Abstract
The human protein interaction network will offer global insights into the molecular organization of cells and provide a framework for modeling human disease, but the network's large scale demands new approaches. We report a set of 7000 physical associations among human proteins inferred from indirect evidence: the comparison of human mRNA co-expression patterns with those of orthologous genes in five other eukaryotes, which we demonstrate identifies proteins in the same physical complexes. To evaluate the accuracy of the predicted physical associations, we apply quantitative mass spectrometry shotgun proteomics to measure elution profiles of 3013 human proteins during native biochemical fractionation, demonstrating systematically that putative interaction partners tend to co-sediment. We further validate uncharacterized proteins implicated by the associations in ribosome biogenesis, including WBSCR20C, associated with Williams–Beuren syndrome. This meta-analysis therefore exploits non-protein-based data, but successfully predicts associations, including 5589 novel human physical protein associations, with measured accuracies of 54±10%, comparable to direct large-scale interaction assays. The new associations' derivation from conserved in vivo phenomena argues strongly for their biological relevance.
Keywords: interactions, mass spectrometry, networks, proteomics, systems biology
Introduction
Although considerable progress has been made in mapping the protein interaction network of yeast (Ito et al, 2000, 2001; Uetz et al, 2000; Ho et al, 2002; Gavin et al, 2006; Krogan et al, 2006), only minimal progress has been made on the interaction networks of higher eukaryotes, due primarily to their scale: for the ∼20 000–25 000 human proteins, we expect a network of roughly 1–400 000 interactions (Hart et al, 2006). Among the few methods scaleable to this size, the yeast two-hybrid assay has proven the most successful, with maps of ∼20 000 interactions in fly (Giot et al, 2003), ∼4000 in worm (Li et al, 2004), and more recently, assays of ∼2800 and ∼3200 human protein interactions (Rual et al, 2005; Stelzl et al, 2005). Direct mapping of protein complexes by mass spectrometry has also contributed another ∼5000 interactions (Ewing et al, 2007). After including previously known human protein interactions (Bader et al, 2003; Lehner and Fraser, 2004; Peri et al, 2004; Joshi-Tope et al, 2005; Ramani et al, 2005), the human protein interaction map is currently perhaps 10–30% complete (Hart et al, 2006). It is therefore important to identify and employ methods for discovering interacting proteins without exhaustive experimental measurement of all pairs of proteins under each relevant condition or assay.
Proteins are evolved to interact under specific conditions in the cell, with the cell correspondingly optimized to facilitate these events, e.g. by expressing mRNAs before proteins are required, coordinating the expression of interacting partners, directing proteins to appropriate locations for their interactions, and so on. In this way, in vivo protein interactions are accompanied by corollary events that can be used to identify biologically relevant physical interaction partners.
We took advantage of two such corollary data types, the tendency for interacting proteins to have correlated mRNA expression patterns and the evolutionary conservation of such patterns, to identify new human protein interactions. It is well established that genes whose mRNA expression patterns are correlated across many diverse conditions can often be inferred to ‘work together', i.e. to be functionally coupled (Eisen et al, 1998; Marcotte et al, 1999; Stuart et al, 2003; Lee et al, 2004a; Segal et al, 2004). Analyses of co-expression patterns of orthologous genes have shown that the conserved correlation in expression can also be used to transfer functional information across species (Teichmann and Babu, 2002; Stuart et al, 2003; van Noort et al, 2003; Bergmann et al, 2004; Snel et al, 2004). Transcriptional co-expression patterns have proved useful for inferring physical protein interactions (e.g. Deane et al, 2002; Jansen et al, 2003), with strongly co-expressed mRNAs more likely to indicate long-lived interactions (Ge et al, 2001; Jansen et al, 2002; Simonis et al, 2006). In general, we do not expect transcriptional data to distinguish between direct protein binding and membership in the same protein complex, and we term all such cases physically associated proteins.
To exploit these trends, we applied a supervised algorithm to discover physical associations among human proteins based upon the co-expression of their mRNAs and that of their orthologs in five organisms (the mustard plant Arabidopsis thaliana, the mouse Mus musculus, the fly Drosophila melanogaster, the nematode Caenorhabditis elegans, and the yeast Saccharomyces cerevisiae). By this approach, we mapped 7000 predicted human protein physical associations, of which 5589 are new to this analysis.
Results
Predicting physically associated proteins from patterns of conserved co-expression
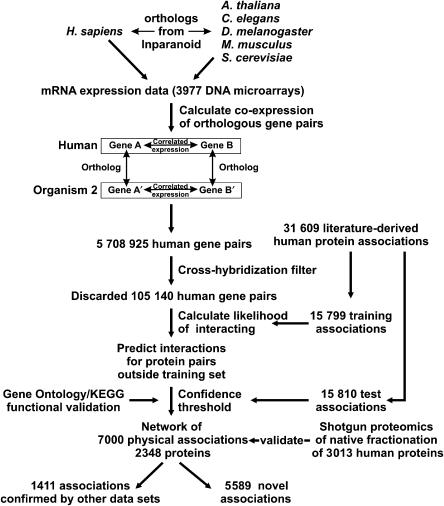
Figure 1 illustrates the overall method. We first identified orthologs for human genes in five other organisms using the InParanoid algorithm (Remm et al, 2001). We then compared the correlation in mRNA expression of each pair of human genes with the correlations in expression of each of their corresponding ortholog pairs from five organisms, in all calculating mRNA expression correlations for 5 708 925 human gene pairs on the basis of 3977 DNA microarrays. After removing 105 140 gene pairs likely to cross-hybridize on the microarrays (see Materials and methods) and filtering pairs with nonsignificant correlations, we employed a supervised algorithm on these data to identify those patterns of conserved co-expression (CCE) diagnostic of physical protein associations, based upon the correlations observed for known protein interactions versus random protein pairs. By searching for additional gene pairs exhibiting these patterns, we identified new associations.
Figure 1.
Overview of the analysis. From gene expression data for pairs of human genes and their orthologs, we identify proteins most likely to physically associate. For each pair of human genes, we compare the correlation in their mRNA expression patterns with the correlation in expression of their corresponding ortholog pairs, searching for patterns of conserved co-expression strongly enriched among physically associated proteins. By filtering the data to remove spurious associations (e.g. from microarray cross-hybridization and non-conserved expression regulation) and testing the associations against known human protein interactions and annotations, we predict 7000 human physical protein associations.
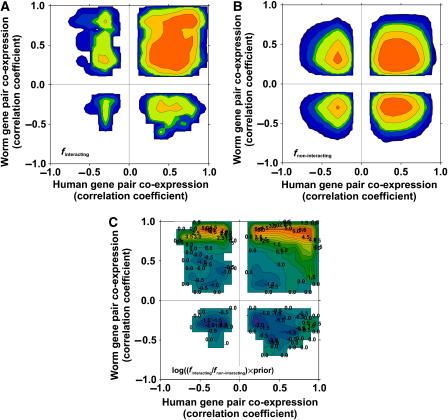
Figure 2 plots the derivation of the relationship between CCE and the tendency to be in the same physical complex, relying in this case on the comparison of human and C. elegans mRNA expression data. Briefly, the distribution of mRNA co-expression relationships was measured for 1769 gene pairs whose corresponding proteins are known to physically associate (Ramani et al, 2005), serving as positive training examples (Figure 2A); these 1769 pairs represent the subset of known human protein associations in the training set that also occur in the human–worm co-expression data sets. Likewise, the distribution was measured for 642 295 gene pairs that are in the physical interaction training set but are not known to physically associate, serving as a negative training set (Figure 2B). Therefore, the log ratio of these two plots, corrected by prior expectation, represents the log likelihood for protein pairs to physically associate given any particular pattern of co-expression conservation (Figure 2C):
Figure 2.
Predicting physically associated proteins from patterns of conserved co-expression. (A) Distribution of mRNA co-expression patterns of 1769 pairs of proteins that physically associate; (B) the distribution of co-expression patterns of 642 295 protein pairs that are not known to physically associate. By comparing the two distributions, we identify patterns that indicate the tendency to physically associate. In all panels, the x axis indicates the correlation of mRNA expression profiles of human gene pairs and the y axis the expression correlation of corresponding ortholog pairs in C. elegans. In (A, B), the z axis (represented as contours from purple (low) to red (high); white indicates zero) indicates the fraction of human gene pairs in either the true-positive (A) or -negative (B) set having a correlation ‘x' with C. elegans orthologs having a correlation ‘y'. (C) Log likelihood that human protein pairs with a given conserved co-expression pattern will physically interact, calculated as the logarithm of the ratio of the two distributions, corrected by prior expectation, and ranging from blue (negative) to red (positive) is plotted; white indicates zero. Contours are labeled with values of the log likelihood score.
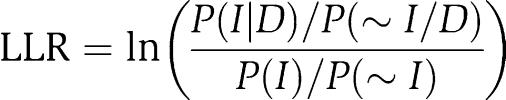 |
where P(I∣D) and P(∼I∣D) are the frequencies of positive (I) and negative (∼I) training associations observed in the data set (D), respectively, while P(I) and P(∼I) represent the overall frequencies of positive and negative training associations, respectively. This score indicates how likely two proteins are to physically associate given their specific mRNA co-expression conservation in these data. The training set includes both direct interactions and protein pairs belonging to the same complex; we therefore consider this approach to support the more general case, i.e. proteins belonging to the same complex whether or not directly interacting. Note that the highest scores do not necessarily occur in the extreme top right corner of Figure 2C; lower counts of both positive and negative examples in the extreme corner, as well as filtration of highly correlated gene pairs where they may suffer from DNA microarray cross-hybridization (see Materials and methods), results in the highest scores occurring at correlation coefficients less than one.
