipids between mitochondria and the ER can occur through membrane association (Achleitner et al, 1999), it is possible that IPCs could be transported as well.
In the mitochondrial membrane, the degradation of IPCs into phytoceramides may have important consequences for the membrane potential and the generation of ATP. In our system, the reduction of ATP level in the absence of Isc1 suggests a potential role of Isc1 in mitochondria in the regulation of ATP synthesis.
In conditions in which Ipc1 is downregulated, yeast cells also need additional time to adapt but this initial adaptation appears to be independent of phytoceramide C26. Indeed, the level of phytoceramide C26 does not decrease upon downregulation of Ipc1 but, instead, increases at low pH. Thus, it is proposed that Ipc1 regulates growth at low pH and Pma1 activity through the formation of IPC and/or DAG. IPC is a complex sphingolipid that is important for the integrity of the plasma membrane, whereas DAG produced by Ipc1 is important for the activation of Pkc1 and the consequent cell wall integrity (Heung et al, 2004, 2005). Cell wall integrity, regulated by the Ipc1–DAG–Pkc1 pathway, is required for the proper function of the laccase enzyme in Cn (Heung et al, 2005) and it may also be important for the proper localization and/or oligomerization of Pma1 in the plasma membrane.
On the other hand, a relationship between Pma1 biogenesis and lipid synthesis is indicated by a number of observations. Lipids containing C26 fatty acids, either phytoceramide or glycerophospholipid bound, are important for stable biogenesis of Pma1 (Toulmay and Schneiter, 2006, 2007). Phytoceramide synthesis is also required for oligomerization and raft association of Pma1 in the ER (Lee et al, 2002; Wang and Chang, 2002), and oligomerization of Pma1 could be important for stabilization of the protein once it has reached the cell surface (Wang and Chang, 2002). Other studies also suggest that the synthesis of complex sphingolipids is critically important for Pma1 biogenesis (Gaigg et al, 2005). Indeed, in Sc, a decrease in proton extrusion by Pma1 is observed in the absence of complex sphingolipids, suggesting that IPC, MIPC, and M(IP)2C may regulate Pma1 function (Achleitner et al, 1999). The fact that Isc1 and Ipc1 activities are required for Pma1 stability is supported by the observation that, in conditions in which Isc1 is deleted or Ipc1 downregulated, the mutant is hypersensitive to ebselen and the KM of Pma1 significantly increased. Our results are in concordance with yeast studies by Patton et al (1992), in which sphingolipid deficiency causes a deficiency in net proton extrusion under acidic pH. More recent studies show that if Pma1 is not directly complexed with very long-chain phytoceramide, its function may be lost and the cells are unable to grow at low pH (Gaigg et al, 2006).
Based on our simulations, the model suggests that Pma1 activity decreases when Isc1 is deleted or Ipc1 is downregulated, and our experimental findings demonstrated this model prediction. Specifically, the model suggests that Isc1 and Ipc1 regulate Pma1 through different mechanisms: Isc1 through phytoceramide C26 and Ipc1 through IPC and/or DAG. Under acidic conditions, the cells require additional production of phytoceramide C26. This is readily achievable by metabolizing IPC-C26 into phytoceramide C26 by the action of Isc1. Under conditions in which Isc1 is deleted, the cells need more time to adapt to the acidic environment and to grow successfully. Eventually they do, perhaps through the overproduction of specific subspecies of DAG, which was observed at 24 and 48 h when Isc1 is deleted. This overproduction of DAG and the consequent stabilization of cell wall integrity by Pkc1 may eventually compensate for the lack of phytoceramide C26 and the dysfunction of Pma1.
Under conditions in which Ipc1 is downregulated, the cells also need more time to adapt to the acidic environment before they can restore growth. In these cases, IPC and DAG levels decrease and phytoceramide level increases (Heung et al, 2005). Interestingly, phytoceramide C18 increases at neutral pH, whereas phytoceramide C26 increases at 48 h of incubation at low pH (Table IV). This increase could reflect a compensatory mechanism triggered by the Ipc1 mutant strain to restore growth under low pH. Indeed, phytoceramide C26 increases only at low but not neutral pH. Perhaps, Isc1, which according to our data is the key regulator of phytoceramide C26 level, compensates by increasing its activity in conditions where Ipc1-downregulated cells suffer from the lack of IPC and/or DAG. The model also predicts an increase in PHS during Ipc1 downregulation at mid- to late-log phase and under acidic external conditions (data not shown). Indeed, accumulation of PHS was also observed experimentally when Ipc1 was downregulated (data not shown), suggesting that other mechanisms for the Ipc1–Pma1 regulation may exist in addition to IPC and/or DAG.
