Abstract
Eukaryotic transcription involves the synergistic interaction of many different proteins. However, the question remains how eukaryotic promoters achieve ultrasensitive or threshold responses to changes in the concentration or activity of a single transcription factor (TF). We show theoretically that by recruiting a histone-modifying enzyme, a TF binding non-cooperatively to a single site can change the balance between opposing positive feedback loops in histone modification to produce a large change in gene expression in response to a small change in concentration of the TF. This mechanism can also generate bistable promoter responses, allowing a gene to be on in some cells and off in others, despite the cells being in identical conditions. In addition, the system provides a simple means by which the activities of many TFs could be integrated at a promoter.
Keywords: epigenetics, histone modification, signal integration, threshold, transcription factor
Introduction
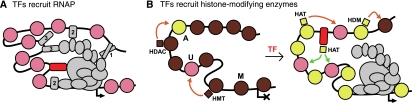
Ultrasensitivity, or threshold behaviour, where small changes in a stimulus near some threshold value produce a large change in a response, but large changes in the stimulus far from the threshold produce small changes in the response (i.e., a sigmoidal stimulus–response curve), is an important property of many biological systems Koshland et al, 1982; Ferrell, 1996. Ultrasensitivity can function to dampen irrelevant fluctuations in the signal, yet allows a decisive response when needed. Prokaryotic transcription factors (TFs) often have ultrasensitive activating or repressing effects by virtue of cooperative binding of the TF to multiple DNA sites. Although eukaryotic transcription is a highly synergistic process, involving the cooperative action of many different DNA-associated proteins (Carey, 1998; Naar et al, 2001; Figure 1A), this synergy does not in itself provide a mechanism for ultrasensitive responses to changes in concentration or activity of a single TF. Such ultrasensitivity can be produced, as in bacteria, by direct cooperative binding of the TF due to favourable contacts between the TF protomers bound at different sites (Burz et al, 1998; Figure 1A, TF1). Cooperative binding can also be indirect. TF protomers bound at different sites may be able to simultaneously make favourable contacts with some common target such as proteins of the transcriptional apparatus (Carey, 1998; Figure 1A, TF2). Alternatively, in collaborative competition, binding of the TF to one site competes with a nucleosome that can block binding of the TF to other sites (Miller and Widom, 2003; Figure 1A, TF3). However, it is not clear how common these cooperative binding mechanisms are in eukaryotes. Here, we propose a mechanism for generating high ultrasensitivity that does not require cooperative TF binding, but rather involves nucleosome modifications.
Figure 1.
Models of regulation by TFs. (A) TFs (rounded boxes) bind to DNA and interact with components of the transcriptional apparatus (ovals). Ultrasensitivity in the response to the TF can arise through cooperative binding of the TF to multiple sites mediated by direct contact (TF1), simultaneous contact with the same target complex (TF2), or by collaborative exclusion of nucleosomes (TF3). Binding of a monomeric TF (red) to a single site cannot provide ultrasensitivity. (B) A TF affects promoter activity by binding to DNA and recruiting a histone-modifying enzyme (squares) that alters the modification state of the local nucleosomes (circles). If modified nucleosomes themselves recruit enzymes that foster the same modification on other nucleosomes (orange arrows), in a positive feedback loop (Dodd et al, 2007), then a TF that binds non-cooperatively can produce an ultrasensitive response.
