Abstract
Self-incompatibility in Brassica is controlled by a single multi-allelic locus (S locus), which contains at least two highly polymorphic genes expressed in the stigma: an S glycoprotein gene (SLG) and an S receptor kinase gene (SRK). The putative ligand-binding domain of SRK exhibits high homology to the secretory protein SLG, and it is believed that SLG and SRK form an active receptor kinase complex with a self-pollen ligand, which leads to the rejection of self-pollen. Here, we report 31 novel SLG sequences of Brassica oleracea and Brassica campestris. Sequence comparisons of a large number of SLG alleles and SLG-related genes revealed the following points. (i) The striking sequence similarity observed in an inter-specific comparison (95.6% identity between SLG14 of B. oleracea and SLG25 of B. campestris in deduced amino acid sequence) suggests that SLG diversification predates speciation. (ii) A perfect match of the sequences in hypervariable regions, which are thought to determine S specificity in an intra-specific comparison (SLG8 and SLG46 of B. campestris) and the observation that the hypervariable regions of SLG and SRK of the same S haplotype were not necessarily highly similar suggests that SLG and SRK bind different sites of the pollen ligand and that they together determine S specificity. (iii) Comparison of the hypervariable regions of SLG alleles suggests that intragenic recombination, together with point mutations, has contributed to the generation of the high level of sequence variation in SLG alleles. Models for the evolution of SLG/SRK are presented.
Keywords: self-incompatibility, SRK
Self-incompatibility (SI) systems in flowering plants prevent self-fertilization and maintain genetic variation in a population, which may confer a selective advantage during evolution. In the Cruciferae, this phenomenon is controlled by a single multi-allelic locus (S locus), which determines the specificity of the pollen–stigma interaction. The specificity expressed by pollen depends on the S genotype of the pollen parent rather than that of the pollen grain itself. This type of SI is designated as sporophytic SI. In many cases both S alleles in a heterozygote are expressed (codominant), but it is also common for one S allele to be dominant to the other (1). To date, 50 S haplotypes in Brassica oleracea (2) and 30 in Brassica campestris (3) have been identified. One of the most interesting problems to be solved has been how SI plants differentiate between such a large number of haplotypes at the same locus.
B. oleracea and B. campestris have apparently identical SI systems. In both species the S locus is a gene complex composed of at least two genes (4): the S glycoprotein gene (SLG) and the S receptor kinase gene (SRK). SLG genes encode highly polymorphic secretory glycoproteins synthesized predominantly in the stigma. SLG proteins have 12 conserved cysteine residues and several potential N-linked glycosylation sites. SLG genes are about 1.3 kb in length and contain no introns. With respect to sequence similarity, they can be classified into two groups: class I and class II (5). Class I SLG genes exhibit about 65% homology in deduced amino acid sequences to class II SLG genes. Interestingly, all of the class II S haplotypes identified thus far are pollen-recessive, whereas all of the class I S haplotypes are dominant (1, 5). Two SLG-like genes unrelated to the S locus have been identified, and they are designated as SLR1 and SLR2. SLR2 has high homology to class II SLG (about 85% in deduced amino acid sequence). The SRK protein is a membrane glycoprotein (6, 7), the cytoplasmic domain of which has intrinsic serine/threonine protein kinase activity, and which is also localized in the stigma. Interestingly, its extracellular domain, which is supposed to be a ligand-binding domain, exhibits high homology to SLG and is also polymorphic. This S domain is encoded in the first exon of the SRK gene. Genetic analysis using self-compatible mutants revealed that both SLG and SRK are essential to the SI response (8, 9).
By analogy with mammalian receptor kinases, SRK is thought to be a ligand-activated protein kinase, and it is plausible that activation of SRK by a self-pollen ligand leads to the rejection of self-pollen (4). SLG is thought to be a soluble receptor from its homology to the putative ligand-binding domain of SRK and perhaps binds to the same ligand as SRK. The S-specific pollen rejection requires that the pollen ligand should also be highly polymorphic; therefore, it is likely to be a protein. Recently, an S-locus gene that is expressed in anthers and is perhaps highly polymorphic was isolated from an S2 B. oleracea plant (10). The product of this gene is one of the candidates for the pollen ligand. It is supposed that coevolution of the three genes occurred during the process of S locus evolution.