The analysis of which genes and enzymes associated with sphingolipid and other pathways are most important for fungal cell homeostasis under certain growth conditions may have important medical implications. For instance, one could evaluate the level of expression of a specific gene or enzyme in fungal cells isolated from patients and establish to what degree the in vivo expression would differ in different patients. During infection, Cn cells may reside extracellularly and intracellularly, and the percentage of Cn cells in each compartment is most likely to vary for a number of reasons (e.g., expression of specific fungal virulence factors, such as capsule, melanin, glucosylceramide, antiphagocytic protein 1 (App1), phospholipase B1 (Plb1), urease, availability of host cells for intracellular growth, and the status of the host immune response). In cases of host immunodeficiency, Cn cells may prefer to reside in one compartment (e.g., intracellular) instead of the other (extracellular), and gene expression and/or protein activities in intracellular Cn may be completely different compared to extracellular Cn (Fan et al, 2005). Thus, if information related to a specific gene expression or protein activity could be evaluated by a mathematical model in cells recovered from a patient, then we could predict the overall fitness of the fungus during a particular time of infection. For instance, overexpression of Isc1 or an elevated level of phytoceramide C26 would predict an intracellular localization of Cn, whereas overexpression of glucosylceramide synthase and downregulation of Ipc1 and/or Isc1 would predict an extracellular localization. Such an analysis might have important implications for the outcome of the infection because we know that the different localization (intracellular versus extracellular) of fungal cells may affect the pathogenesis of Cn infection (Rittershaus et al, 2006; Shea et al, 2006) and its susceptibility to antifungal treatment. For instance, chloroquine increases the efficacy of fluconazole by alkalinizing the intracellular compartment and favoring the extrusion of Cn cells into the extracellular space (Mazzolla et al, 1997; Khan et al, 2004; Alvarez and Casadevall, 2006; Ma et al, 2006). Thus, mathematical models accounting for Cn gene expression profiles and enzymatic activities at a specific time of the infection may help to define the context (host immunity) and the space (extracellular or intracellular) in which the infection occurs in a specific patient. It is through a combination of targeted, reductionistic molecular biology and system-wide computational biology that this goal may be achieved.
Materials and methods
Biochemical systems theory
BST has been reviewed numerous times in the recent literature (Voit, 2000; Torres and Voit, 2002), and we will discuss only its most salient features. The starting point is the description of temporal changes in each of the dependent variables Xi in a biochemical system with the general system equation
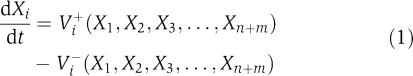 |
Here, Vi+ and Vi− are positive valued functions that may involve some or all of the system variables X1, X2, …, Xn+m. The first n variables typically change over time, whereas the remaining m are considered constant during each mathematical experiment, but may change from one experiment to the next. The key feature of BST is the representation of the functions Vi+ and Vi− as products of power-law functions (Savageau, 1969a, 1969b). In the so-called S-system form, which we will use here, the dynamics of each dynamic variable is thus formulated as
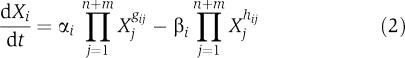 |
The parameters αi and βi are non-negative rate constants, and gij and hij are real-valued kinetic orders. The parameters αi and gij are associated with the aggregate rate law Vi+ for synthesis or augmentation of Xi, whereas βi and hij are associated with the aggregate rate law Vi− for degradation or elimination of Xi.
Details of model design, estimation of parameter values, and conversions between this and other model types are presented in Supplementary information. Simulations, stability and sensitivity analysis, as well as the assessment of gains were executed with the free software PLAS© (Ferreira, 2000). Results that are not of prime interest here are presented in Supplementary information. Suffice it to say that the model has a stable steady state, that it is overall very robust, and that no obvious reasons to worry about its properties were detected in extensive diagnostic studies.