We similarly analyzed co-expression patterns of human gene pairs with orthologs from four other organisms (A. thaliana, M. musculus, D. melanogaster, and S. cerevisiae), analyzing 3977 DNA microarray experiments in all. From each analysis (Figure 2C and Supplementary Figure 1) strongly co-expressing human genes with co-expressing orthologs are generally likely to encode physically associated proteins. The highest likelihoods of associating occur when the mRNA expression patterns of both human gene pairs and their orthologs are positively correlated, with odds of associating approaching 460:1 for C. elegans, 200:1 for A. thaliana, 100:1 for M. musculus, 25:1 for D. melanogaster, and 400:1 for S. cerevisiae. These learned relationships between mRNA expression profiles and physical associations were then applied to protein pairs not in the training set, thereby assigning a likelihood of physically associating to each untested protein pair. Each human gene pair discovered has at least one log likelihood score, to a maximum of five, from which the highest score was identified; pairs were ranked based on this score, then evaluated as a function of their rank.
Validation of predicted physical protein associations using known interactions
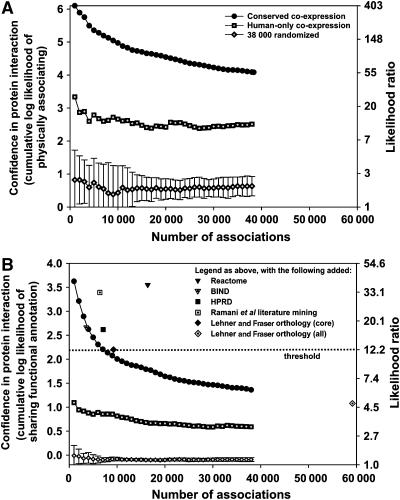
As this assay relies upon indirect evidence, it is critical that putative physical associations discovered by this approach be carefully evaluated. We devised six tests for the enrichment of true physical associations, including direct experimental assay of physical association and of four proteins' functions suggested by the associations. First and most critically, to verify the accuracy of the co-expression-derived associations, we measured their likelihood to physically associate using an independent test set of 15 810 known physical associations, including both direct interactions and complexes (Ramani et al, 2005). Figure 3A shows that the CCE associations are highly enriched for true physical associations, varying from a likelihood ratio of ∼60:1 to as high as ∼400:1 of correctly capturing true physical associations. Importantly, the CCE pairs score ∼25–200 times higher than randomized pairs of the same proteins, as well as associations derived in the same manner but using only human (not ortholog) DNA microarray data (Figure 3A). Therefore, the data from orthologs enriched the signal for human physical protein associations considerably beyond the human data alone.
Figure 3.
Two measurements of the quality of the derived physical protein associations. (A) The cumulative log likelihood ratio (LLR) of physically associating, measured with an independent test set of 15 810 human protein physical associations, plotted as a function of the number of associations. The CCE associations are significantly more enriched for known physical associations than randomized protein pairs or those derived only from human mRNA co-expression. The left y axis indicates the LLR score for the associations based on comparison to the known interaction test set; the right y axis indicates the corresponding likelihood ratio. Associations were ranked by confidence (see Materials and methods) and binned into sets of 1000 associations per bin for analysis. (B) The tendency for putative interaction partners to participate in the same pathway. The left y axis indicates the cumulative LLR (and the right y axis the corresponding likelihood ratio) for interaction partners to belong to the same pathway, using the same log likelihood framework as in (A), but employing as a positive test set the ∼1.5 million human protein pairs defined in the GO and KEGG databases as belonging to the same pathway. As in (A), CCE associations are comparable in quality to literature associations and score significantly higher than randomized associations and those derived using only human expression data.
Second, we examined the functional relationships between the putative interaction partners. For this test, we compared the Gene Ontology (GO) and KEGG pathway database annotations of interacting partners, using a log likelihood framework (Lee et al, 2004b; Ramani et al, 2005) and testing the performance of the mapped associations with that of literature physical interactions (Figure 3B) (Bader et al, 2003; Peri et al, 2004; Joshi-Tope et al, 2005; Ramani et al, 2005). Literature associations score in the range of log likelihood ratio (LLR)=2.6–3.6, indicating high consistency with GO/KEGG annotation. As expected, randomized interactions score near zero, and interactions derived from human-only co-expression data score lower (LLR=0.59–1.09). The CCE associations are comparable to the literature associations. Using interactions transferred from other organisms (orthology core set (Lehner and Fraser, 2004); LLR=2.2) to define a threshold of minimum acceptable quality and choosing associations (in bins of 1000) exceeding this threshold, we obtain 7000 associations from the present analysis, and all subsequent tests were performed on this set. These associations have a minimum likelihood of 9:1 (90%) of belonging to the same GO/KEGG pathways. For consistency, subsequent tests include comparisons to the top-scoring 7000 associations derived from human-only mRNA co-expression, as well as to networks generated from the same proteins found in the CCE associations, but connected by 7000 random interactions (N=10 random networks).
Validation of predicted physical protein associations by mass spectrometry
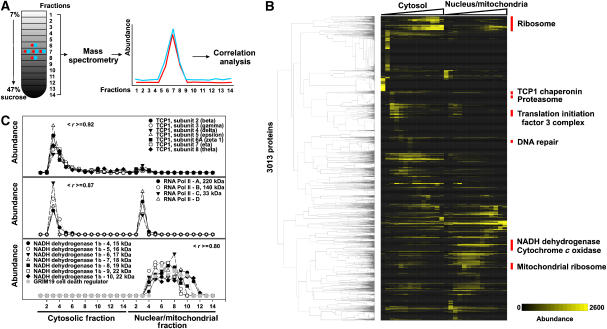
We next used quantitative mass spectrometry to test for physical associations between CCE partners (Figure 4A). Performing purifications under native conditions known to keep protein complexes intact (Dignam et al, 1983), HeLa cells were lysed, the cytoplasmic and nuclear/mitochondrial fractions were separated, and their respective contents were fractionated biochemically on two sucrose density gradients. Proteins were quantified in each fraction by mass spectrometry. In all, 3013 proteins were quantified across 14 cytoplasmic and 14 nuclear/mitochondrial sucrose density gradient fractions (Figure 4B). As proteins in the same complex should generally co-sediment, we expected physically associated proteins to often have correlated elution profiles.
Figure 4.
Mass spectrometry evidence for physical associations among 3013 proteins identified from HeLa cells. (A) HeLa cells were lysed under native conditions that maintained protein complexes intact, the nuclei/mitochondria were separated from the cytoplasm, and the two were fractionated by sucrose density gradient ultracentrifugation, collecting 14 fractions from each of the two gradients. Each fraction was analyzed by quantitative shotgun proteomics, resulting in an elution profile for each of 3013 proteins across the gradients. Proteins in the same physical complex tend to exhibit correlated elution profiles, as shown in (B) for major complexes following hierarchical clustering of the proteins by their elution profiles (labeling several sets of proteins notably enriched for interaction partners from the indicated pathways), and in (C) for three specific examples of known protein complexes. Abundance in (B) is calculated as the frequency of MS/MS spectral counts in a given fraction per protein × 10 000. Examples in (C) are labeled with the average pairwise Pearson correlation coefficient (< r >) among the profiles.
Analysis of known protein complexes verified that components of a complex tended to co-elute (Figure 4C; additional controls in Supplementary Figures 3–5). For example, components of the TCP1 chaperone complex show strongly correlated elution profiles, as do core components of RNA polymerase II; the latter profiles are distinct from the former. Likewise, components of the NADH dehydrogenase 1b complex show strongly correlated elution profiles, eluting entirely in the nuclear/mitochondrial fraction (Figure 4C). As an example of the utility of this approach, the protein GRIM-19, initially identified as a regulator of cell death induced by interferon-beta and retinoic acid, was later identified to be a subunit of the NADH dehydrogenase complex 1 (Fearnley et al, 2001); this association is clearly evident in the co-elution of GRIM-19 with other components of this complex.
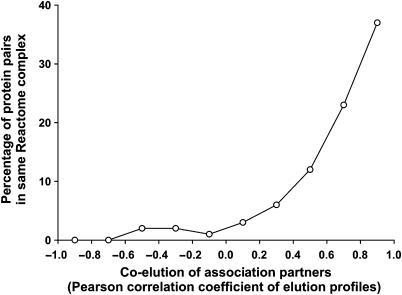
More systematically, positive control human protein interaction partners known from literature (Joshi-Tope et al, 2005) show highly correlated elution profiles (Figure 5), unlike negative control random pairs (see histograms in Figure 6A). For cases in which both interaction partners were observed in the mass spectrometry experiment, 63% of the positive control pairs exhibited Pearson correlation coefficients >0.4, indicating a false-negative rate for identifying physical associations using the mass spectrometry-based elution profiles of 37% at this correlation threshold (28% if considering correlation coefficients >0.2). This agrees with the expectation that not all interacting proteins will co-sediment, with a probable bias toward stable complexes. Similarly, for proteins in the positive control set, protein pairs with the most correlated elution profiles showed ∼40% probability of being in the same physical complex (Figure 5). Thus, correlated elution across these fractions is a strong indicator of direct physical association. As this assay is not used independently for discovery, but is confirmatory in nature, the false-negative and true-positive rates are sufficient for evaluating CCE associations.
Figure 5.
Enrichment of known complexes among co-eluting proteins. Proteins co-eluting across both sucrose gradient experiments are highly likely to belong to the same physical complex, as demonstrated by considering the subset of proteins in known human protein complexes (from Reactome; Joshi-Tope et al, 2005) that are also identified in the mass spectrometry experiments, then calculating the percentage of these protein pairs belonging to the same Reactome complex as a function of the correlation in their elution profiles. With increasing correlation, we observe strongly increasing probability of belonging to the same physical protein complex. Proteins with the most correlated elution profiles across the 28 experiments are ∼40% likely to belong to the same protein complex.