We have successfully amplified class I SLG by PCR with specific primers and detected polymorphism among SLG alleles by PCR-restriction fragment length polymorphism (11). Cloning PCR products and sequencing them enabled us to analyze a large number of DNA sequences of class I SLG alleles from B. oleracea and B. campestris. In this paper, we report novel class I SLG sequences of 31 S haplotypes from B. oleracea and B. campestris. The SI recognition mechanism in Brassica and the evolution of S haplotypes are discussed.
MATERIALS AND METHODS
The S tester lines of B. oleracea and B. campestris were derived from the S allele reference collections that are maintained at Horticulture Research International and Tohoku University, respectively (2, 3). Genomic DNA extraction, PCR amplification, cloning of PCR products, and sequencing were conducted as described elsewhere (11). To avoid errors that may have occurred during the PCR process, two or three independent clones were sequenced. All of the sequence analyses were conducted with genetyx software, version 8.0 (Software Kaihatu, Tokyo). References for other sequences are: SLR1Bol(S22) (12), SLR1Bol (S29) (13), SLR1Bol (S63) (14), NS1Bca (15), NS3Bca (16), SLR2Bol (S2) (17), SLG8Bca (18), SLG9Bca (19), SLG12Bca (20), SLG2Bol (21), SLG3Bol (6), SLG5Bol (22), SLG6Bol (23), SLG13Bol (23), SLG29Bol (13), SRK8Bca (24), SRK9Bca (19), SRK12Bca (24), SRK2Bol (21), SRK3Bol (6), SRK6Bol (25), and SRK29Bol (26). Because the previously submitted sequence of SLG5Bol (22) was not full length, its 5′ region was complemented by the sequence of the PCR product amplified with the class II-specific primer set (11). The phylogenetic trees were generated by the neighbor joining method (27). Variation among regions in evolutionary history was examined as described by Obata et al. (28) and Takahata et al. (29).
RESULTS
Sequence Comparison Among Class I SLG Alleles.
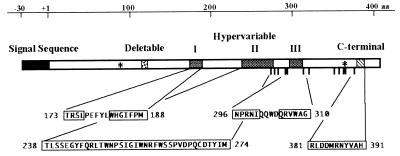
The nucleotide sequences of 31 novel class I SLG alleles from B. oleracea and B. campestris were determined, and their deduced amino acid sequences were compared with known class I SLGs. The conserved potential N-linked glycosylation sites among class I SLGs have been identified in previous papers (20, 25). Our data show that some of them are not conserved. There are only two perfectly conserved potential N-linked glycosylation sites: at positions 90 and 359 aa (of SLG6Bol SLG6 of B. oleracea). These sites are also conserved in class II SLG, SLR1, SLR2 (4), and SRK (24). Eleven perfectly conserved and one almost conserved cysteine residues (20, 23) were recognized. Deletions were often observed in the region from 115 to 120 aa. This region is highly asparagine-rich. Moreover, three hypervariable regions were recognized: 173–188 aa, 238–274 aa, and 296–310 aa. They were designated as the hypervariable regions I, II, and III, respectively (Fig. 1). The hypervariable region I is separated into two parts by five conserved amino acid residues. The hypervariable region II contains the first conserved cysteine residue and is the longest of the three regions. The hypervariable region III is sandwiched between the fifth and sixth conserved cysteine residues and is also divided into two parts by four conserved amino acid residues. These features were common to both species and the S domain of SRK. Although a C-terminal variable region (381–391 aa) was also recognized, this region contained few nonsynonymous nucleotide substitutions that would alter the property of polypeptides. The three hypervariable regions, but not necessarily all of them, may be involved in determining S specificity of SLG and SRK (18), whereas the C-terminal variable region is probably not.
Figure 1.