Strains and growth conditions
Cn var. grubii serotype A strain H99 (WT), Cn GAL7:IPC1 (Luberto et al, 2001) strain, Δisc1 mutant, and the Δisc1+ISC1 (Δisc1REC) strains (Shea et al, 2006) were used in this study. Strains were routinely grown in yeast extract/peptone/dextrose (YPD) (Difco) medium buffered to pH 7.0 or 4.0 with 25 mM HEPES. Yeast nitrogen base (YNB) was composed of 6.7 g/l yeast nitrogen base (Difco) with amino acids and 5 g/l glucose. To downregulate Ipc1, the GAL7:IPC1 strain was grown on glucose medium as previously described (Luberto et al, 2001; Mare et al, 2005).
Extraction and analysis of yeast sphingolipids
Strains were grown in YPD at pH 7.0 for 24 h, washed twice with sterile distilled water, and inoculated into fresh YPD pH 7.0 or 4.0 media. At appropriate time points (0, 6, 12, 24, and 48 h), cells were collected by centrifugation at 3000 r.p.m. for 10 min, washed twice with sterile distilled water, and the pellets were stored at −80°C. Neutral lipids were extracted by the method of Bligh and Dyer (1959). An aliquot of the extraction (300 μl) was used for phosphorous determination (Perry et al, 2000). Internal standards were added to the remaining aliquots, and sphingolipids were extracted in a one-phase neutral organic solvent (propan-2-ol/water/ethyl acetate, 30:10:60, v/v). Samples were then analyzed by a Surveyor/TSQ 7000 liquid chromatography–MS system. Lipids were qualitatively defined by parent-ion scanning for known fragments characteristic for a specific sphingolipid class, including sphingoid bases, ceramides, and phytoceramides. For all experiments, source ion optics was adjusted to accomplish desolvation of ions while minimizing fragmentation of ions in the inlet region of the mass spectrometer (Pettus et al, 2004). Samples were quantitatively analyzed on the basis of calibration curves generated with synthetic standards. The mass of each species was normalized to phosphorous levels of each sample.
Growth inhibition
Experiments to determine MICs and MFCs were performed by the broth microdilution method according to the recommendations of the National Committee for Clinical Laboratory Standards (NCCLS and Stardards, 1997). Cn strains were grown in YPD (pH 7.0) for 24 h at 30°C. Cells were washed twice with sterile deionized water, and the density was adjusted to a final concentration of 2.5 × 103 cells/ml in YNB with 25 mM HEPES at a pH of 7.0 or 4.0. Ebselen was obtained from Sigma. The MIC was defined as the lowest drug concentration in which a visual turbidity of ⩽80% inhibition was observed compared to that produced by the growth control. One hundred microliter aliquots from wells with growth inhibition were plated onto YPD agar plates. The lowest concentration that yielded three or fewer colonies was recorded as the MFC.
Measurement of ATP level
Intracellular ATP concentrations were determined by using a bioluminescent ATP assay kit (Sigma), which measures the conversion of luciferin to light by firefly luciferase in the presence of ATP. The light emitted is proportional to the ATP present and was measured at 490 nm using a luminometer (Berthold Australia Pty Ltd, Bundoora, Australia). Cn WT, Δisc1, Δisc1REC, or GAL7:IPC1 strains were grown in 40 ml of YNB containing 2% glucose with 25 mM HEPES (pH 7.0 or 4.0) for 12, 24, or 48 h. Cells were washed twice with sterile deionized water. A pellet containing 5 × 108 cryptococcal cells was flash-frozen and stored at −80°C. Pellets were resuspended in 800 μl of 50 mM HEPES (pH 7.75) and 200 μl DMSO. Acid-washed glass beads were added and cells were homogenized five times for 45 s each using the Bead-Beader-8, with samples being kept on ice for at least 1 min between homogenization cycles. After centrifugation at 2500 g for 10 min at 4°C, the supernatant was collected to determine ATP concentrations according to the manufacturer's instructions. Protein concentration of an aliquot of the supernatant was measured by the Bradford method and was used to normalize ATP determination to control for the extraction process.