Many eukaryotic TFs act, at least in part, by recruiting histone-modifying enzymes to the promoter region Figure 1B. These enzymes include histone acetyltransferases and deacetylases (HATs and HDACs; Struhl, 1998; Naar et al, 2001; Eberharter and Becker, 2002), methyltransferases (HMTs; Demers et al, 2007) and demethylases (HDMs; Swigut and Wysocka, 2007). This recruitment can result in changes to the modification state of nearby nucleosomes (Vignali et al, 2000). In turn, the histone modifications carried by the nucleosomes located in the vicinity of the promoter affect transcription, although in ways that are not yet well understood. Histone modifications change the DNA-binding properties of the nucleosomes, affecting competition with other DNA-binding proteins (Anderson et al, 2001), and also can affect the binding of other proteins, including those of the transcription apparatus, to the nucleosomes themselves (Naar et al, 2001; Taverna et al, 2007; Vermeulen et al, 2007). Again, recruitment of a particular histone-modifying enzyme should not in itself provide an ultrasensitive transcriptional response, because changes in the localized activity of the enzyme should be linear with respect to changes in the DNA site occupancy of the TF that recruits it.
However, it has been proposed that nucleosome modifications can sustain positive feedback loops, where nucleosomes carrying a particular modification recruit enzymes capable of maintaining the same modification state among neighbouring nucleosomes (Grunstein, 1998; Turner, 1998; Schreiber and Bernstein, 2002). We show that such feedback loops can convert linear changes in DNA occupation by a TF into a highly ultrasensitive transcriptional response.
Results and discussion
The model
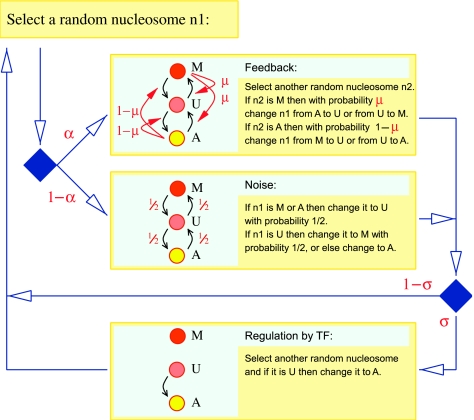
We previously introduced a simplified numerical model of positive feedback in nucleosome modification for investigation of epigenetic cell memory (Dodd et al, 2007). The model considers a patch of N nucleosomes carrying mutually exclusive modifications, for example, methylation or acetylation of a specific lysine residue (such as lysine 9 of histone H3), giving three types of nucleosome with regard to this modification: methylated (M), unmodified (U) or acetylated (A). Nucleosome interconversions can occur by a positive feedback process, where acetylated nucleosomes recruit specific HDMs and HATs, while methylated nucleosomes recruit HDACs and HMTs that can act on any other nucleosome in the patch (Figure 2). As well as these local positive feedback reactions, non-feedback or ‘noisy' interconversions can occur independently of the local nucleosomes, for example, through the action of non-recruited enzymes or enzymes bound to nucleosomes outside the patch. The relative strengths of the feedback and noise processes is parameterized by α (Figure 2), with the feedback-to-noise ratio, F defined as F=α(1−α). We found that with increasing N and F, the patch of nucleosomes becomes increasingly bistable, existing in either a highly acetylated or a highly methylated state with rare transitions between the states. Bistability also required the positive feedback reactions leading to one modification to be of similar efficiency to those leading to the opposing modification (Dodd et al, 2007).
Figure 2.
Model for positive feedback in nucleosome modifications, with regulation by a TF that recruits a histone-modifying enzyme. The system is comprised of N nucleosomes of three types: M, U and A, representing nucleosomes that are methylated, unmodified or acetylated at a specific histone residue. Interconversions occur by modification and demodification reactions (black arrows) that can be stimulated (red arrows) by M or A nucleosomes anywhere in the system (Dodd et al, 2007). Bias in the positive-feedback reactions is introduced by the parameter μ, and the strength of the TF-stimulated modification reaction (in this case histone acetylation) is set by σ. The behaviour of the system is examined by iteration of the sequence of reactions, as indicated by the flowchart. Diamonds are points where alternative reactions are chosen, based on the indicated probabilities. Thus, in each time step, there is a probability σ that a positive-feedback reaction is attempted, or else a ‘noise' reaction is attempted. In each time step, there is also a probability σ that a TF-stimulated acetylation is attempted.