Structure of the SLG in B. oleracea and B. campestris. As an example, the deduced amino acid sequence of SLG6Bol is shown. Shaded boxes represent the hypervariable regions I, II, and III. Filled, stippled, and hatched boxes show signal sequence, the deletable region, and the C-terminal variable region, respectively. ∗, Indicates perfectly conserved potential N-linked glycosylation sites. Boldface bars are the 12 conservative cysteine residues. Variable regions are boxed.
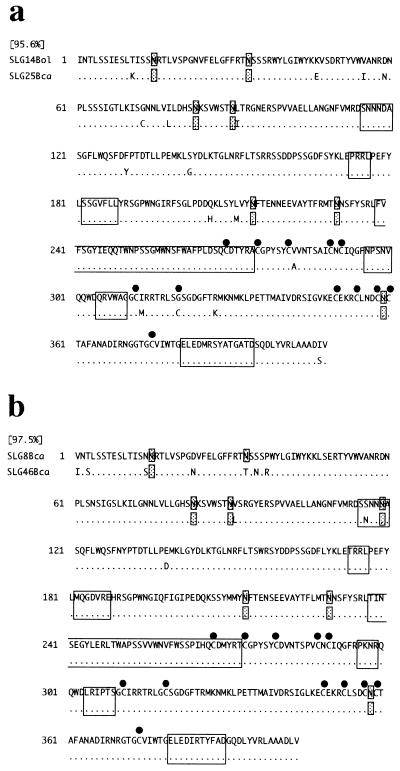
The similarity of class I SLGs to a class II SLG (SLG2Bol) ranged from 64 to 69% in the deduced amino acid sequence of the mature protein region. In comparisons between class I SLGs, similarities from 73 to 97.5% were observed. The highest similarity was observed between SLG14Bol and SLG25Bca (SLG25 of B. campestris; 95.6%, Fig. 2a), SLG46Bol and SLG63Bol (97.0%, data not shown), and SLG8Bca and SLG46Bca (97.5%, Fig. 2b). The sequences of the three hypervariable regions matched perfectly in SLG14Bol/SLG25Bca and SLG8Bca/SLG46Bca, whereas SLG46Bol/SLG63Bol contained two different amino acid residues in the hypervariable region II. Although the positions of the potential N-linked glycosylation sites are identical in SLG14Bol/SLG25Bca and SLG46Bol/SLG63Bol, SLG46Bca did not have the one that occurs at position 33 aa in SLG8Bca.
Figure 2.
Comparisons of deduced amino acid sequence between markedly similar SLG pairs. Shaded “N” and filled circles show the positions of potential N-linked glycosylation sites and conserved cysteine residues, respectively. Boxed regions represent the deletable region, the hypervariable regions, and the C-terminal variable region. Dots indicate identical amino acid residues. (a) SLG14Bol and SLG25Bca. (b) SLG8Bca and SLG46Bca.
Molecular Phylogeny of SLG and SLG-Related Sequences.
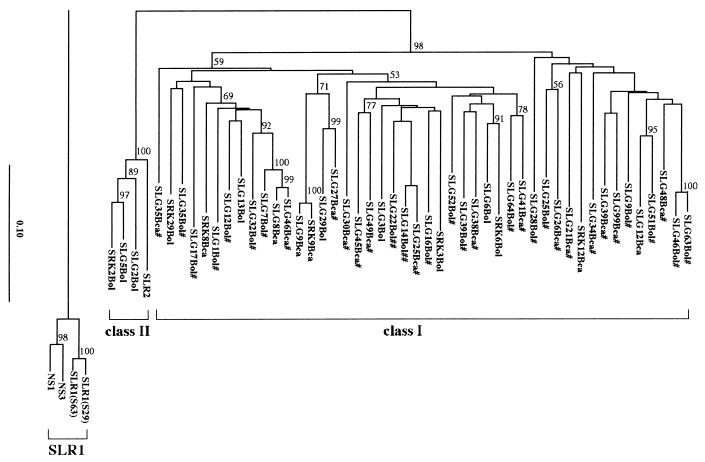
The phylogeny of SLG and SLG-related sequences including SLR1, SLR2, and the S domain of SRK was constructed using the deduced amino acid sequences of a mature protein region (Fig. 3). They clustered into three groups as shown previously (30): an SLR1 group, a class I SLG group, and a class II SLG group. The class II SLG group included SLR2. SRK did not form an independent cluster; class I SRK belonged to the class I SLG group, and class II SRK belonged to the class II SLG group. In some cases, the most similar sequence to an SRK was not the SLG of the same S haplotype. SLG alleles of B. oleracea and those of B. campestris did not form independent clusters. A number of class I SLG alleles of B. oleracea were positioned closest to one of B. campestris alleles. This observation indicates that their divergence occurred prior to speciation as suggested previously (18).