Pma1 activity
Plasma membranes were isolated from Cn WT, Δisc1, Δisc1-Rec, and GAL7:IPC1 strains grown in 250 ml YNB containing 2% dextrose with 25 mM HEPES (pH 4 or 7) for 24 h. The membranes for each strain were isolated by the procedure described by Wang et al (1996). Specifically, the cells were resuspended in 25 ml homogenization buffer consisting of 100 mM Tris–HCl, pH 7.5, 5 mM EDTA, 5 mg/ml bovine serum albumin, and 1 mM phenylmethylsulfonyl fluoride, which was added just before use. The cells were passed through a French pressure cell at 20 000 p.s.i. at 4°C. The lysate was adjusted to pH 7 with 1 M HCl and centrifuged at 10 000 g for 10 min. The resulting supernatant was centrifuged at 100 000 g for 60 min and the pellet was resuspended in 3 ml extraction buffer consisting of 10 mM HEPES–KOH, pH 7.0, 100 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol, and 0.45% (w/v) glycerol by sonication for 3 × 15 s (15% amplitude). Deoxycholate (10% w/v stock) was added to the suspension on ice to a final concentration of 0.5% w/v. This suspension was then centrifuged at 150 000 g for 1 h. The pellet was washed by resuspension (via sonication) in extraction buffer (3 ml) and centrifuged at 150 000 g for 1 h. The final pellet was resuspended in 1.5 ml extraction buffer. Protein determination was carried out using the Bradford method with bovine serum albumin as the standard. ATPase assays were conducted in 96-well microtiter plates similar to the method described by Wang et al (1996). Briefly, a 125 μl assay mixture contained 10 mM MES–Tris, pH 6.5, 0–14 mM MgSO4, 25 mM NH4Cl, 0–14 mM ATP, and 0.6–1 μg membrane protein. Samples were incubated at 30°C for 15 min and inorganic phosphate released from ATP was determined by the addition of 125 μl of phosphate developing reagent, consisting of 0.6 M H2SO4, 9% L-ascorbic acid, and 0.9% ammonium molybdate. The absorbance at 820 nm was determined after 10 min incubation at room temperature. The amount of phosphate liberated was estimated by using a linear standard curve in the range 0–100 μM NaH2PO4. All KM and Vmax values were obtained by determining ATP hydrolysis as a function of substrate concentration (0–14 mM for both ATP and MgSO4) and the data were fit to the Michaelis–Menten equation.
Supplementary Material
Supplementary Information
Acknowledgments
We are grateful to Drs Alicja Bielawska, Jacek Bielawski, Zdzislaw Szulc, and the Lipidomics Core Facility for sphingolipid analysis. We also thank all members of Del Poeta's and Luberto's laboratories for sharing data and materials. This work was supported in part by the Burroughs Welcome Fund, in part by Grants AI56168 and AI72142 (to MDP) from the National Institutes of Health, in part by RR17677 Project 2 (to MDP) and Project 6 (to CL) from the Centers of Biomedical Research Excellence Program of the National Center for Research Resources, in part by the National Science Foundation/EPSCoR Grant EPS-0132573 to CL, and in part by NIH C06 RR015455 from the Extramural Research Facilities Program of the National Center for Research Resources. Dr M Del Poeta is a Burroughs Wellcome New Investigator in Pathogenesis of Infectious Diseases.
References
- Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G (1999) Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem 264: 545–553 [DOI] [PubMed] [Google Scholar]
- Ahn AC, Tewari M, Poon CS, Phillips RS (2006a) The clinical applications of a systems approach. PLoS Med 3: e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn AC, Tewari M, Poon CS, Phillips RS (2006b) The limits of reductionism in medicine: could systems biology offer an alternative? PLoS Med 3: e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhter S, McDade HC, Gorlach JM, Heinrich G, Cox GM, Perfect JR (2003) Role of alternative oxidase gene in pathogenesis of Cryptococcus neoformans. Infect Immun 71: 5794–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez M, Casadevall A (2006) Phagosome extrusion and host-cell survival after Cryptococcus neoformans phagocytosis by macrophages. Curr Biol 16: 2161–2165 [DOI] [PubMed] [Google Scholar]
- Alvarez-Vasquez F, Sims KJ, Cowart LA, Okamoto Y, Voit EO, Hannun YA (2005) Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature 433: 425–430 [DOI] [PubMed] [Google Scholar]
