Abstract
We purified from pea (Pisum sativum) tissue an ≈40 kDa reversibly glycosylated polypeptide (RGP1) that can be glycosylated by UDP-Glc, UDP-Xyl, or UDP-Gal, and isolated a cDNA encoding it, apparently derived from a single-copy gene (Rgp1). Its predicted translation product has 364 aminoacyl residues and molecular mass of 41.5 kDa. RGP1 appears to be a membrane-peripheral protein. Immunogold labeling localizes it specifically to trans-Golgi dictyosomal cisternae. Along with other evidence, this suggests that RGP1 is involved in synthesis of xyloglucan and possibly other hemicelluloses. Corn (Zea mays) contains a biochemically similar and structurally homologous RGP1, which has been thought (it now seems mistakenly) to function in starch synthesis. The expressed sequence database also reveals close homologs of pea Rgp1 in Arabidopsis and rice (Oryza sativa). Rice possesses, in addition, a distinct but homologous sequence (Rgp2). RGP1 provides a polypeptide marker for Golgi membranes that should be useful in plant membrane studies.
Plant cell wall polysaccharides, with the exception of cellulose and callose, are synthesized in Golgi dictyosomes and then exported via secretory vesicles to the cell wall by exocytosis (1). Synthases that produce a number of the Golgi-synthesized polysaccharides have been detected and partially characterized from various plant species (2). However, progress toward purification and identification of the polypeptides responsible for these synthase activities has been slow compared with that for plasma membrane-localized synthases for cellulose (β-1,4-glucan) (reviewed in ref. 3) and callose (β-1,3-glucan) (ref. 4 and references cited therein). We previously searched for pea membrane polypeptides that might be covalently labeled by UDP-[14C]Glc under the reaction conditions required by Golgi-localized glucan synthase-I (GS-I), an enzyme that forms the β-1,4-glucan backbone of hemicellulosic xyloglucan (5). A polypeptide doublet of molecular mass ≈40 kDa was rapidly labeled in the presence of the cofactors of GS-I (Mg2+ or Mn2+) and appeared, from density gradient centrifugation, to be associated with Golgi membranes (6). The glycosylation is reversible, suggesting that the doublet has enzymatic properties. The reported evidence (6) and additional information given here indicates that the two members of the doublet are different size forms of the same polypeptide, hereafter called reversibly glycosylated polypeptide-1 (RGP1).
This paper reports purifying RGP1 to homogeneity, raising an antiserum against the purified polypeptide, and using this (i) to immunocytochemically localize RGP1 intracellularly and (ii) to isolate a corresponding cDNA, which we have characterized and compared with homologous sequences in the expressed sequence tag (EST) database.
MATERIALS AND METHODS
Purification of RGP1.
Peas, Pisum sativum L. cv. early Alaska, were grown for 8 days under dim red light (6). Tissue from the 2 cm of stem just below the apical hook was homogenized and centrifuged as described (6), except that the post-1,000 × g supernatant was centrifuged at 120,000 × g. The supernatant was brought to 40% saturation with (NH4)2SO4 and, after 2 hr in ice, was centrifuged 20 min at 30,000 × g. The pellet was resuspended in 50 mM Mops (pH 7.0) and dialyzed against this buffer overnight, 15 mM MnCl2 was added, and, after ≈12 hr in ice, the mixture was centrifuged 20 min at 30,000 × g. The supernatant was passed through 5 ml of UDP-glucuronic acid agarose (Sigma) equilibrated with resuspension buffer containing 5 mM MnCl2. The column was washed with this buffer until the A280 of the effluent approached zero. RGP1 was eluted with resuspension buffer containing 1 mM EDTA and no Mn2+.
Glycosylation Assay.
RGP1 was incubated normally for 5 min at 22°C with UDP-[14C]Glc or another labeled UDP sugar in 50 mM Mops buffer (pH 7.0) containing 5 mM MnCl2, terminated by adding 80% ethanol, and collecting insoluble material on a glass fiber paper disc, as described for GS-I assay (7). This assay is satisfactory for glycosylation of purified RGP1 because (in contrast to membrane preparations) with RGP1 alone no polysaccharide is formed from UDP-Glc or other glycosyl donors.
Peptide Sequencing.