Figure 3.
Neighbor joining tree of SLG and SLG-related sequences. #, Indicates SLG sequences reported in this paper. ##, Indicates sequences for which our data show some differences from the sequences reported previously (17, 28). Our data were used in the analysis. Amino acid sequences of the putative mature protein regions of SLG, SLR1, SLR2, and S domain of SRK were used for the analysis. SLR1 group was used as an outgroup. The numbers besides the branches are the bootstrap values (%) with 1000 repeats. The scale shows amino acid substitutions/site.
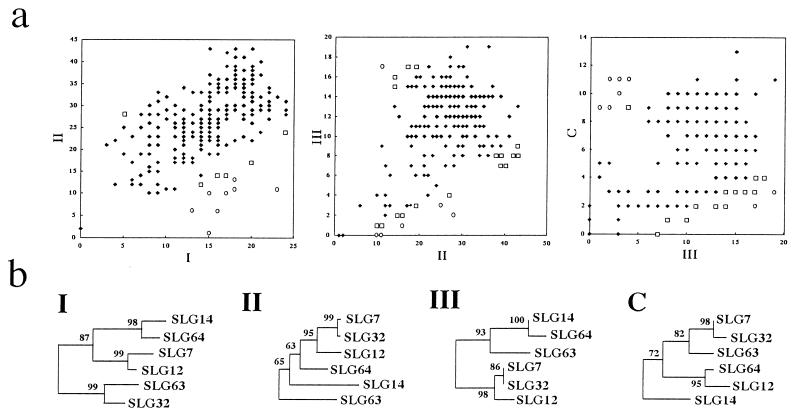
To examine whether or not different regions of SLG alleles have taken the same evolutionary pathway, we performed the following analysis. In the absence of recombination, two regions of the same gene share their evolutionary history. Different regions may have different substitution rates, but they are expected to accumulate substitutions at a constant relative rate. Unusual relative numbers of substitutions between the regions in particular pairs of sequences may indicate that the regions compared have different evolutionary histories. Under this concept, we compared the number of nucleotide substitutions in the hypervariable regions I, II, and III and the C-terminal variable region between different SLG alleles. We computed the mean pairwise number of substitutions of a specified region (d̄1) as well as the expected proportion (p̄1) of d̄1 relative to that in the adjacent two regions (d̄1 + d̄2). Assuming that d̄1 for each given pair of sequences follows the binomial distribution with parameters p̄1 and the mean total number of substitutions in the two regions, we evaluated statistically whether d̄1 among d̄1 + d̄2 can occur. If the two regions compared for a pair of SLG alleles have the same phylogenetic relationship, the number of nucleotide substitutions in the two regions should show a good correlation. We observed several exceptions to this in a comparison of the hypervariable regions I/II, II/III, and III the C-terminal variable region of B. oleracea (Fig. 4a).
Figure 4.
(a) Pairwise distribution of the number of substitutions in two different regions of class I SLG alleles in B. oleracea. All pairwise comparisons are shown. Open boxes and open circles indicate pairs of sequences showing a significant deviation from the expected binomial distribution (P < 0.05 and P < 0.01, respectively). (b) Neighbor-joining trees using nucleotide sequences of the hypervariable regions I, II, III, and the C-terminal variable region of SLG7, SLG12, SLG14, SLG32, and SLG63. SLG2 was used as an outgroup (not shown in Fig. 4b). The numbers besides the branches are the bootstrap values (%) with 1,000 repeats.