- Alvarez-Vasquez F, Sims KJ, Hannun YA, Voit EO (2004) Integration of kinetic information on yeast sphingolipid metabolism in dynamical pathway models. J Theor Biol 226: 265–291 [DOI] [PubMed] [Google Scholar]
- Berne S, Sepcic K, Anderluh G, Turk T, Macek P, Poklar Ulrih N (2005) Effect of pH on the pore forming activity and conformational stability of ostreolysin, a lipid raft-binding protein from the edible mushroom Pleurotus ostreatus. Biochemistry 44: 11137–11147 [DOI] [PubMed] [Google Scholar]
- Birbes H, El Bawab S, Hannun YA, Obeid LM (2001) Selective hydrolysis of a mitochondrial pool of sphingomyelin induces apoptosis. FASEB J 15: 2669–2679 [DOI] [PubMed] [Google Scholar]
- Birbes H, Luberto C, Hsu YT, El Bawab S, Hannun YA, Obeid LM (2005) A mitochondrial pool of sphingomyelin is involved in TNFalpha-induced Bax translocation to mitochondria. Biochem J 386: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ (1959) A rapid method for total lipid extraction and purification. Can J Biochem Physiol 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Buede R, Rinker-Schaffer C, Pinto WJ, Lester RL, Dickson RC (1991) Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J Bacteriol 173: 4325–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR (1998) Cryptococcus neoformans, pp 381–405. Washington, DC: ASM Press [Google Scholar]
- Chang A, Slayman CW (1991) Maturation of the yeast plasma membrane [H+]ATPase involves phosphorylation during intracellular transport. J Cell Biol 115: 289–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson RC, Lester RL (1999) Metabolism and selected functions of sphingolipids in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta 1438: 305–321 [DOI] [PubMed] [Google Scholar]
- Durand T, Delmas-Beauvieux MC, Canioni P, Gallis JL (1999) Role of intracellular buffering power on the mitochondria-cytosol pH gradient in the rat liver perfused at 4 degrees C. Cryobiology 38: 68–80 [DOI] [PubMed] [Google Scholar]
- Fan W, Kraus PR, Boily MJ, Heitman J (2005) Cryptococcus neoformans gene expression during murine macrophage infection. Eukaryot Cell 4: 1420–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmesser M, Tucker S, Casadevall A (2001) Intracellular parasitism of macrophages by Cryptococcus neoformans. Trends Microbiol 9: 273–278 [DOI] [PubMed] [Google Scholar]
- Ferreira A (2000) Power Law Analysis and Simulation (PLAS), http://www.dqb.fc.ul.pt/docentes/aferreira/plas.html [Google Scholar]
- Fronzes R, Chaignepain S, Bathany K, Giraud MF, Arselin G, Schmitter JM, Dautant A, Velours J, Brethes D (2003) Topological and functional study of subunit h of the F1Fo ATP synthase complex in yeast Saccharomyces cerevisiae. Biochemistry 42: 12038–12049 [DOI] [PubMed] [Google Scholar]
- Funato K, Riezman H (2001) Vesicular and nonvesicular transport of ceramide from ER to the Golgi apparatus in yeast. J Cell Biol 155: 949–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futerman AH, Hannun YA (2004) The complex life of simple sphingolipids. EMBO Rep 5: 777–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaigg B, Timischl B, Corbino L, Schneiter R (2005) Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. J Biol Chem 280: 22515–22522 [DOI] [PubMed] [Google Scholar]
- Gaigg B, Toulmay A, Schneiter R (2006) Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. J Biol Chem 281: 34135–34145 [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C (2000) Ceramide in the eukaryotic stress response. Trends Cell Biol 10: 73–80 [DOI] [PubMed] [Google Scholar]
- Hechtberger P, Zinser E, Saf R, Hummel K, Paltauf F, Daum G (1994) Characterization, quantification and subcellular localization of inositol-containing sphingolipids of the yeast, Saccharomyces cerevisiae. Eur J Biochem 225: 641–649 [DOI] [PubMed] [Google Scholar]
- Heidler SA, Radding JA (2000) Inositol phosphoryl transferases from human pathogenic fungi. Biochim Biophys Acta 1500: 147–152 [DOI] [PubMed] [Google Scholar]
- Heung LJ, Kaiser AE, Luberto C, Del Poeta M (2005) The role and mechanism of diacylglycerol-protein kinase C1 signaling in melanogenesis by Cryptococcus neoformans. J Biol Chem 280: 28547–28555 [DOI] [PubMed] [Google Scholar]