Figure 6.
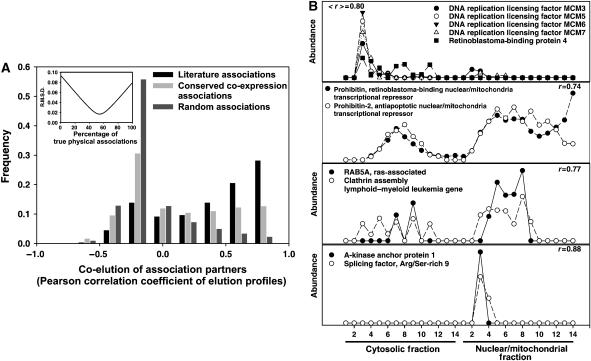
Validation of the CCE associations by mass spectrometry. (A) Between 49 and 59% of the 7000 CCE associations correspond to true physical associations, as estimated with shotgun proteomics elution profiles. The extent of co-elution of positive control (literature; Joshi-Tope et al, 2005), negative control (random), and CCE associations were calculated as the Pearson correlation coefficients between interaction partners' elution profiles, defining a correlation coefficient histogram for each set of associations. The proportion of true positives in the 7000 CCE associations was estimated by fitting the CCE correlation coefficient histogram as a linear mixture of the control histograms, with the true-positive rate corresponding to the percentage of the positive control histogram providing the best fit (inset). (B) Specific examples of correlated elution profiles for CCE partners, supporting the physical association of these protein pairs.
Although individual associations could be validated in this manner, by instead examining the aggregate distribution of elution profile correlation coefficients, we could directly estimate the error rate of the CCE associations. We calculated histograms of Pearson correlation coefficients from pairwise comparisons of elution profiles for CCE protein pairs, for protein pairs known from literature (Joshi-Tope et al, 2005) to be in the same complexes, and from random pairs of proteins (Figure 6A). We then fit the CCE histogram as a linear mixture of the positive and negative control histograms; the proportions that give the best fit thereby provided an estimate of the relative proportions of true and false associations in the CCE set. From this analysis, we estimate that 49–59% of the CCE associations correspond to true physical associations (Figure 6A, inset).
Comparing the CCE associations and the shotgun proteomics elution profiles reveals many interesting associations. For example, known complexes are correctly recovered, as for the DNA replication licensing factors MCM3, 5, 6, and 7, or for components of the proteasome. Figure 6B shows the example of the proteins prohibitin and prohibitin-2, known to form a large complex on the mitochondrial membrane that acts to suppress apoptosis, but which also shuttles to the nucleus in an estrogen-receptor-dependent manner and acts to repress transcription (Kasashima et al, 2006). New associations are also revealed: we observed a predicted physical association between MCM3 and MCM6 with the retinoblastoma-binding protein 4 (RBBP4). RBBP4 is known to participate in several chromosome replication and chromatin remodeling complexes, among them the chromatin assembly factor CAF-1 and a DNA replication-dependent chromatin assembly complex (Verreault et al, 1996). The CCE associations, supported by mass spectrometry, suggest direct physical association of RBBP4 with the replication initiation complex as well.
Figure 6B illustrates two other such examples: first, we predict the ras oncogene-related small GTPase RAB5A, an essential component of receptor-mediated endocytosis (Bucci et al, 1992), to associate with the clathrin assembly lymphoid–myeloid leukemia gene (CALM), a protein that helps recruit clathrin to endocytic vesicles. The CALM gene is a recurring site for chromosomal translocations in acute myeloid and mixed lineage leukemias (Wechsler et al, 2003). Physical association with RAB5A highlights a possible functional connection between these two endocytic components and is interesting in light of the leukemogenesis potential of chromosomal translocations involving CALM.
Likewise, we predict the A-kinase anchor protein AKAP1 to be associated with the splicing factor SFRS9. AKAP1 is primarily involved in anchoring protein kinases, phosphatases, and a phosphodiesterase to specific cellular locations, but also contains KH and Tudor domains, motifs for single-strand RNA binding (Trendelenburg et al, 1996), which help target AKAP1 to well-defined nuclear foci in an RNA-dependent manner (Rogne et al, 2006). The association with SFRS9, which among other functions is involved in both constitutive and alternative splicing and can be specifically localized with other RNA processing proteins to nuclear stress bodies (Denegri et al, 2001), suggests that AKAP1 may also have a role in these processes or in mRNA localization, perhaps integrating RNA processing with signaling.
Quantitative estimates of interaction accuracy
We further validated predicted physical associations by additional approaches. For each of these tests, we defined a standard curve relating a quantitative property of a protein pair with the tendency of the pair to physically associate, constructing the curve from control mixtures of literature (positive) and randomized (negative) physically associating proteins (Figure 7A). A set of all true-positive physical associations typical of those used to construct the curve scores high on these tests (∼100% for each standard curve); the addition of random interactions degrades the performance. The relationship between each test's performance and inclusion of false positives is unambiguous and well behaved, as judged by the quality of the standard curves, agreement between the tests, and performance curves from mixtures of known accuracy (Supplementary Figure 6). It is important to note that there are possible sources of bias for each test; however, taken as a whole, the tests are strong indicators for the enrichment of true physical associations.
Figure 7.
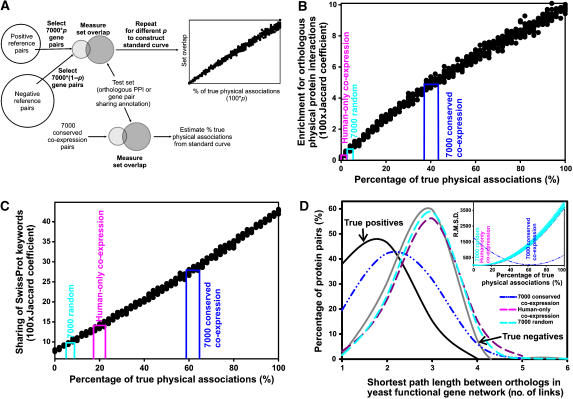
Three additional estimates of the proportion of true physical associations in the CCE pairs. (A) Overview of the method: 500 control sets of 7000 associations each (filled circles), composed of varying (but known) proportions of true-positive and true-negative associations, were tested either for overlap with orthologous protein interactions (in B) or for sharing of functional annotation (in C), generating a standard curve. From this curve and similar measurements on the CCE associations, the percentage of true physical associations can be estimated. (B) Accuracy estimates from comparison to physical protein interactions between orthologous protein pairs measured in model organisms. A standard curve that relates an interaction set's enrichment with orthologous interactions to its percentage of true-positive physical associations was constructed by measuring the Jaccard coefficients between control sets of known proportions of positive (Joshi-Tope et al, 2005) and negative (random) physical associations and an independent set of physical interactions derived from yeast (Ito et al, 2000, 2001; Uetz et al, 2000; Ho et al, 2002; Gavin et al, 2006; Krogan et al, 2006), C. elegans (Li et al, 2004), and fly (Giot et al, 2003). From this curve and the overlap measured for the CCE associations, we estimated that 37–41% of the CCE associations correspond to true physical associations, considerably higher than for randomized sets of the 7000 interactions (plotted as the mean of 10 trials) and the top 7000 associations were derived using only human mRNA expression data. (C) A standard curve based on overlap of SwissProt keywords suggests that 59–68% of CCE associations correspond to physical associations. (D) Accuracy was estimated from comparison to a probabilistic yeast gene network (Lee et al, 2007). The distances between yeast orthologs of interacting human proteins were measured in the yeast network as the minimum number of interactions separating each pair of proteins. The resulting histogram of distances is plotted for each association set tested and for positive and negative control sets. Note that the distribution from CCE associations resembles the positive control set. The percentage of true and false positives in the CCE associations was estimated by fitting the distribution as a linear mixture of the positive and negative distributions (inset), minimizing the least squares criterion (r.m.s.d.; root mean square deviation); 63±3% of the 7000 CCE associations correspond to true physical associations by this test. Shuffling the interactions among the same proteins lowers the accuracy to 6±3% by this test. Error bars on the randomized association set indicate ±1 s.d. for N=10 random trials.
First, true human physical protein associations should be enriched for physical interactions among orthologous proteins in model organisms. We generated control association sets with known error rates by randomly selecting sets of 7000 interactions with varying proportions of true-positive and true-negative associations, ranging from 0% true positive (all 7000 interactions chosen from the true-negative set) to 100% true positive (all 7000 interactions chosen from the literature set), repeating the randomization 10 times. We measured the overlap of each control set with a benchmark set of 19 119 human protein pairs whose worm, yeast, or fly orthologs have been observed to interact by yeast two-hybrid (Ito et al, 2000, 2001; Uetz et al, 2000; Giot et al, 2003; Li et al, 2004) or affinity purification/mass spectrometry assays (Ho et al, 2002; Gavin et al, 2006; Krogan et al, 2006). Figure 7B shows the resulting standard curve that relates enrichment for orthologous interactions to percentage of true physical associations. On the basis of standard curve, we estimated that ∼37–41% of the CCE pairs correspond to true physical associations, somewhat lower than the value by mass spectrometry co-sedimentation but considerably higher than both randomized pairs and pairs derived from only human co-expression data.