RGP1 protein was precipitated with 9 vol of 9:1 (vol/vol) methanol:chloroform, resuspended in 2 M urea/100 mM NH4HCO3. To 20 μg of protein, 1 μg of trypsin (Boehringer Mannheim) suspended in 0.01% (wt/vol) trifluoroacetic acid (TFA) was added and kept overnight at 37°C, 2.5% TFA was added and, after centrifugation, the supernatant was fractionated by HPLC on a C-4 column (250 mm × 5 μm; Vydac, Hesperia, CA) with a gradient of 3.5–70% (vol/vol) acetonitrile in water with 0.1% TFA in the water and 0.025% TFA in the acetonitrile solution. A212 was recorded on-line and peaks were collected manually. Material from two clean-looking peaks was pooled and rechromatographed; the resulting peaks were sequenced by the Stanford Protein and Nucleic Acids Facility.
Antiserum.
RGP1 purified as described above was further purified by SDS/PAGE and electroelution and antiserum was raised as described for other proteins in ref. 4 except that twice as much RGP1 was used for each injection. After the first bleeding following the third injection, booster shots of 50–100 μg RGP1 in incomplete adjuvent were given at approximately 10-week intervals, between which the rabbit was bled twice.
Immunoglobulins were isolated from antiserum as described (4). Anti-RGP1 antibodies were isolated from antiserum by adsorption to RGP1 protein expressed in Escherichia coli from cloned Rgp1 cDNA, using the method of Johnson et al. (8).
Electrophoresis and Blotting.
SDS/PAGE was performed as previously described (6) and the separated proteins were blotted onto nitrocellulose sheets. Western blotting was performed as described (4), but with a 1:100,000 dilution of the anti-RGP1 antiserum in TBSN buffer (50 mM Tris⋅Cl, pH 7.5/125 mM NaCl/0.3% Nonidet P-40).
Genomic DNA was isolated from pea stem tissue as described (9). Southern blot analysis was performed by the method of Athma and Peterson (10) as described in ref. 11, using cDNA as a probe. Northern blot analysis was carried out as described (12) using whole cDNA as a probe. RNA was prepared using the RNeasy kit from Qiagen (Chatsworth, CA).
Gene Cloning.
A pea cDNA library from mRNA from the subapical zone of 7–8-day-old etiolated pea seedlings was constructed by Julie Palmer in Winslow Briggs’ laboratory (Department of Plant Biology, Carnegie Institution of Washington, Stanford, CA). Sequences from this library were cloned into the expression vector λ uni-ZAP XR (Stratagene), between EcoRI and XhoI cloning sites. E. coli cells (XL-1 Blue, Stratagene) were plated with ≈50,000 plaque-forming units of constructs per 150-mm plate and incubated at 42°C for 4 hr. Nitrocellulose sheets (BA-S 85, Schleicher & Schuell) treated with 10 mM isopropyl β-d-thiogalactoside and dried were laid over plaque-bearing plates, incubated at 37°C for 4 hr, the blots removed, washed in TBSN, and developed as with Western blots but using 1:10,000 dilution of anti-RGP1 antiserum. Positive plaques were cored, vortexed in 50 mM Tris⋅Cl (pH 7.5)/10 mM MgCl2, kept in ice overnight, replated at a density of 200–300 plaque-forming units per 100-mm plate, and screened in the same way. Phage from positive plaques were subjected to in vivo excision as recommended by Stratagene. The phagemid-containing E. coli cells (SOLR strain, Stratagene) were grown in Terrific broth (12) with 50 μg/ml ampicillin. DNA was prepared by the alkaline lysis method (12). Sequencing was performed by the Iowa State University Biomolecular Resource Center (Ames). Sequence analysis was performed with the Wisconsin Sequence Genetics Computer Group Analysis Package (GCG, Madison).
Electron Microscopy.
Pea stem subapical tissue was prepared by rapid freeze-fixation and freeze-substitution as described (13), except omitting OsO4 and embedding in LR White following the manufacturer’s recommendations (Ted Pella, Redding, CA). Ultrathin sections on formvar-coated gold grids were floated for 1 hr on 100 mM Tris⋅Cl (pH 7.5), 0.45 M NaCl, 0.5% Tween 20, 0.1% NaN3 (TBST) containing 5% BSA (TBST/B) followed by 10 min on TBST, then 1 hr on 20 μl of anti-RGP1 antiserum diluted 1:50 with TBST/B. After two washes in TBST they were treated for 1 hr with goat anti-rabbit IgG conjugated to 15 nm colloidal gold particles (Ted Pella) diluted 1:20 with TBST/B, washed twice with TBST, rinsed in distilled water, stained in 2% aqueous uranyl acetate followed by lead citrate, and viewed with a Phillips 400 transmission electron microscope at 60 keV.