Here, we make two changes to this model to investigate gene activation or repression by a TF that recruits a histone-modifying enzyme. First, we allow the relative strengths of the positive feedback reactions in one direction (e.g. towards A) to be of a different strength to the reactions in the other direction (e.g. towards M). The balance of these reactions is expressed by the parameter μ (Figure 2). If μ deviates significantly from 0.5, indicating an imbalance, then the patch of nucleosomes is only stable in one modification state (Dodd et al, 2007). We imagine that for any particular mutually exclusive modification (e.g. the M and A in Figure 2), the concentrations of modifying enzymes, their affinities for the recruiting nucleosomes and their enzymatic activities will often differ so that one modification will be favoured, possibly in a location-dependent manner, so that a particular patch of nucleosomes will tend to exist by default in a particular modification state.
Second, we add a TF that binds to the DNA within the nucleosome patch and recruits a histone-modifying enzyme, bringing the enzyme into proximity with the nucleosomes in the patch and facilitating their modification (Figure 1B). We do not consider any other action of the TF (e.g. direct interaction with the transcription complex). The strength of the TF-stimulated reaction is given by σ, which sets an additional rate for the U to A transition; a larger σ makes it proportionally more likely to change a U to an A anywhere in the system (Figure 2). We imagine that increases in σ result from increases in the activity or concentration of the TF that, in turn, increase occupation of its DNA-binding site or its ability to recruit the modifying enzyme.
Ultrasensitive responses
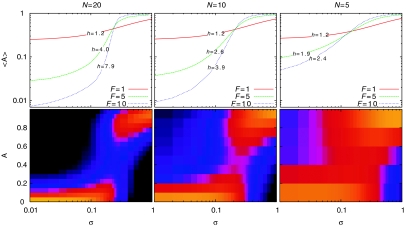
In Figure 3, we show the results of iterating the reaction steps of Figure 2 when, by default, the patch contains a low proportion of A-type nucleosomes due to an imbalance in the feedback reactions due to μ=2/3 (i.e., M-favouring reactions are twice as likely as A-favouring reactions). As the strength of the TF-stimulated HAT reaction is increased (σ increased from 0 to 1), the average steady-state fraction of acetylated nucleosomes, 〈A〉, increases to the point where A nucleosomes dominate the patch. Depending on the parameters, the model is able to give a range of ultrasensitive responses of 〈A〉 to increases in σ (Figure 3, upper panels), as quantified by the Hill coefficient h, with h>1 indicating ultrasensitivity (Koshland et al, 1982; Ferrell, 1996). If promoter activity is taken to be proportional to 〈A〉 and σ is taken to be proportional to TF activity, such that the only source of non-linearity in the system is the nucleosome modification reactions, then it is clear that this mechanism alone can generate an ultrasensitive response of promoter activity to TF activity.
Figure 3.
Ultrasensitivity and bistable expression in response to TF stimulation. The steps in Figure 2 were iterated for systems with various numbers of nucleosomes N, feedback-to-noise ratios F=α(1−α), and different levels of recruited HAT activity σ. The system was biased towards methylation by setting μ=2/3 (see Figure 2). The upper panels show the average fraction of acetylated nucleosomes 〈A〉 versus σ. The lower panels show the probabilities (lighter colours for higher) for the system to exist with a particular fraction of A nucleosomes A for the cases where F=5. The h values were obtained by fitting the curves to 〈A〉−A0∞σh/(k+σh), varying A0, K and h.
The ultrasensitivity arises from the positive feedback in the modification reactions. When there is a high fraction of one nucleosome type, increases in the minority modification due to σ are strongly resisted by the dominating activity of the majority type. However, as the threshold (M∼A) is approached, the resistance decreases, because the opposing feedback reactions are more balanced, allowing changes in A to occur easily. The strength of the positive feedback increases with F and N and thus these parameters control the ultrasensitivity (Figure 3, top panels). In principle, this could allow the ultrasensitivity of the response of a promoter to a TF to be tuned by adjusting the strength of the positive-feedback modifications relative to noise (F) or by alterations to the size of the nucleosome patch (N). A Java applet is available (http://www.cmol.nbi.dk/models/epitrans/transcript.html) for exploration of these relationships.