As physically associated proteins often share similar functional annotation (von Mering et al, 2002), we also created a standard curve based upon the agreement of interaction partners' functional annotation, relating agreement of SwissProt keywords (Wu et al, 2006) to the percentage of true physical associations (Figure 7C). For each control set, we measured the average keyword overlap across 882 SwissProt keywords between the interaction partners. Keyword overlap varied from ∼5% for the true-negative set to ∼42% for the true-positive set. From this curve and measurements of the keyword overlap by the 7000 interactions, we estimate ∼59–68% of the CCE set represent physical associations. Measurements of the percentage overlap of GO ‘biological process' and KEGG pathway annotations result in comparable values (∼50–53%).
Finally, legitimately associated proteins should be closer in a gene network (i.e. separated by fewer interactions) than random pairs. For each putative physical association, we calculated the distance between the genes' yeast orthologs in a functional gene network (Lee et al, 2007). We compared the distribution of path lengths to distributions from positive and negative control sets. Figure 7D shows that the interactions from CCE have a path length distribution more similar to the positive control set than the negative set, indicating strong enrichment for true-positive associations among the 7000 interactions. We fit the distribution of path lengths from the 7000 CCE associations as a linear combination of the positive and negative control distributions (as in Deane et al, 2002). The proportions from the best fit (Figure 7D) provide an estimate of the percentage of true physical associations. This approach estimates the CCE set at ∼63±3% true physical associations.
Table I summarizes the measurements of physical association, along with comparisons to the randomized and human-only co-expression control sets. Estimates vary only minimally with changes in parameters (e.g. using percentage keyword overlap for Figure 7B versus Jaccard coefficient) or choice of control sets (e.g. employing alternative literature positive control sets for Figures 3 or 7; see Supplementary Table 2). Although individual tests may show some bias, we expect these biases to average out across the five tests; in fact, the estimates are similar across the five tests. These measures demonstrate that CCE associations are, on average, reasonably accurate (54±10% true physical associations) and biologically relevant, are comparable in accuracy to direct large-scale experimental assays (Rual et al, 2005; Stelzl et al, 2005), and are significantly more enriched for physical associations than random controls.
Table 1.
Proportions of true physical associations measured for the CCE pairs and two control sets, using the methods of Figures 3, 6 and 7
| Percentage of true physical associations as measured by |
Average of five tests (±s.d.) | |||||
|---|---|---|---|---|---|---|
| Shotgun proteomics co-elution | Worm/fly/yeast physical interaction | GO/KEGG overlap | SwissProt keyword overlap | Yeast network path length | ||
| 7000 conserved co-expression | 49–59 | 37–41 | 50–53 | 59–68 | 63±3 | 54±10 |
| Human-only co-expression (top 7000) | 18–28 | 1–2 | 3–6 | 12–22 | 9±3 | 15±9 |
| 7000 randomized | 0–5 | 2–5 | 7–10 | 3–8 | 6±3 | 5±2 |
Ranges of values are derived by comparison to the corresponding standard curves. Estimates of variance (±s.d.) for the path length method and ranges for the co-elution method are average values derived from analysis of control mixtures of known proportions of true and false positives.
To summarize benchmark support for individual CCE associations, we calculated a ‘Binary Interaction Overlap Score (BIOS)' (Stelzl et al, 2005) for each association (Supplementary Figure 10). By this measure, 4354 (62%) of the 7000 associations have at least one line of additional evidence supporting them. Scores are reported in the supporting data file.
Detailed evaluation of ribosome biogenesis proteins
To experimentally evaluate the quality of hypotheses arising from the CCE associations, and given a statistical enrichment for proteins of ribosome biogenesis (see below), we analyzed proteins predominantly linked to proteins of ribosome biogenesis, a pathway involving several hundred proteins yet still incomplete (Granneman and Baserga, 2004). We chose four proteins with yeast orthologs for direct validation: (i) WBSCR20C, named for Williams–Beuren syndrome chromosome region 20C, shares high sequence similarity with duplicate genes WBSCR20A and WBSCR20B. This gene is deleted in Williams–Beuren syndrome, a multi-system developmental disorder caused by deletion of genes at the 7q11.23 locus (Doll and Grzeschik, 2001). WBSCR20C encodes a conserved Nol1/Nop2/Sun family protein domain and is also a member of the COG0144 protein family, other members of which are tRNA and rRNA cytosine-C5-methylases involved in translation, ribosomal structure, and biogenesis. YNL022C, the yeast ortholog of WBSCR20C, is also uncharacterized. (ii) BCCIP, or ‘BRCA2 and CDKN1A interacting protein', is an evolutionarily conserved nuclear protein with multiple protein interaction domains. This protein may be an important cofactor for BRCA2 in tumor suppression (Lu et al, 2005) and a modulator of CDK2 kinase activity via p21 (Meng et al, 2004). The yeast ortholog of this protein (Bcp1p) is an essential nuclear protein involved in nuclear export of lipid kinase Mss4p (Audhya and Emr, 2003). (iii) EPRS is predicted by sequence to be a multi-functional aminoacyl-tRNA synthetase. Its yeast ortholog YHR020W is essential, with sequence similarity to proline-tRNA ligase, but otherwise uncharacterized. (iv) LYAR is a nucleolar zinc-finger-containing protein (Su et al, 1993) whose yeast ortholog YCR087C-A is nucleolar (Huh et al, 2003), but uncharacterized.
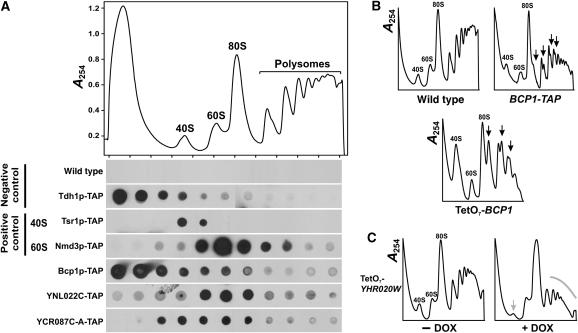
We tested the ribosomal processing phenotypes of yeast strains with tetracycline-controlled downregulatable alleles of the genes (Mnaimneh et al, 2004). Two of the strains (TetO7-BCP1 and TetO7-YHR020W) show clear ribosomal processing defects upon downregulation of the genes (Figure 8B and C). From polysome profiles, Bcp1p (corresponding to human protein BCCIP) appears to participate in 60S ribosomal subunit biogenesis; loss of the protein results in the reduction in the 60S peak relative to the 40S peak. YHR020W (corresponding to human protein EPRS) appears to participate in 40S biogenesis, resulting in a decreased 40S/60S ratio when depleted. We also tested each of the four proteins for co-sedimentation with the 40S, 60S, or 80S monoribosomes, which would provide additional support for the proteins' participation in ribosome processing. From crude cell lysates of yeast strains expressing TAP-tagged versions of each protein (Ghaemmaghami et al, 2003), we size-fractionated ribosomal subunits, free ribosomes, and polysomes using sucrose gradients. Three of the proteins (YCR087C-A, YNL022C, and Bcp1p) showed clear association with 40S and 60S subunits (Figure 8A), with Bcp1p and YCR087C-A associated with both 40S and 60S, and YNL022C showing preferential co-sedimentation with the 60S subunits. Mass spectrometry of untagged YHR020W, analyzing yeast lysate with the approach of Figure 4 (data not shown), indicates that YHR020W also co-sediments with 60S ribosomal subunits (Z Li and EM Marcotte, unpublished data). The Bcp1p-TAP co-sedimentation is less definitive than the controls; however, the polysome profile of the TAP-tagged Bcp1p strain indicates that the TAP tag itself disrupts Bcp1p activity (Figure 8B), causing a 60S ribosomal biogenesis defect and definitively implicating the protein in this process. The human BCCIP protein was also found by mass spectrometry to co-sediment with free cytosolic 40S and 60S ribosomal subunits (Supplementary Figure 4), raising the possibility of a role in ribosome recycling or nuclear export. All four genes assayed could therefore be implicated in ribosomal biogenesis.
Figure 8.
Experimental evidence supporting the network-based association of four proteins with ribosome biogenesis. (A) Co-sedimentation of TAP-tagged proteins (Ghaemmaghami et al, 2003) with ribosomal subunits. (Top) An extract of wild-type yeast cells was fractionated on a 7–47% sucrose gradient, monitoring absorbance at 254 nm. Labeled peaks indicate the 40S and 60S subunits, 80S monoribosomes, and polysomes. (bottom) Immunoblots of sucrose gradient fractions indicate the distributions of TAP-tagged proteins. YNL022C and YCR087C-A co-sediment with 60S, and both 40S and 60S ribosomal subunits, respectively, as can be seen by comparison with the sedimentation of Tsr1p-TAP, known to associate with 40S subunits (Gelperin et al, 2001), and Nmd3p-TAP, known to associate with 60S subunits (Ho and Johnson, 1999), as well as by contrast to the sedimentation of the unrelated negative control protein Tdh1p-TAP and with the background signal from wild-type cells lacking TAP-tagged proteins. Bcp1p co-sediments with 40S and 60S subunits to a lesser extent than the controls; however, this behavior apparently stems from destabilization of the protein by the TAP tag, as shown in (B). (B) Polysome profiles of cells depleted (by doxycycline-controlled downregulation; Mnaimneh et al, 2004) for Bcp1p or of cells expressing Bcp1p-TAP both show increased 40S/60S ratios and formation of aberrant ribosome halfmers (black arrows), implicating Bcp1p in 60S subunit biogenesis. (C) Polysome profiles of cells depleted for YHR020W show diminished 40S peaks and polysome peaks after doxycycline incubation (+DOX), suggesting participation of YHR020W in 40S biogenesis and possibly translation initiation.