- Heung LJ, Luberto C, Plowden A, Hannun YA, Del Poeta M (2004) The sphingolipid pathway regulates protein kinase C 1 (Pkc1) through the formation of diacylglycerol (DAG) in Cryptococcus neoformans. J Biol Chem 279: 21144–21153 [DOI] [PubMed] [Google Scholar]
- Khan MA, Jabeen R, Mohammad O (2004) Prophylactic role of liposomized chloroquine against murine cryptococcosis less susceptible to fluconazole. Pharm Res 21: 2207–2212 [DOI] [PubMed] [Google Scholar]
- Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH, Bielawski J, Obeid LM, Hannun YA (2007) Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim Biophys Acta 1768: 2849–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M, Hashida-Okado T, Yasumoto R, Gomi K, Kato I, Takesako K (1999) An aureobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphorylceramide (IPC) synthase activity. Mol Gen Genet 261: 290–296 [DOI] [PubMed] [Google Scholar]
- Lee MC, Hamamoto S, Schekman R (2002) Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. J Biol Chem 277: 22395–22401 [DOI] [PubMed] [Google Scholar]
- Levine TP, Wiggins CAR, Munro S (2000) Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell 11: 2267–2281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Zhang X, Sumanasekera C, Lester RL, Dickson RC (2005) Signalling functions for sphingolipid long-chain bases in Saccharomyces cerevisiae. Biochem Soc Trans 33: 1170–1173 [DOI] [PubMed] [Google Scholar]
- Luberto C, Hannun YA (1999) Sphingolipid metabolism in the regulation of bioactive molecules. Lipids 34: S5–11 [DOI] [PubMed] [Google Scholar]
- Luberto C, Toffaletti DL, Wills EA, Tucker SC, Casadevall A, Perfect JR, Hannun YA, Del Poeta M (2001) Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev 15: 201–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Croudace JE, Lammas DA, May RC (2006) Expulsion of live pathogenic yeast by macrophages. Curr Biol 16: 2156–2160 [DOI] [PubMed] [Google Scholar]
- Mao C, Obeid LM (2000) Yeast sphingosine-1-phosphate phosphatases: assay, expression, deletion, purification, and cellular localization by GFP tagging. Methods Enzymol 311: 223–232 [DOI] [PubMed] [Google Scholar]
- Mao C, Xu R, Bielawska A, Obeid LM (2000) Cloning of an alkaline ceramidase from Saccharomyces cerevisiae. An enzyme with reverse (CoA-independent) ceramide synthase activity. J Biol Chem 275: 6876–6884 [DOI] [PubMed] [Google Scholar]
- Mare L, Iatta R, Montagna MT, Luberto C, Del Poeta M (2005) APP1 transcription is regulated by IPC1–DAG pathway and is controlled by ATF2 transcription factor in Cryptococcus neoformans. J Biol Chem 280: 36055–36064 [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Reed JC (2000) Mitochondria-dependent apoptosis and cellular pH regulation. 7: 1155–1165 [DOI] [PubMed]
- Mazzolla R, Barluzzi R, Brozzetti A, Boelaert JR, Luna T, Saleppico S, Bistoni F, Blasi E (1997) Enhanced resistance to Cryptococcus neoformans infection induced by chloroquine in a murine model of meningoencephalitis. Antimicrob Agents Chemother 41: 802–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuiston TJ, Haller C, Del Poeta M (2006) Sphingolipids as targets for microbial infections. Mini Rev Med Chem 6: 671–680 [DOI] [PubMed] [Google Scholar]
- NCCLS, Standards NCfCL (1997) Reference Method for Broth Dilution Susceptibility Testing of Yeasts. Approved Standard M27-A. Wayne, PA: National Committee for Clinical Laboratory Standards [Google Scholar]
- Patton JL, Srinivasan B, Dickson RC, Lester RL (1992) Phenotypes of sphingolipid-dependent strains of Saccharomyces cerevisiae. J Bacteriol 174: 7180–7184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR (2005) Cryptococcus neoformans: a sugar-coated killer with designer genes. FEMS Immunol Med Microbiol 45: 395–404 [DOI] [PubMed] [Google Scholar]
- Perlin DS, Seto-Young D, Monk BC (1997) The plasma membrane H(+)-ATPase of fungi. A candidate drug target? Ann NY Acad Sci 834: 609–617 [DOI] [PubMed] [Google Scholar]
- Perona R, Portillo F, Giraldez F, Serrano R (1990) Transformation and pH homeostasis of fibroblasts expressing yeast H(+)-ATPase containing site-directed mutations. Mol Cell Biol 10: 4110–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry DK, Bielawska A, Hannun YA (2000) Quantitative determination of ceramide using diglyceride kinase. Methods Enzymol 312: 22–31 [DOI] [PubMed] [Google Scholar]