To evaluate the power of the binomial test, we carried out computer simulations (29). A random bifurcated tree was generated and the number of substitutions on each branch was determined in proportion to the branch length under a given number (d̄) of substitutions in the whole tree. We examined the proportion of allelic pairs that do not follow the binomial distribution under free recombination and complete linkage at 95% significance. Free recombination is simulated by two random trees with two different means, whereas complete linkage is simulated by a single tree with the two means. According to the generated genealogy, we calculated all pairwise distances and examined the P of the proportion of deviating allelic pairs in 1000 replications.
This probability strongly depends on the value of d̄. When d̄ was less than 10, the proportion of deviating allelic pairs could not be used to distinguish between free recombination and complete linkage. On the other hand, when d̄ was greater than 10, more than 20% of the allelic pairs deviated from the binomial distribution with P > 0.95 under free recombination, whereas less than 5% of allelic pairs did so under the complete linkage (data not shown). The observed values of d̄ in the hypervariable regions I, II, and III are more than 10. The proportion of the deviating allelic pairs for the region I/II is 6.1% and is 10.8% for the region II/III. These values suggest that these hypervariable regions are neither in free recombination nor complete linkage.
The variable regions of six SLG alleles that showed significant deviations (at the 5% level) in the above analysis were chosen to examine their phylogenetic relationship by the neighbor joining method (Fig. 4b). High bootstrap values (more than 90%) support the notion SLG32 is similar to SLG63 in hypervariable region I, whereas SLG32 is similar to SLG7 in hypervariable regions II and III and the C-terminal variable region. Also, SLG14 and SLG64 share similar sequences in hypervariable regions I and III, whereas SLG64 is similar to SLG12 in the C-terminal variable region. These observations suggest intragenic recombination in the evolution of SLG alleles.
DISCUSSION
Are SLG and SRK of the Same S Haplotype Highly Related in Their Sequences?
Stein et al. (25) found that SLG and the S domain of SRK within a haplotype showed extremely high similarity. It had been thought that an SLG would be more closely related in its sequence to the SRK of the same haplotype than to any other SLG because SLG and SRK of the same haplotype would have the same S specificity. This observation was based on the data of S2 and S6 haplotypes of B. oleracea, which differ considerably, because one is a class II and the other a class I S haplotype. To date, many SLGs and SRKs have become available for analysis. Fig. 3 shows that although some SLG/SRK pairs are similar, not all of them are closely similar. In fact, some comparisons between SLG and SRK of the same S haplotypes showed considerable differences in the three hypervariable regions, which are thought to be involved in the determination of S specificity. For example, SLG3Bol/SRK3Bol have 45.5% amino acid identity in hypervariable region I, 70.3% in hypervariable region II, and 63.6% in hypervariable region III, whereas SLG3Bol/SLG22Bol have 63.6% amino acid identity in hypervariable region I, 78.4% in hypervariable region II, and 63.6% in hypervariable region III. Even in the most similar pair, SLG9Bca and SRK9Bca, three amino acid differences occurred in hypervariable region III. This observation seems to contradict the idea that SLG and SRK of the same S haplotype must have the same specificity. To resolve this apparent contradiction and preserve the concept that the hypervariable regions could be the binding sites for the polymorphic pollen ligand (18), we theorize that SLG and SRK bind to different sites of the same pollen ligand.
Striking Sequence Similarity in Inter- and Intra-Specific Comparisons.