Discussion
Characteristics of the newly mapped associations
We have described the prediction of 7000 human protein physical associations from indirect transcriptional evidence, as well as measurement of overall error rates and validation of specific associations. We further examined the associations for novelty, functional bias, and evidence for stable protein complexes. First, we compared the predicted interaction set directly to the existing human protein interaction data sets. Roughly 20% of the CCE associations can be directly verified from previously known interactions, while ∼80% are new. Our analysis bears some relation to one reported by Stuart et al, which analyzed CCE, although not for the purpose of discovering physical interactions. However, we obtain a largely non-overlapping set of associations, sharing only 12% of associations (Supplementary Table 1). Differences arise primarily because we are explicitly learning physical associations using a supervised training framework; other differences include the choice of expression data, the methods for defining orthologs, the criterion used to define co-expression (we set a statistical significance threshold on the correlation coefficient; Stuart et al use correlation coefficients >0.2), and our removal of potential cross-hybridization artifacts, all of which contribute to producing largely distinct sets of associations. Only three CCE interactions are shared with large-scale yeast two-hybrid analyses of human proteins (Rual et al, 2005; Stelzl et al, 2005), 15 with affinity purification/mass spectrometry analysis (Ewing et al, 2007), 195 with a previous computational analysis (Rhodes et al, 2005), and 211 with interactions inferred from other organisms in the OPHID database (Brown and Jurisica, 2005). These comparisons are summarized in Supplementary Table 1. In all, 5589 of the 7000 associations predicted in this analysis were not identified in previous high-throughput human protein interaction screens.
Besides simply being novel associations, 80% of the interaction partners (66% of annotated interaction partners) share neither KEGG nor GO annotations. While this extent of annotation sharing across the set of partners is sufficient to imply high confidence associations (Table I), these results indicate that the inferred associations extend beyond trivial identification of new associations among proteins already known to be in the same pathways.
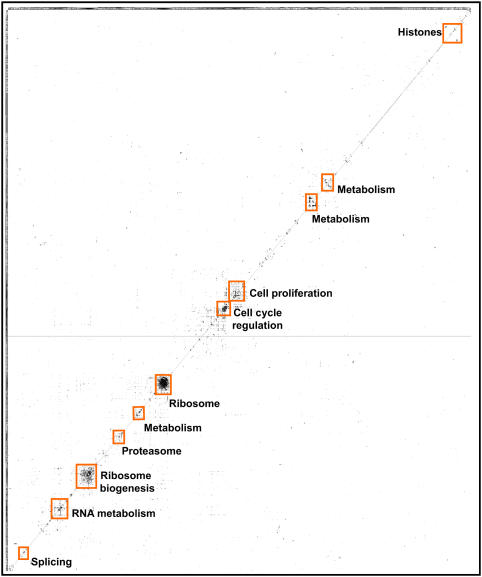
We examined the functions of the 2348 proteins in the set of 7000 associations (Supplementary Figure 7), using for this purpose the proteins' KOG annotations (Tatusov et al, 2003). We find the dominant class of proteins to be those for whom only general function is known (224 proteins); followed by 180 proteins participating in post-translational modifications, protein turnover, and chaperones; 163 in signal transduction; 141 in translation, ribosomal structure, and biogenesis; 141 in transcription; 117 of RNA processing and modification; and 87 of unknown function. Therefore, the proteins are not dominated by a single structure (e.g. the ribosome), but are generally informative for major cellular systems and uncharacterized proteins. Nonetheless, some specific functional biases occur among the CCE proteins (Supplementary Figures 8 and 9), mostly notable a statistical enrichment for proteins of membrane-bound organelles (e.g. nucleus, mitochondria, nucleolus, etc.), presumably reflecting evolutionary conservation of these proteins and their regulation, favoring detection by the CCE method. Likewise, proteins of RNA metabolism are enriched, especially ribosome biogenesis, with accompanying enrichment for nucleotide-binding protein domains. The overall trends among the proteins can be seen in a plot of the CCE associations, clustered by the spectrum of associations per protein (Figure 9), which shows that although several major clusters exist, many associations are binary protein pairs that are not otherwise seen to exist in larger assemblies, and thus lie far from the diagonal of the clustergram.
Figure 9.
The 7000 associations discovered by CCE. The data are plotted as a matrix showing associations (filled entries) among 2348 proteins (rows and columns) after hierarchically clustering (Eisen et al, 1998) proteins by their association vectors. Clustering of proteins into complexes is clear in the marked boxes. The majority of associations are distributed among smaller clusters of proteins, with many occurring between pairs of proteins not participating in larger cliques. Clusters were drawn with TreeView (Eisen et al, 1998).
This clustering, along with the 1411 associations overlapping other data sets, provides some insight into the nature of the CCE associations. Both direct interactions, especially among members of larger complexes, as well as co-complex physical associations are observed. For example, interactions are observed between alpha- and beta-tubulin, which assemble into a heterodimer; SNRPE and SNRPF, known to bind directly in the core complex of spliceosomal U1, U2, U4, and U5 snRNPs (Camasses et al, 1998); and the E2 and E3 subunits of pyruvate dehydrogenase, which interact directly. A comparison of the 7000 CCE associations with experimentally determined protein interactions among components of the 20S proteasome, as determined from the X-ray crystal structure (Groll et al, 1997), reveals four interactions between proteins that directly contact each other in the proteasome (PSMA2–PSMA6, PSMB3–PSMB2, PSMA4–PSMB2, and PSMA1–PSMB1), and nine interactions between proteins that assemble into the same physical complex, but do not directly contact each other (PSMA3–PSMA4, PSMA6–PSMB2, PSMA4–PSMA6, PSMA6–PSMB3, PSMA2–PSMB1, PSMA2–PSMA1, PSMA2–PSMB2, PSMA6–PSMA5, and PSMA5–PSMB2). Therefore, as expected, both direct binding and co-complex interactions can be found among the 7000 associations.
In fact, the CCE pairs are strongly statistically enriched for co-complex associations typical of affinity purification/mass spectrometry interaction assays. If we consider only the subset of 2138 CCE pairs (out of the 7000) in which both proteins have yeast orthologs, 118 of these can be verified by the MIPS database as belonging to the same yeast complex (using the ‘hand-curated' set of protein complexes; Guldener et al, 2005). This value is 21 standard deviations (P<10−98) above the mean of random trials: randomizing the 7000 interactions and repeating the comparison gives a mean of 23±4.5 confirmed interactions for N=100 random trials. Similar enrichment can be seen by comparison with yeast protein complexes defined by affinity purification/mass spectrometry (Gavin et al, 2006; Krogan et al, 2006): 74 of the 2138 CCE pairs can be confirmed, 8 standard deviations (P<10−15) above the mean of random trials (29±5.5, N=100). Likewise, 392 of the 2138 CCE pairs can be confirmed by comparison to the full-matrix form of these data (i.e. considering both bait–prey and prey–prey interactions), 10 standard deviations (P<10−25) above the mean of random trials (214±17, N=100).
Finally, we looked for organismal bias among the CCE pairs, examining which model organism contributed the top LLR score for each interaction (Supplementary Table 3). The most associations were contributed from the comparison of human and C. elegans expression, accounting for 2949 of the 7000 associations, and the least (158) from mouse. The low number contributed by comparison to mouse may suggest the importance of employing more distant orthologs, especially to non-mammalian animals, in identifying interactions by this approach, but more probably stems from characteristics of the data employed, such as the smaller number of mouse microarray experiments analyzed (Supplementary Table 4).
One interesting aspect of the CCE assay is that it intrinsically samples all pairs of genes that are measured on the DNA microarrays. This has the effect of increasing the numbers of proteins for which interactions are observed, and thereby decreasing the number of interactions per protein (7000 interactions for 2348 proteins ∼3 interactions per protein, somewhat lower than the 5–15 interactions per protein observed in other data sets (Ramani et al, 2005)).
Limitations, false positives, and potential improvements
Given the derivation of CCE pairs from transcriptional evidence, there are important features and limitations to note. First, strong co-expression tends to coincide with stable, rather than transient, physical association (Jansen et al, 2002), and we expect CCE pairs to reflect this trend, with a correspondingly higher false-negative rate for transient interactions. Second, based on our measured error rates, there are still appreciable false-positive associations, although the false-positive and -negative error rates are comparable to the only direct experimental approaches—yeast two-hybrid assays and mass spectrometry of cloned, epitope-tagged human proteins—that have been applied to map physical associations on this scale. However, CCE false positives have unusual properties. As the CCE pairs were the highest scoring (top 0.1%) of >5 million tested gene pairs, the association partners are strongly co-regulated in an evolutionarily conserved manner, and thus are highly likely to function together, even if not physically associated. Finally, algorithmic improvements, such as better orthology assignment and alternative supervised learning frameworks, and application to additional DNA microarray data, e.g. tissue- and cell-type-specific data to learn tissue- and cell-type-specific associations, are certain to reveal new associations when applied in the general framework we have described. Thus, we expect new CCE associations can be identified by modifications to this method.
Similarly, the mass spectrometry data used to test the CCE associations have some important features and limitations. Primarily, co-sedimentation alone is not proof of physical association—it is possible for unrelated complexes to co-sediment—as reflected in the measured true-positive and false-negative rates for associations inferred solely from these data. These sedimentation-derived associations should thus not be viewed as standalone. However, as a benchmark applied in the manner we present (e.g. analyzed in aggregate form), or when considered in combination with other data, such as incorporated into the BIOS scores of the CCE associations, we find the mass spectrometry data to be extremely valuable. We suggest that benchmarks of this sort could be of great utility for evaluating physical complexes determined by other methods, and could be generally adopted for measuring assay accuracy.