RESULTS
Purification of RGP1.
Although we discovered RGP1 as a membrane-associated pair of glycosylatable polypeptides (6), we noted in that report that these polypeptides occur also in the soluble fraction of pea homogenates. In this work we purified RGP1 to apparent homogeneity, from the soluble fraction, by affinity chromatography on UDP-glucuronic acid agarose (Fig. 1). Because UDP-glucuronate is coupled by carbodiimide linkage to the agarose of this matrix, it is equivalent to UDP-Glc agarose (14). RGP1 binds to the matrix in the presence of Mn2+ and can be eluted by EDTA, as was an earlier purified UDP-Glc glucosyltransferase (14). No detectable proteins from pea other than RGP1 bound to and eluted from the column under these conditions.
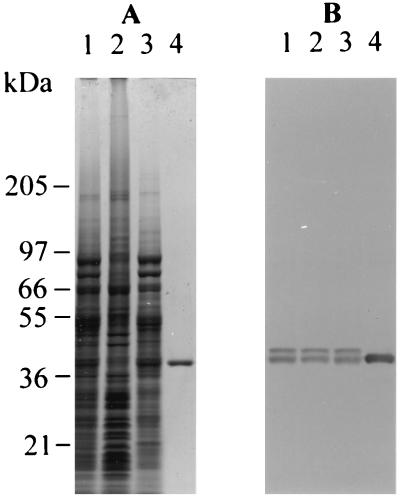
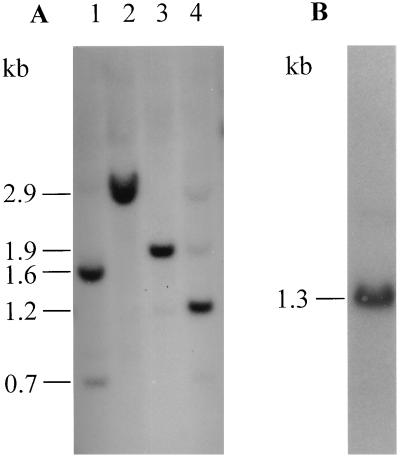
Figure 1.
Coomassie blue-stained SDS gel of cell fractions (A) and corresponding Western blot probed with anti-RGP1 antiserum (B). Lane 1, initial homogenate; lane 2, pellet from 120,000 × g centrifugation of homogenate; lane 3, supernatant from the preceding step; lane 4, affinity-purified RGP1. For Coomassie blue staining, 15 μg of total protein was loaded in lanes 1–3 and 1 μg in lane 4; half as much protein was loaded for Western blotting.
Although the smaller and larger members of the RGP1 doublet occur in about equal amounts in the starting material (ref. 6 and Fig. 1), when the affinity purification was conducted as described, the smaller member was obtained exclusively (Fig. 1). If the ammonium sulfate precipitation step was omitted from that procedure, however, a minor proportion of the larger member accompanied the smaller (not shown). Since the larger member persists unaltered in unfractionated homogenates and in purified preparations that contain it over periods much longer than are needed to carry out the purification, we think that the affinity matrix must have a higher affinity for the smaller than the larger member and thus selects in favor of the smaller member during purification.
Characterization of Purified RGP1.