Although Figure 3 shows activation by recruitment of a modifying enzyme that adds a modification (Figure 2), recruitment of an enzyme that removes the opposing modification produces a similar response. Of course, the system is not limited to activation by a TF. If the modification favoured by the TF has an inhibitory effect on the promoter, then an ultrasensitive repressive response can be produced.
Comparison with experimental data
The nucleosome modification model is easily able to reproduce observed in vivo ultrasensitivities. Rossi et al (2000) found Hill coefficients (h) of 1.6–3.2 for doxycycline regulation of promoters activated by Tet repressor–VP16 fusions, while Ajo-Franklin et al (2007) found h values of 2.7–3.6 for promoter control by VP16-derived activation domains fused to ZifH or LexA DNA-binding domains. Because multiple adjacent activator DNA-binding sites were used, it is possible (but was not tested) that the ultrasensitivities were caused by cooperative DNA binding. However, the responses may also be explained by our model, as the VP16 activation domain has been shown to interact with HATs, and when fused to the Gal4 DNA-binding domain can target HAT activity to nucleosomes around the Gal4-binding site (Vignali et al, 2000). In these experiments, the two models could be tested by measuring ultrasensitivity with the same number of activator binding sites but placed at separate locations, which should disrupt cooperative binding, but not our mechanism.
The model can also explain the results of Carey et al (1990), in which an ultrasensitive promoter response was seen with respect to increasing copies (in tandem) of a binding site for a Gal4–VP16 activator. Because all the binding sites were saturated with TF, Carey and co-workers ruled out cooperative binding and proposed that the ultrasensitivity was due to 5–10 TF molecules simultaneously contacting the transcription machinery. We find it sterically implausible that the transcription machinery has so many sites simultaneously available to adjacently bound VP16 moities, while the results are easily explained by VP16 at each saturated binding site recruiting additional HAT activity, adding to σ.
Conditions in the model that give high ultrasensitivity also produce bistability. The lower panels of Figure 3 show the probability distributions of the fraction of A nucleosomes at various σ. In the high ultrasensitivity case (left panel), there are some σ values for which the promoter can exist with high probability in either a high- or a low-A state. Such bistability cannot arise from cooperative TF binding, even at high ultrasensitivities, as without positive feedback there can be only one promoter activity for each TF activity. Bistability by our mechanism may help explain observations of bimodal gene expression, where in a population of cells, a promoter can be fully on in some cells and fully off in the others, despite the cells being in the same environment (Fiering et al, 2000; Rossi et al, 2000). Stable bimodal expression can result from the combination of high ultrasensitivity and differences in TF levels between cells, as long as these differences span the off–on transition region and are reasonably persistent. In contrast, bimodal expression in our model can occur even when promoters are exposed to identical conditions, also explaining bimodal promoter activity within different nuclei in the one cell (Newlands et al, 1998) or even in the same nucleus (Riviere et al, 1998).
Adaptability to complex regulation
We note also that the proposed mechanism provides a straightforward means by which the activities of many TFs can be integrated at the promoter. If each TF recruits a histone-modifying enzyme that affects the balance between a particular pair of modifications (i.e., stimulates one of the four modification reactions; Figure 2), then the activities of these TFs combine to produce a single output. An analysis of the way in which these signals combine is beyond the scope of this report; however, our preliminary investigations indicate that the integration is not simply additive and depends on which combination of reactions is stimulated.