Conclusions
The scale of the human interactome appears to be beyond any individual technique; a combination of complementary approaches will be needed to map the complete human protein–protein interaction network. Although current methods for mapping interactions focus largely on direct experimental observations, sufficient functional genomics data exist that physical protein associations can also be indirectly identified from these data. We demonstrate that these approaches can be comparable in scale and quality, both in terms of false-positive and false-negative rates, to the current largest scale experimental screens. Finally, as CCE-based physical protein association mapping is based on conserved in vivo phenomena, this approach is likely to specifically discover associations relevant to in vivo biology.
Materials and methods
Mapping of orthologs
Orthologs were obtained from the InParanoid database (Remm et al, 2001) as SwissProt identifiers for human proteins and their orthologs from five other organisms (A. thaliana, C. elegans, D. melanogaster, M. musculus and S. cerevisiae). Using ID-Serve (http://bioinformatics.icmb.utexas.edu/idserve) and organism-specific databases, the SwissProt identifier for each gene was mapped to alternate identifiers: LocusLink identifiers (human), common names (M. musculus), WormBase identifiers (C. elegans), Locus codes (A. thaliana), Flybase gene identifiers (D. melanogaster), and standardized gene names (S. cerevisiae). Supplementary Table 5 lists the numbers of orthologous genes analyzed.
mRNA expression data
All mRNA expression data (Supplementary Table 4) were obtained from the Stanford Microarray Database (Ball et al, 2005). It has previously been shown that extraction of co-expression relationships is improved by restricting comparisons to similar conditions and experiments (Lee et al, 2004a, 2004b; Segal et al, 2004). We therefore divided the available 1922 human DNA microarray experiments into 11 categories of experiments, as assigned by the Stanford Microarray Database, and restricted comparisons to experiments in the same category. Expression data for other organisms were treated as single categories. Each of the microarray expression vectors was mean centered (row and column) and normalized before carrying out correlation analysis.
Calculation of co-expression
For each pair of human genes, as well as for their corresponding orthologs, the Pearson correlation coefficient was computed between the mRNA expression vectors. For each gene pair, this gives 11 measurements of correlation corresponding to the 11 categories of human expression data sets and up to 5 for the correlation between the orthologs in the other organisms. Paralogs (as defined by InParanoid) were excluded from being compared to each other, as they tend to have similar expression profiles and thus high correlation, which we empirically observe to substantially increase the false-positive rate. The significance of each correlation was computed based on t-test statistics as
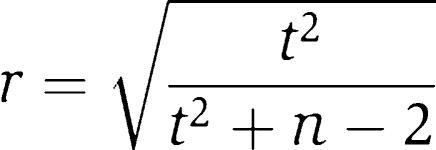 |
where r is the minimum significant correlation for n values in the two expression vectors being compared and t is the t-test value at a probability of P⩽0.01 from a t-test table. Only statistically significant correlation coefficients were retained, thereby accounting for variability in the sparseness of expression vectors. For example, using expression vectors of 100 experiments with only 50 data points available for both genes being compared, the absolute value of correlation must be >0.36 for the comparison to be statistically significant at P⩽0.01.
Removal of cross-hybridization artifacts
Cross-hybridization occurs when an mRNA probe binds to a non-cognate spot on the microarray instead of its perfect complement spot. This creates both false positives (due to additional signal at incorrect positions on the array) and false negatives (due to reduced signal in correct positions). Although cross-hybridization is well established in spotted cDNA-based DNA microarray experiments (Kane et al, 2000; Murray et al, 2001; Xu et al, 2001), there are no universal standards for filtering such effects. In this analysis, we expected that cross-hybridizing gene pairs would appear to have similar expression patterns and therefore contribute false positives to our analysis.
To filter out these potentially spurious interactions arising from cross-hybridization, we established a threshold for excluding cross-hybridization based upon analysis of the hybridization of four yeast genes (YPL274W, YLR467W, YIR039C, and YKL224) to their homologs on a yeast DNA microarray. The four genes were chosen such that BLAST-based comparisons of the genes' DNA sequences to other genes in the yeast genome yielded hits with percent identities to the query sequence in the range of 50–100% and BLAST E-values ⩽10−4. The four query genes were amplified using standard PCR techniques and primers to flanking DNA, labeled with Cy5, mixed with Cy3-labeled reference DNA (Carlson, 2002), and hybridized to a yeast cDNA microarray containing ∼12 000 spots comprising all the yeast genes and intergenic regions (Carlson, 2002; Hahn et al, 2004; Kim and Iyer, 2004). Standard microarray analysis was carried out to quantify hybridization strength as the mean of ratios of Cy5/Cy3 fluorescence intensities across spots. By plotting hybridization strength against the DNA sequence identity of the genes (Supplementary Figure 2), we identified an operational threshold of BLAST E-value ⩽10−4 and DNA sequence identity ⩾70% within the aligned regions. Gene pairs that exceed this threshold (with either the human or model organism gene pair DNA sequences) were likely to cross-hybridize and were excluded from further analysis. This filter removes 47 145 protein pairs from the plant–human analysis, 37 519 from the worm–human, 26 724 from the fly–human, 39 286 from the mouse–human, and 2193 from the yeast–human analysis. This filtration preferentially removes many false-positive interactions, as the average expression correlation of the filtered pairs was significantly higher than for the remaining pairs (e.g. the average expression correlation in the human–plant analysis was 0.28, while the average for the filtered pairs was 0.56), with the maximum expression correlation among the removed pairs equal to 1.0 for all comparisons.
Training to extract physical protein associations
We used the 31 609 human protein interactions from Ramani et al (2005) as the physical association benchmark. The associations were randomly separated into testing and training data sets (15 810 and 15 799 associations, respectively). For each of the five human gene pair/ortholog gene pair sets, the maximum expression correlation of the human genes from the 11 data sets was plotted along the x axis and the correlation of the orthologous genes plotted along the y axis (as in Figure 2). The fraction of gene pairs that showed a particular expression pattern was measured in bins of 0.1 × 0.1 units. Two-dimensional histograms were calculated for interacting proteins and for non-interacting proteins in the training set. The logarithm of the ratio of the histograms at a given position in the plot, corrected by the background likelihood of physical associations in the training set, gives the log likelihood estimate of physical association conditioned on the degree of co-expression of the human genes and their orthologs in that organism. To minimize possible errors due to orthology assignments, we further considered only counts in the upper right-hand quadrant of each analysis, corresponding to gene pairs for which the human and other organismal experiments describe similar co-expression trends. Protein pairs outside of the training set were then assigned log likelihood scores according to their expression patterns in these data sets. Similar analyses were performed for associations derived from comparison of human expression data with each of the four other organism-specific data sets, associating the maximum score from these five analyses as each protein pairs' estimated likelihood of associating physically. (The maximum score outperformed the naïve Bayes sum of scores, suggesting that the five scores are not independent.) The 7000 top-scoring associations are listed in Supplementary information.
The human-only co-expression control set was generated by considering only the human DNA microarray data, ignoring contributions from other organisms and lifting the requirement for each member of a gene pair to have orthologs in the same second organism. Putative associations were identified as for the CCE case, but instead using the log likelihood framework to relate the correlation coefficients across only the human DNA microarray experiments to the likelihood of physically associating. All other calculations were performed identically to the CCE case, including calculation of correlation coefficients, significance testing of correlations, calculation of likelihood values, selection of priors, and filtration for cross-hybridization.
Testing for enrichment of known physical associations
We measured enrichment for known physical associations using the independent test set of 15 810 physical associations and the same LLR framework used to initially derive the CCE associations. The 15 810 associations formed the positive test set; the negative test set was defined as all pairs of proteins chosen from the 15 810 associations set, omitting the 15 810 associations themselves. The prior odds ratio of interacting (P(I)/P(∼I)) equaled the ratio of positive to negative test set examples (0.00085). For each query association network being tested (or for a given bin of 1000 associations selected from a rank-ordered list), we measured the fraction of query set associations shared with the positive test set (P(I∣D)), as well as the fraction shared with the negative test set (P(∼I∣D)). The posterior odds ratio was calculated as P(I∣D)/P(∼I∣D), and the LLR calculated as indicated in the main text, equal to the posterior odds ratio divided by the prior odds ratio. For the purposes of Figure 3A, the log likelihood was calculated in a cumulative manner (i.e. aggregating successive bins of 1000 associations for analysis.).
Testing for functional similarity
We measured functional similarity of interacting protein pairs by using the gene annotation information obtained from GO (Ashburner et al, 2000) process level 8 annotation and KEGG pathway annotation (Kanehisa et al, 2004). These databases provide specific pathway and biological process annotations for 7390 human genes, assigning them into 155 KEGG pathways (at the lowest level of KEGG) and 1356 GO pathways (at level 8 of the GO biological process annotation). Interactions were first rank-ordered by confidence scores. For each successive bin of 1000 interactions, the functional similarity was calculated in a cumulative manner by counting the number of pairs in that bin or previous bins that shared a functional annotation, dividing this by the number of pairs that did not share functional annotation, and correcting by the prior probability of annotated pairs sharing annotation (0.0589).
Construction of standard curves for estimating percentages of physical associations
Standard curves were constructed as described in the main text. Positive control sets for Figure 7B and C were selected from the hand-curated protein complex assignments of Reactome (Joshi-Tope et al, 2005). For the analysis of Figure 7B, we restricted the analysis to the portion of each data set for which both interacting proteins have orthologs among the yeast, worm, or fly proteins sampled by the benchmark assays (i.e. considering only the subspace of interactions spanned by the assay bait–prey pairs). For the standard curves of both Figure 7B and C, the derived percentages of physical associations do not strongly depend upon the sizes of the data sets or control sets, only upon their tendencies to share orthologous interactions or functional annotations (data not shown). Ranges of accuracies were derived directly from the standard curve (i.e. as empirically measured from replicate analysis of control mixtures of true- and false-positive interactions).