From the great similarity between SLG13Bol and SLG8Bca (89.6% in the amino acid sequence of the putative mature protein), Dwyer et al. (18) suggested that SLG allele divergence predated the speciation of B. oleracea and B. campestris. The ancient derivation of S haplotypes of Brassica was suggested also by statistical analysis (31). In our data, even greater similarity was observed between SLG14Bol and SLG25Bca (95.7%, Fig. 2a), providing further support for this idea. It is noteworthy that the amino acid sequences in the hypervariable regions of this pair are identical, whereas the SLG13Bol/SLG8Bca pair contains some differences. SLG14Bol/SLG25Bca may recognize the same pollen ligand. This could be confirmed by inter-specific crosses between B. oleracea and B. campestris.
Surprisingly, we also observed striking similarity in intra-specific comparisons. In particular, SLG8Bca and SLG46Bca exhibited 97.5% similarity in their amino acid sequences, and the three hypervariable regions matched perfectly (Fig. 2b). This observation seems to contradict the concept that the hypervariable regions of SLG are the sole determinants of S specificity. The loss of the carbohydrate moiety at position 33 aa in SLG46Bca might affect S specificity. It is also conceivable that amino acid residue(s) outside the three hypervariable regions influence S specificity. Alternatively, differences in SRK may permit B. campestris S8 and S46 to determine distinct specificities even though their SLGs are identical. This suggests that S specificity is determined by the combination of SLG and SRK. If SLG and SRK recognize different sites on the same pollen ligand, as we suggested earlier, the active receptor kinase complex could involve binding of the pollen ligand by SLG and SRK in concert.
Evolution of S Haplotypes.
Fisher (32) was the first to propose the idea that recombination may have been involved in the evolution of S alleles. In the gametophytic SI system, Clark and Kao (33) suggested that the evolution of the S-RNase gene, the determinant of S specificity in Solanaceae, did not involve intragenic recombination. On the other hand, Nasrallah and Nasrallah (4) claimed that “an ancient evolutionary history” of SLG alleles had been “influenced by intragenic recombination.” The present study provides the first evidence suggesting that intragenic recombination has occurred between SLG alleles. The generation of a large number of SLG alleles may have been caused not only by point mutation but also by intragenic recombination. This evolutionary process is similar to the diversification of the major histocompatibility complex class I genes in mammals (34), but the diversification of the SI system would have involved the coevolution of three genes: the soluble receptor gene, the receptor kinase gene, and their ligand gene. The coevolution of SLG and SRK might include gene conversion between them (19, 35) and the duplication of the S domain of SRK (36).
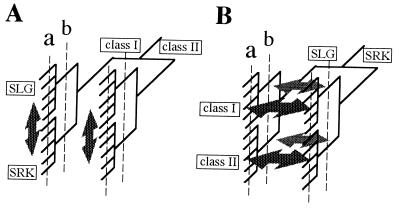
The time of divergence of SLG and SRK is an interesting question in relation to the evolutionary history of the S locus in Brassica. As mentioned earlier, SLG and SRK of the same S haplotype belong to the same class. This observation suggests that the common ancestor of SLG and SRK diverged into SLG and SRK after the class I/class II divergence (Fig. 5A) or that SLG and SRK diverged first and SLG and SRK belonging to the same class had accumulated mutations as well as maintaining their similarity through gene conversion until many S haplotypes arose (Fig. 5B). In model A, the question arises as to whether the SLG/SRK divergence occurred before or after differentiation among S haplotypes within each class. Because the latter requires that SLG/SRK divergence occurred independently in every S haplotype, we suppose that it occurred before the differentiation of S haplotypes within each class. In either model, an explanation for why SLG and SRK did not cluster independently in the phylogenetic tree would be that gene conversion between SLG and SRK had occurred after their divergence.
Figure 5.
Two models for the evolution process of SLG and SRK alleles. Arrows indicate gene conversion between SLG and SRK. Broken lines indicate possible stages at which the S haplotypes acquired the function of self-incompatibility (see text).