Finally, the evolution of a system consisting of indirectly acting TF factors should be less constrained than for systems requiring direct contact with the transcription complex. Evolution of the promoter to respond to a new TF would simply require the formation of a binding site in the DNA within the nucleosome patch. In contrast, evolution of a new regulation by a TF that acts by recruiting or stabilizing RNAP binding is likely to be limited by the stereochemical constraints of linking a DNA-binding site and an available contact site on the transcriptional apparatus.
Thus, positive-feedback loops in nucleosome modification can, in theory, provide not only a mechanism for long-term epigenetic memory (Dodd et al, 2007), but also a powerful system for controlling the way in which a promoter responds to and integrates multiple signals. Our modelling highlights the importance of further experimental work to substantiate and characterize such feedback loops.
Materials and methods
The model outlined in Figure 2 is simulated as an ongoing sequence of events (time steps) that take place in a system consisting of N nucleosomes (sites). Each site can be in one of the three states: M, U or A. There are no intermediates between these states; we disregard nucleosome heteromodification (possible as any nucleosome has two copies of any specific histone residue). Furthermore, we only allow transitions between A and U, and U and M, that is, no direct transitions between A and M. Furthermore, in the standard model, we disregard any positional effects, implying that the state of the system is completely characterized by the fraction of M sites (M) and the fraction of A sites (A). Given M and A the fraction of U sites is U=N−A−M.
The dynamics of the system is determined by two intrinsic parameters, F=α(1−α)>0 and μ ε[0,1] plus one external parameter σ that quantifies the strength of the TF.
The system dynamics is simulated in time steps (Figure 2).
First, we consider the TF-independent move, extending the previous model (Dodd et al, 2007) to take into account asymmetry between the strength of the recruited enzymes facilitating more Ms and the one facilitating more As. This asymmetry is large when the parameter μ is close to 0 or 1.
At each time step, one first selects a random nucleosome n1 among the 1,2…N nucleosomes in the system. Then:
- One selects a random number r from a uniform distribution over the interval [0,1]. If r<α, one attempts a move that simulates the feedback from the system. This is done by selecting another random nucleosome n2 from anywhere in the system and letting this attempt to mediate a change in n1:
- If n2 is in the M state, one with probability μ changes n1 one step towards M. This means that an n1 in the A state makes a transition to the U state, whereas an n1 in the U state makes a transition to the M state. If n1 was already in the M state, no change is made.
- If n2 is in the A state, one with probability 1−μ changes n1 one step towards A. This means that an n1 in the M state makes a transition to the U state, whereas an n1 in the U state makes a transition to the A state. If n1 was already in the A state, no change is made.
- If n2 is in the U state, no change is made.
If r⩾α, one makes a noise move, where the change in n1 is independent of the rest of the system. When n1 is in the M or A state, it is changed to U with probability 1/2. When n1 is in the U state, it is changed to M with probability 1/2, or else it is changed to the A state.
The above description of a single time-step involves some steps that take place with probability μ<1 and 1−μ<1. To simulate an event taking place with probability μ, we select a random number x from a uniform distribution between 0 and 1, and do the change if and only if x<μ.
To simulate the action of a TF that recruits a HAT, we at each time-step supplement the above action and:
Select a random number xε[0,1]. If x<σ, we randomly select one of the N nucleosomes in the system. If this nucleosome is in the U state, it is changed to the A state. If the nucleosome is in any other state then no change is made.
A small σ simulates a situation where the TF is not bound to its operator very often, and therefore, only rarely influences the system. A large σ corresponds to the case where the TF often recruits a HAT that makes an attempted acetylation somewhere in the system.
The model is sufficiently iterated over many time-steps to allow the system to explore all its possible states. The data in Figure 3 are collected from 109 iterations.
The program code, in C++, is given in the Supplementary information. An applet is provided at http://www.cmol.nbi.dk/models/epitrans/transcript.html.
Supplementary Material
Supplementary information
Acknowledgments
We thank Genevieve Thon for discussions and comments on the manuscript. The Center for Models of Life www.cmol.nbi.dk, is funded by the Danish National Research Foundation, MAM is also supported by the Velux foundation. IBD is funded by the NIH and the ARC.