For the linear mixture model of Figure 7D, positive control associations were taken from Joshi-Tope et al (2005), only considering genes with yeast orthologs, and negative control associations taken as pairs of human genes from the positive control set that have yeast orthologs but do not have recorded interactions. To minimize possible circularity, we removed all functional linkages from the yeast network that were derived only from mRNA co-expression data. The variances associated with accuracy estimates in Figure 7D were derived from 10 replicate analyses of mixtures of known proportions of true- and false-positive interactions (Supplementary Figure 6).
Binary interaction overlap score
To further assign confidence to each association, we have adopted the BIOS of Stelzl et al (2005): based upon the benchmark sets (Table I), we assign each association +1 if the protein pair is observed in the physical interaction benchmark, +1 for sharing GO/KEGG keywords, +1 for sharing SwissProt keywords, +1 for sharing KOG annotation, +1 for being observed in the orthologous interaction benchmark, +1 for having a correlation coefficient >0.4 in the mass spectrometry elution profile experiments, and +1 for having yeast orthologs that are either directly connected or one link separated in the yeast functional network benchmark (with expression-only and orthology-derived links omitted). Each association is thus scored from 0 to 7 based on additional support for that association; the BIOS scores generally correlate with the LLR scores (Supplementary Figure 10) and are reported in the supporting data file.
Human cell culture and mass spectrometry
HeLa S3 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum at 37°C with 5% CO2. At about 80% confluency, cells were treated with 100 μg/ml emetine for 10 min and harvested by scraping. Cells were centrifuged at 500 g for 10 min, washed three times with cold PBS buffer, and resuspended in five packed cell volumes of cold lysis buffer (10 mM Tris pH 7.4, 20 mM KCl, 5 mM MgCl2). After swelling on ice for 10 min, cells were centrifuged at 500 g for 10 min and resuspended in one packed cell volume cold lysis buffer supplemented with 1 × protease inhibitor cocktail (Roche) and 100 μg/ml emetine. After lysing the cells with a dounce homogenizer, nuclei were collected by centrifuging at 1000 g for 10 min. The supernatant was centrifuged at 15 000 g for 10 min to obtain the cytosolic fraction. Nuclei were suspended in lysis buffer and lysed by sonication, collecting the clarified supernatant after centrifugation at 15 000 g for 10 min.
The cytosolic and the nuclear fractions were each loaded onto continuous 7–47% sucrose gradients in lysis buffer. After a 2.5-h spin at 40 000 r.p.m. in a Beckman SW40 rotor, the sucrose gradient was fractionated using an ISCO gradient fractionation system. Proteins from each fraction were precipitated with 10% cold trichloroacetic acid (TCA) and washed with 100% cold acetone. The protein pellets were suspended in 100 mM pH 8.0 Tris buffer and digested with sequencing grade trypsin (Sigma). For each fraction, tryptic peptides were loaded onto a reverse-phase C18 column and washed with 95% water, 5% acetonitrile, and 0.1% formic acid. Peptides were eluted with a 240-min gradient from 5 to 40% acetonitrile and analyzed online with a nanoelectrospray ionization (300 nl/min flow rate) LTQ-Orbitrap hybrid mass spectrometer (Thermo Electron) using data-dependent precursor ion selection. Each parent ion mass spectrum (MS) was analyzed at high resolution (100 000) with the Orbitrap; the top seven MS peaks were fragmented by helium collision-induced dissociation at 35 eV, analyzing the resulting MS/MS spectra with the LTQ. Approximately 35 000 MS/MS spectra were collected per fraction. Spectra were searched against the set of NCBI human protein sequences using TurboSequest (Bioworks v.3.2, Thermo Electron). Proteins from each fraction were identified at a 5% false detection rate using Peptide/ProteinProphet (Keller et al, 2002; Nesvizhskii et al, 2003). The spectral count (number of total observations of MS/MS spectra from a given protein in a given fraction) was used as an estimate of protein abundance (Liu et al, 2004), dividing the spectral count of a protein ( × 10 000) by the sum of spectral counts for all proteins identified in that fraction. Protein elution profiles are provided as Supplementary information.
Yeast media and strains
All yeast strains were cultured in YPD (1% yeast extract, 2% peptone, and 2% dextrose) at 30°C. Tetracycline promoter-controlled essential gene haploid MATa strains (Mnaimneh et al, 2004) and TAP-tagged haploid MATa strains (Ghaemmaghami et al, 2003) were obtained from Open Biosystems.
Polysome profile analysis
All yeast strains were cultured to OD600 0.3–0.5. For tetracycline promoter-controlled alleles, overnight cultures were diluted to OD600 0.01, 10 μg/ml doxycycline (Fisher Scientific) was added into the media, and cells were grown to OD600 0.3–0.5. Cycloheximide (100 μg/ml) (Sigma) was added to each culture. Cultures were immediately cooled with ice, and all subsequent steps were performed on ice or at 4°C. Each cell pellet was washed once with lysis buffer (20 mM Tris pH 7.4, 20 mM KCl, 5 mM MgCl2, 100 μg/ml cycloheximide, 12 mM β-mercaptoethanol, 2 μg/ml leupeptin, 2 μg/ml aprotinin, 1 μg/ml bestatin, and 1 μg/ml pepstatin A) without protease inhibitors (MP Biomedicals Inc.). The cells were pelleted, resuspended in one volume lysis buffer, and lysed with glass beads. Crude lysates were centrifuged at 15 000 g for 10 min. Fifteen OD260 units of each supernatant were loaded onto continuous 12 ml 7–47% sucrose gradients in polysome lysis buffer without protease inhibitors, as in Baim et al (1985). After a 2.5-h spin at 40 000 r.p.m. in a Beckman SW40 rotor, the sucrose gradient was fractionated and absorbance at 254 nm was measured. For TAP-tagged strains, fractions were collected, and proteins were precipitated with 10% cold TCA and washed with 100% cold acetone.
Immunoblotting
Precipitated proteins were resuspended in 20 μl Laemmli buffer and 2 μl of each sample was deposited onto nitrocellulose membrane. TAP-tagged proteins were detected with PAP antibody (Rockland Immunochemicals Inc.) and chemiluminescence (ECL; Amersham Biosciences).
Supplementary Material
Supplementary Figures
Supplementary Tables
Supplementary Information 1
Supplementary Information 2
Acknowledgments
We thank Insuk Lee for critical comments, Vishy Iyer and members of his lab for help with DNA microarray analysis, Arlen Johnson and Nai Jung Hung for help with polysome profile analysis, and Scott Stevens for critical comments. This study was supported by grants from the NSF (IIS-0325116, EIA-0219061), NIH (GM06779-01, GM076536-01), Welch (F1515), and a Packard Fellowship (EMM).