- Pettus BJ, Kroesen BJ, Szulc ZM, Bielawska A, Bielawski J, Hannun YA, Busman M (2004) Quantitative measurement of different ceramide species from crude cellular extracts by normal-phase high-performance liquid chromatography coupled to atmospheric pressure ionization mass spectrometry. Rapid Commun Mass Spectrom 18: 577–583 [DOI] [PubMed] [Google Scholar]
- Portillo F, de Larrinoa IF, Serrano R (1989) Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett 247: 381–385 [DOI] [PubMed] [Google Scholar]
- Portillo F, Eraso P, Serrano R (1991) Analysis of the regulatory domain of yeast plasma membrane H+-ATPase by directed mutagenesis and intragenic suppression. FEBS Lett 287: 71–74 [DOI] [PubMed] [Google Scholar]
- Rittershaus PC, Kechichian TB, Allegood J, Merrill AHJ, Hennig M, Luberto C, Del Poeta M (2006) Glucosylceramide is an essential regulator of pathogenicity of Cryptococcus neoformans. J Clin Invest 116: 1651–1659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Takakuwa N, Ohnishi M, Oda Y (2006) Presence of glucosylceramide in yeast and its relation to alkali tolerance of yeast. Appl Microbiol Biotechnol 69: 1–7 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1969a) Biochemical systems analysis. I. Some mathematical properties of the rate law for the component enzymatic reactions. J Theor Biol 25: 365–369 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1969b) Biochemical systems analysis. II. The steady-state solutions for an n-pool system using a power-law approximation. J Theor Biol 25: 370–379 [DOI] [PubMed] [Google Scholar]
- Savageau MA (1976) Biochemical Systems Analysis. A Study of Function and Design in Molecular Biology. Reading, MA: Addison-Wesley [Google Scholar]
- Serrano R (1988) Structure and function of proton translocating ATPase in plasma membranes of plants and fungi. Biochim Biophys Acta 947: 1–28 [DOI] [PubMed] [Google Scholar]
- Serrano R, Kielland-Brandt MC, Fink GR (1986) Yeast plasma membrane ATPase is essential for growth and has homology with (Na++K+), K+- and Ca2+-ATPases. Nature 319: 689–693 [DOI] [PubMed] [Google Scholar]
- Shea J, Kechichian TB, Luberto C, Del Poeta M (2006) The cryptococcal enzyme inositol phosphosphingolipid-phospholipase C (Isc1) confers resistance to the antifungal effects of macrophages and promotes fungal dissemination to the central nervous system. Infect Immun 74: 5977–5988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soteropoulos P, Vaz T, Santangelo R, Paderu P, Huang DY, Tamas MJ, Perlin DS (2000) Molecular characterization of the plasma membrane H(+)-ATPase, an antifungal target in Cryptococcus neoformans. Antimicrob Agents Chemother 44: 2349–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres NV, Voit EO (2002) Pathway Analysis and Optimization in Metabolic Engineering. Cambridge, UK: Cambridge University Press [Google Scholar]
- Toulmay A, Schneiter R (2006) A two-step method for the introduction of single or multiple defined point mutations into the genome of Saccharomyces cerevisiae. Yeast 23: 825–831 [DOI] [PubMed] [Google Scholar]
- Toulmay A, Schneiter R (2007) Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie 89: 249–254 [DOI] [PubMed] [Google Scholar]
- Vaena de Avalos S, Okamoto Y, Hannun YA (2004) Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J Biol Chem 279: 11537–11545 [DOI] [PubMed] [Google Scholar]
- Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA (2005) The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem 280: 7170–7177 [DOI] [PubMed] [Google Scholar]
- Voit EO (2000) Canonical modeling: review of concepts with emphasis on environmental health. Environ Health Perspect 108 (Suppl 5): 895–909 [DOI] [PubMed] [Google Scholar]
- Wang G, Tamas MJ, Hall MJ, Pascual-Ahuir A, Perlin DS (1996) Probing conserved regions of the cytoplasmic LOOP1 segment linking transmembrane segments 2 and 3 of the Saccharomyces cerevisiae plasma membrane H+-ATPase. J Biol Chem 271: 25438–25445 [DOI] [PubMed] [Google Scholar]
- Wang Q, Chang A (2002) Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proc Natl Acad Sci USA 99: 12853–12858 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information