References
- Ajo-Franklin CM, Drubin DA, Eskin JA, Gee EP, Landgraf D, Phillips I, Silver PA (2007) Rational design of memory in eukaryotic cells. Genes Dev 21: 2271–2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JD, Lowary PT, Widom J (2001) Effects of histone acetylation on the equilibrium accessibility of nucleosomal DNA target sites. J Mol Biol 307: 977–985 [DOI] [PubMed] [Google Scholar]
- Burz DS, Rivera-Pomar R, Jackle H, Hanes SD (1998) Cooperative DNA-binding by Bicoid provides a mechanism for threshold-dependent gene activation in the Drosophila embryo. EMBO J 17: 5998–6009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M (1998) The enhanceosome and transcriptional synergy. Cell 92: 5–8 [DOI] [PubMed] [Google Scholar]
- Carey M, Lin YS, Green MR, Ptashne M (1990) A mechanism for synergistic activation of a mammalian gene by GAL4 derivatives. Nature 345: 361–364 [DOI] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M (2007) Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell 27: 573–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd IB, Micheelsen MA, Sneppen K, Thon G (2007) Theoretical analysis of epigenetic cell memory by nucleosome modification. Cell 129: 813–822 [DOI] [PubMed] [Google Scholar]
- Eberharter A, Becker PB (2002) Histone acetylation: a switch between repressive and permissive chromatin. EMBO Rep 31: 224–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell JE Jr (1996) Tripping the switch fantastic: how a protein kinase cascade can convert graded inputs into switch-like outputs. Trends Biochem Sci 21: 460–466 [DOI] [PubMed] [Google Scholar]
- Fiering S, Whitelaw E, Martin DI (2000) To be or not to be active: the stochastic nature of enhancer action. BioEssays 22: 381–387 [DOI] [PubMed] [Google Scholar]
- Grunstein M (1998) Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell 93: 325–328 [DOI] [PubMed] [Google Scholar]
- Koshland DE Jr, Goldbeter A, Stock JB (1982) Amplification and adaptation in regulatory and sensory systems. Science 217: 220–225 [DOI] [PubMed] [Google Scholar]
- Miller JA, Widom J (2003) Collaborative competition mechanism for gene activation in vivo. Mol Cell Biol 23: 1623–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naar AM, Lemon BD, Tjian R (2001) Transcriptional coactivator complexes. Annu Rev Biochem 70: 475–501 [DOI] [PubMed] [Google Scholar]
- Newlands S, Levitt LK, Robinson CS, Karpf AB, Hodgson VR, Wade RP, Hardeman EC (1998) Transcription occurs in pulses in muscle fibers. Genes Dev 12: 2748–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere I, Sunshine MJ, Littman DR (1998) Regulation of IL-4 expression by activation of individual alleles. Immunity 9: 217–228 [DOI] [PubMed] [Google Scholar]
- Rossi FM, Kringstein AM, Spicher A, Guicherit OM, Blau HM (2000) Transcriptional control: rheostat converted to on/off switch. Mol Cell 6: 723–728 [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Bernstein BE (2002) Signaling network model of chromatin. Cell 111: 771–778 [DOI] [PubMed] [Google Scholar]
- Struhl K (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12: 599–606 [DOI] [PubMed] [Google Scholar]
- Swigut T, Wysocka J (2007) H3K27 demethylases, at long last. Cell 131: 29–32 [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14: 1025–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM (1998) Histone acetylation as an epigenetic determinant of long-term transcriptional competence. Cell Mol Life Sci 54: 21–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT (2007) Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 131: 58–69 [DOI] [PubMed] [Google Scholar]
- Vignali M, Steeger DJ, Neely KE, Workman JL (2000) Distribution of acetylated histones resulting form Gal4-VP16 recruitment of SAGA and NuA4 complexes. EMBO J 19: 2629–2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information