A classical focus of investigation concerns the origin of the physiological expression of self-incompatibility (37, 38). Multiple haplotypes are required for SI expression. Furthermore, because a functional Brassica S haplotype requires both SLG and SRK, SI expression could only have occurred after the SLG/SRK divergence. With these restrictions, two possibilities are conceivable: that SI arose after differentiation among S haplotypes within each class (Fig. 5A, line a) or that SI arose just after the SLG/SRK divergence within each class (Fig. 5B, line b). The latter hypothesis may appear unlikely because reproduction requires at least three S haplotypes under sporophytic SI with codominant determination of specificity. SI could have been first expressed in a two-haplotype system causing incomplete suppression of selfing, with differentiation among haplotypes and enhancement of SI expression evolving subsequently (38, 39). Alternatively, if the ancestor of the class I S haplotypes was dominant and the ancestor of the class II S haplotypes recessive, then a single fully functional haplotype from each class could together permit reproduction, as is the case in Primula (40). This scenario provides an explanation for the existence in Brassica of two distinct classes of S haplotypes that differ in dominance expression in the determination of specificity.
Acknowledgments
We thank Dr. Dave Astley for providing plant materials. We express our gratitude to Prof. Naoyuki Takahata for his suggestions for statistical analyses and to Dr. Marcy K. Uyenoyama for her critical review of the manuscript. This work was supported by a grant from the Science and Technology Agency of Japan.
ABBREVIATION
- SI
self-incompatibility
Footnotes
References
- 1.Thompson K F, Taylor J P. Heredity. 1966;21:345–362. [Google Scholar]
- 2.Brace J, King G J, Ockendon D J. Sex Plant Reprod. 1994;7:203–208. [Google Scholar]
- 3.Nou I S, Watanabe M, Isogai A, Hinata K. Sex Plant Reprod. 1993;6:79–86. [Google Scholar]
- 4.Nasrallah J B, Nasrallah M E. Plant Cell. 1993;5:1325–1335. doi: 10.1105/tpc.5.10.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nasrallah J B, Nishio T, Nasrallah M E. Annu Rev Plant Physiol Plant Mol Biol. 1991;42:393–422. [Google Scholar]
- 6.Delorme V, Giranton J-L, Hatzfeld Y, Friry A, Heizman P, Ariza M J, Dumas C, Gaude T, Cock J M. Plant J. 1995;7:429–440. doi: 10.1046/j.1365-313x.1995.7030429.x. [DOI] [PubMed] [Google Scholar]
- 7.Stein J C, Dixit R, Nasrallah M E, Nasrallah J B. Plant Cell. 1996;8:429–445. doi: 10.1105/tpc.8.3.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasrallah M E, Kandasamy M K, Nasrallah J B. Plant J. 1992;2:497–506. [Google Scholar]
- 9.Nasrallah J B, Rundle S J, Nasrallah M E. Plant J. 1994;5:373–384. [Google Scholar]
- 10.Boyes D C, Nasrallah J B. Plant Cell. 1995;7:1283–1294. doi: 10.1105/tpc.7.8.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishio T, Kusaba M, Watanabe M, Hinata K. Theor Appl Genet. 1996;92:388–394. doi: 10.1007/BF00223684. [DOI] [PubMed] [Google Scholar]
- 12.Lalonde B A, Nasrallah M E, Dwyer K G, Chen C-H, Barlow B, Nasrallah J B. Plant Cell. 1989;1:249–258. doi: 10.1105/tpc.1.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trick M, Flavell R B. Mol Gen Genet. 1989;218:112–117. doi: 10.1007/BF00330573. [DOI] [PubMed] [Google Scholar]
- 14.Trick M. Plant Mol Biol. 1990;15:203–205. doi: 10.1007/BF00017746. [DOI] [PubMed] [Google Scholar]