References
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25: 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audhya A, Emr SD (2003) Regulation of PI4,5P2 synthesis by nuclear–cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J 22: 4223–4236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader GD, Betel D, Hogue CW (2003) BIND: the biomolecular interaction network database. Nucleic Acids Res 31: 248–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baim SB, Pietras DF, Eustice DC, Sherman F (1985) A mutation allowing an mRNA secondary structure diminishes translation of Saccharomyces cerevisiae iso-1-cytochrome c. Mol Cell Biol 5: 1839–1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball CA, Awad IA, Demeter J, Gollub J, Hebert JM, Hernandez-Boussard T, Jin H, Matese JC, Nitzberg M, Wymore F, Zachariah ZK, Brown PO, Sherlock G (2005) The Stanford Microarray Database accommodates additional microarray platforms and data formats. Nucleic Acids Res 33: (Database issue) D580–D582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann S, Ihmels J, Barkai N (2004) Similarities and differences in genome-wide expression data of six organisms. PLoS Biol 2: E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KR, Jurisica I (2005) Online predicted human interaction database. Bioinformatics 21: 2076–2082 [DOI] [PubMed] [Google Scholar]
- Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M (1992) The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70: 715–728 [DOI] [PubMed] [Google Scholar]
- Camasses A, Bragado-Nilsson E, Martin R, Seraphin B, Bordonne R (1998) Interactions within the yeast Sm core complex: from proteins to amino acids. Mol Cell Biol 18: 1956–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson MW (2002) Surveying yeast genomic diversity using cDNA microarrays. Masters Thesis, Biomedical Engineering, University of Texas, Austin, pp 60
- Deane CM, Salwinski L, Xenarios I, Eisenberg D (2002) Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics 1: 349–356 [DOI] [PubMed] [Google Scholar]
- Denegri M, Chiodi I, Corioni M, Cobianchi F, Riva S, Biamonti G (2001) Stress-induced nuclear bodies are sites of accumulation of pre-mRNA processing factors. Mol Biol Cell 12: 3502–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll A, Grzeschik KH (2001) Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams–Beuren syndrome. Cytogenet Cell Genet 95: 20–27 [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA 95: 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, Taylor R, Dharsee M, Ho Y, Heilbut A, Moore L, Zhang S, Ornatsky O, Bukhman YV, Ethier M, Sheng Y et al. (2007) Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol Syst Biol 3: 89 10.1038/msb4100134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearnley IM, Carroll J, Shannon RJ, Runswick MJ, Walker JE, Hirst J (2001) GRIM-19, a cell death regulatory gene product, is a subunit of bovine mitochondrial NADH:ubiquinone oxidoreductase (complex I). J Biol Chem 276: 38345–38348 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dumpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M et al. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440: 631–636 [DOI] [PubMed] [Google Scholar]
- Ge H, Liu Z, Church GM, Vidal M (2001) Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet 29: 482–486 [DOI] [PubMed] [Google Scholar]
- Gelperin D, Horton L, Beckman J, Hensold J, Lemmon SK (2001) Bms1p, a novel GTP-binding protein, and the related Tsr1p are required for distinct steps of 40S ribosome biogenesis in yeast. RNA 7: 1268–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS (2003) Global analysis of protein expression in yeast. Nature 425: 737–741 [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, Vijayadamodar G, Pochart P, Machineni H, Welsh M, Kong Y, Zerhusen B, Malcolm R, Varrone Z, Collis A, Minto M et al. (2003) A protein interaction map of Drosophila melanogaster. Science 302: 1727–1736 [DOI] [PubMed] [Google Scholar]
- Granneman S, Baserga SJ (2004) Ribosome biogenesis: of knobs and RNA processing. Exp Cell Res 296: 43–50 [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature 386: 463–471 [DOI] [PubMed] [Google Scholar]
- Guldener U, Munsterkotter M, Kastenmuller G, Strack N, van Helden J, Lemer C, Richelles J, Wodak SJ, Garcia-Martinez J, Perez-Ortin JE, Michael H, Kaps A, Talla E, Dujon B, Andre B, Souciet JL, De Montigny J, Bon E, Gaillardin C, Mewes HW (2005) CYGD: the comprehensive yeast genome database. Nucleic Acids Res 33: D364–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn JS, Hu Z, Thiele DJ, Iyer VR (2004) Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol Cell Biol 24: 5249–5256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart GT, Ramani AK, Marcotte EM (2006) How complete are current yeast and human protein-interaction networks? Genome Biol 7: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho JH, Johnson AW (1999) NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol Cell Biol 19: 2389–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y (2000) Toward a protein–protein interaction map of the budding yeast: a comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. Proc Natl Acad Sci USA 97: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Greenbaum D, Gerstein M (2002) Relating whole-genome expression data with protein–protein interactions. Genome Res 12: 37–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M (2003) A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science 302: 449–453 [DOI] [PubMed] [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D'Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L, Lewis S, Birney E, Stein L (2005) Reactome: a knowledge base of biological pathways. Nucleic Acids Res 33: (Database issue) D428–D432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane MD, Jatkoe TA, Stumpf CR, Lu J, Thomas JD, Madore SJ (2000) Assessment of the sensitivity and specificity of oligonucleotide (50 mer) microarrays. Nucleic Acids Res 28: 4552–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M (2004) The KEGG resource for deciphering the genome. Nucleic Acids Res 32: (Database issue) D277–D280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasashima K, Ohta E, Kagawa Y, Endo H (2006) Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J Biol Chem 281: 36401–36410 [DOI] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kim J, Iyer VR (2004) Global role of TATA box-binding protein recruitment to promoters in mediating gene expression profiles. Mol Cell Biol 24: 8104–8112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davey M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B et al. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440: 637–643 [DOI] [PubMed] [Google Scholar]
- Lee HK, Hsu AK, Sajdak J, Qin J, Pavlidis P (2004a) Coexpression analysis of human genes across many microarray data sets. Genome Res 14: 1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Date SV, Adai AT, Marcotte EM (2004b) A probabilistic functional network of yeast genes. Science 306: 1555–1558 [DOI] [PubMed] [Google Scholar]
- Lee I, Li Z, Marcotte EM (2007) An improved, bias-reduced probabilistic functional gene network of Baker's yeast, Saccharomyces cerevisiae. PLoS ONE 2: e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner B, Fraser AG (2004) A first-draft human protein-interaction map. Genome Biol 5: R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Armstrong CM, Bertin N, Ge H, Milstein S, Boxem M, Vidalain PO, Han JD, Chesneau A, Hao T, Goldberg DS, Li N, Martinez M, Rual JF, Lamesch P, Xu L, Tewari M, Wong SL, Zhang LV, Berriz GF et al. (2004) A map of the interactome network of the metazoan C.elegans. Science 303: 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR III (2004) A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem 76: 4193–4201 [DOI] [PubMed] [Google Scholar]
- Lu H, Guo X, Meng X, Liu J, Allen C, Wray J, Nickoloff JA, Shen Z (2005) The BRCA2-interacting protein BCCIP functions in RAD51 and BRCA2 focus formation and homologous recombinational repair. Mol Cell Biol 25: 1949–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D (1999) A combined algorithm for genome-wide prediction of protein function. Nature 402: 83–86 [DOI] [PubMed] [Google Scholar]
- Meng X, Liu J, Shen Z (2004) Inhibition of G1 to S cell cycle progression by BCCIP beta. Cell Cycle 3: 343–348 [PubMed] [Google Scholar]
- Mnaimneh S, Davierwala AP, Haynes J, Moffat J, Peng WT, Zhang W, Yang X, Pootoolal J, Chua G, Lopez A, Trochesset M, Morse D, Krogan NJ, Hiley SL, Li Z, Morris Q, Grigull J, Mitsakakis N, Roberts CJ, Greenblatt JF et al. (2004) Exploration of essential gene functions via titratable promoter alleles. Cell 118: 31–44 [DOI] [PubMed] [Google Scholar]
- Murray AE, Lies D, Li G, Nealson K, Zhou J, Tiedje JM (2001) DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc Natl Acad Sci USA 98: 9853–9858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, Gandhi TK, Chandrika KN, Deshpande N, Suresh S, Rashmi BP, Shanker K, Padma N, Niranjan V, Harsha HC, Talreja N, Vrushabendra BM, Ramya MA, Yatish AJ, Joy M et al. (2004) Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res 32: (Database issue) D497–D501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince JT, Carlson MW, Wang R, Lu P, Marcotte EM (2004) The need for a public proteomics repository. Nat Biotechnol 22: 471–472 [DOI] [PubMed] [Google Scholar]
- Ramani AK, Bunescu RC, Mooney RJ, Marcotte EM (2005) Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol 6: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm M, Storm CE, Sonnhammer EL (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol 314: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Tomlins SA, Varambally S, Mahavisno V, Barrette T, Kalyana-Sundaram S, Ghosh D, Pandey A, Chinnaiyan AM (2005) Probabilistic model of the human protein–protein interaction network. Nat Biotechnol 23: 951–959 [DOI] [PubMed] [Google Scholar]
- Rogne M, Landsverk HB, Van Eynde A, Beullens M, Bollen M, Collas P, Kuntziger T (2006) The KH-Tudor domain of a-kinase anchoring protein 149 mediates RNA-dependent self-association. Biochemistry 45: 14980–14989 [DOI] [PubMed] [Google Scholar]
- Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S et al. (2005) Towards a proteome-scale map of the human protein–protein interaction network. Nature 437: 1173–1178 [DOI] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A (2004) A module map showing conditional activity of expression modules in cancer. Nat Genet 36: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Simonis N, Gonze D, Orsi C, van Helden J, Wodak SJ (2006) Modularity of the transcriptional response of protein complexes in yeast. J Mol Biol 363: 589–610 [DOI] [PubMed] [Google Scholar]
- Snel B, van Noort V, Huynen MA (2004) Gene co-regulation is highly conserved in the evolution of eukaryotes and prokaryotes. Nucleic Acids Res 32: 4725–4731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksoz E, Droege A, Krobitsch S, Korn B et al. (2005) A human protein–protein interaction network: a resource for annotating the proteome. Cell 122: 957–968 [DOI] [PubMed] [Google Scholar]
- Stuart JM, Segal E, Koller D, Kim SK (2003) A gene-coexpression network for global discovery of conserved genetic modules. Science 302: 249–255 [DOI] [PubMed] [Google Scholar]
- Su L, Hershberger RJ, Weissman IL (1993) LYAR, a novel nucleolar protein with zinc finger DNA-binding motifs, is involved in cell growth regulation. Genes Dev 7: 735–748 [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, Rao BS, Smirnov S, Sverdlov AV, Vasudevan S, Wolf YI, Yin JJ, Natale DA (2003) The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichmann SA, Babu MM (2002) Conservation of gene co-regulation in prokaryotes and eukaryotes. Trends Biotechnol 20: 407–410; discussion 410 [DOI] [PubMed] [Google Scholar]
- Trendelenburg G, Hummel M, Riecken EO, Hanski C (1996) Molecular characterization of AKAP149, a novel A kinase anchor protein with a KH domain. Biochem Biophys Res Commun 225: 313–319 [DOI] [PubMed] [Google Scholar]
- Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403: 623–627 [DOI] [PubMed] [Google Scholar]
- van Noort V, Snel B, Huynen MA (2003) Predicting gene function by conserved co-expression. Trends Genet 19: 238–242 [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B (1996) Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell 87: 95–104 [DOI] [PubMed] [Google Scholar]
- von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P (2002) Comparative assessment of large-scale data sets of protein–protein interactions. Nature 417: 399–403 [DOI] [PubMed] [Google Scholar]
- Wechsler DS, Engstrom LD, Alexander BM, Motto DG, Roulston D (2003) A novel chromosomal inversion at 11q23 in infant acute myeloid leukemia fuses MLL to CALM, a gene that encodes a clathrin assembly protein. Genes Chromosomes Cancer 36: 26–36 [DOI] [PubMed] [Google Scholar]
- Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B (2006) The universal protein resource (UniProt): an expanding universe of protein information. Nucleic Acids Res 34: D187–D191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Bak S, Decker A, Paquette SM, Feyereisen R, Galbraith DW (2001) Microarray-based analysis of gene expression in very large gene families: the cytochrome P450 gene superfamily of Arabidopsis thaliana. Gene 272: 61–74 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables
Supplementary Information 1
Supplementary Information 2