- 15.Isogai A, Yamakawa S, Shiozawa H, Takayama S, Tanaka H, Kono T, Watanabe M, Hinata K, Suzuki A. Plant Mol Biol. 1991;17:269–271. doi: 10.1007/BF00039503. [DOI] [PubMed] [Google Scholar]
- 16.Yamakawa S, Watanabe M, Isogai A, Takayama S, Satoh S, Hinata K, Suzuki A. Plant Cell Physiol. 1993;34:173–175. [PubMed] [Google Scholar]
- 17.Boyes D C, Chen C-H, Tantikanjana T, Esch J J, Nasrallah J B. Genetics. 1991;127:221–228. doi: 10.1093/genetics/127.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dwyer K G, Balent M A, Nasrallah J B, Nasrallah M E. Plant Mol Biol. 1991;16:481–486. doi: 10.1007/BF00024000. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe M, Takasaki T, Toriyama K, Yamakawa S, Isogai A, Suzuki A, Hinata K. Plant Cell Physiol. 1994;35:1221–1229. doi: 10.1093/oxfordjournals.pcp.a078716. [DOI] [PubMed] [Google Scholar]
- 20.Yamakawa S, Shiba H, Watanabe M, Shiozawa H, Takayama S, Hinata K, Isogai A, Suzuki A. Biosci Biotechnol Biochem. 1994;58:921–925. doi: 10.1271/bbb.58.921. [DOI] [PubMed] [Google Scholar]
- 21.Chen C-H, Nasrallah J B. Mol Gen Genet. 1990;222:241–248. doi: 10.1007/BF00633824. [DOI] [PubMed] [Google Scholar]
- 22.Scutt C P, Croy R R D. Mol Gen Genet. 1992;232:240–246. doi: 10.1007/BF00280002. [DOI] [PubMed] [Google Scholar]
- 23.Nasrallah J B, Kao T-H, Chen C-H, Goldberg M L, Nasrallah M E. Nature (London) 1987;326:617–619. [Google Scholar]
- 24.Yamakawa S, Watanabe M, Hinata K, Suzuki A, Isogai A. Biosci Biotechnol Biochem. 1995;59:161–162. doi: 10.1271/bbb.59.161. [DOI] [PubMed] [Google Scholar]
- 25.Stein J C, Howlett B, Boyes D C, Nasrallah M E, Nasrallah J B. Proc Natl Acad Sci USA. 1991;88:8816–8820. doi: 10.1073/pnas.88.19.8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar V, Trick M. Plant J. 1994;6:807–813. doi: 10.1046/j.1365-313x.1994.6060807.x. [DOI] [PubMed] [Google Scholar]
- 27.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Obata Y, Satta Y, Moriwaki K, Shiroishi T, Hasegawa H, Takahashi T, Takahata N. Proc Natl Acad Sci USA. 1994;91:6589–6593. doi: 10.1073/pnas.91.14.6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takahata N, Satta Y, Klein J. Genetics. 1992;130:925–938. doi: 10.1093/genetics/130.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trick M, Heizmann P. Int Rev Cytol. 1992;140:484–524. doi: 10.1016/s0074-7696(08)61107-9. [DOI] [PubMed] [Google Scholar]
- 31.Uyenoyama M K. Genetics. 1995;139:975–992. doi: 10.1093/genetics/139.2.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher R. J Theoret Biol. 1961;1:411–414. [PubMed] [Google Scholar]
- 33.Clark A G, Kao T. Proc Natl Acad Sci USA. 1991;88:9823–9827. doi: 10.1073/pnas.88.21.9823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parham P, Ohta T. Science. 1996;272:67–74. doi: 10.1126/science.272.5258.67. [DOI] [PubMed] [Google Scholar]
- 35.Goring D R, Glavin T R, Schafer U, Rothstein S J. Plant Cell. 1993;5:531–539. doi: 10.1105/tpc.5.5.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tantikanjana T, Nasrallah M E, Stein J C, Chen C-H, Nasrallah J B. Plant Cell. 1993;5:657–666. doi: 10.1105/tpc.5.6.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitehouse H L K. Ann Bot New Series. 1950;14:199–216. [Google Scholar]
- 38.Bateman A J. Heredity. 1952;6:285–310. [Google Scholar]
- 39.Uyenoyama M K. Theor Popul Biol. 1988;34:47–91. doi: 10.1016/0040-5809(88)90035-4. [DOI] [PubMed] [Google Scholar]
- 40.Dowrick V P J. Heredity. 1956;10:219–236. [Google Scholar]