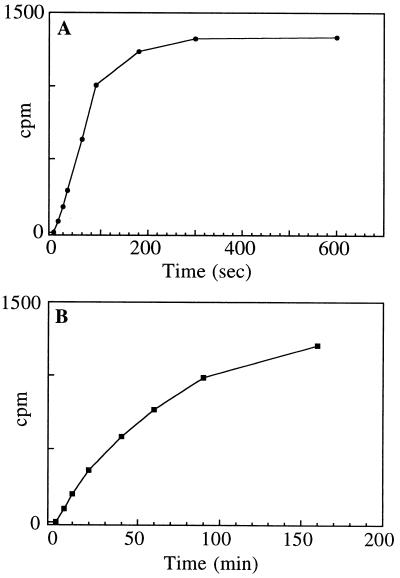
UDP-[14C]Glc glycosylates purified RGP1 under the same conditions (namely, in the presence of Mn2+ or Mg2+) as are needed for glycosylation of membrane-associated RGP1 (6). Glycosylation occurs at least as rapidly (Fig. 2) as in crude membranes (6) despite the absence of other soluble or insoluble proteins in the purified preparations. RGP1 thus evidently is autoglycosylated. As with membrane-associated RGP1 (6), glycosylation by UDP-[14C]Glc is reversible, addition of unlabeled UDP-Glc discharging the label from the polypeptide within minutes (Fig. 3B). Also, in common with membrane-associated RGP1 (6), purified RGP1 can alternatively be glycosylated by UDP-Xyl or UDP-Gal (Fig. 4). Consistent with this, unlabeled UDP-Xyl or UDP-Gal discharge, from RGP1, label previously incorporated from labeled UDP-Glc, just as unlabeled UDP-Glc itself does (Fig. 3B). Also, consistent with these observations, RGP1 glucosylation by labeled UDP-Glc can be inhibited by unlabeled UDP-Xyl or UDP-Gal, but not by UDP-Man, GDP-Glc, ADP-Glc, or TDP-Glc [Fig. 3A; we showed earlier (6) that glucosylation also is not inhibited by GDP-Man]. Evidently, nonuridine sugar nucleotides as well as uridine sugar nucleotides other than those containing Glc, Xyl, or Gal (to the extent tested) do not glycosylate RGP1.
Figure 2.
Time course of incorporation of label from UDP-[14C]Glc into RGP1 (A) at 25°C and (B) at 0°C. After 4 hr at 0°C (not shown), incorporation reached the same value as the final incorporation at 25°C.
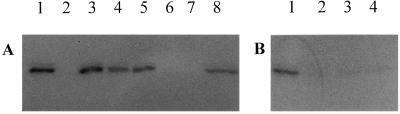
Figure 3.
Fluorograph of the 40-kDa zone of an SDS gel showing (A) the effect of various sugar nucleotides on labeling of purified RGP1 by UDP-[14C]Glc and (B) the activity of certain sugar nucleotides to cause chase-out, from purified RGP1, of label previously incorporated from UDP-[14C]Glc. (A) RGP1 was incubated for 7 min with UDP-[14C]Glc in the absence (lane 1) or presence of 1 mM unlabeled UDP-Glc (lane 2), 1 mM ADP-Glc (lane 3), 1 mM GDP-Glc (lane 4), 1 mM TDP-Glc (lane 5), 1 mM UDP-Gal (lane 6), 1 mM UDP-Xyl (lane 7), or 1 mM UDP-Man (lane 8). (B) After incubating RGP1 with UDP-[14C]Glc for 7 min (lane 1), one of the following unlabeled sugar nucleotides (1 mM) was added and incubation continued for another 7 min before stopping the reaction: lane 2, UDP-Glc; lane 3, UDP-Gal; lane 4, UDP-Xyl.
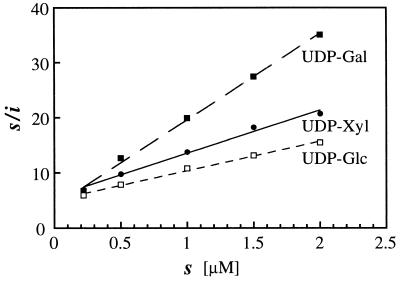
Figure 4.
Hanes plot of steady-state glycosylation (after 10 min incubation at 25°C) of RGP1 by [14C]sugar-labeled UDP-Glc, UDP-Gal, or UDP-Xyl at different concentrations. The ratio (s/i) between glycosyl donor (s) and incorporated glycosyl group (i) concentrations is plotted on the ordinate, against donor concentration (s) on the abscissa. Extrapolating the regression lines to the abscissa gives estimates of 1.05, 0.78, and 0.25 μM for the concentrations of UDP-Glc, UDP-Xyl, and UDP-Gal, giving one-half of maximum glycosylation (s1/2), respectively. The value of s/i where an extrapolated regression line intersects the ordinate is s1/2/imax, where imax is the extent of the process at substrate saturation. From the s1/2 and s1/2/imax values the ratio between the imaxs comes out 10:7:3 for UDP-Glc:UDP-Xyl:UDP-Gal.
Fig. 4 gives Hanes plots for the steady state of glycosylation (plateau of 14C incorporation in Fig. 2A) at different concentrations of labeled UDP-Glc, -Xyl, or -Gal. The linearity of the Hanes plots is consistent with a steady state in which the glycosylation reaction is glycosyl donor-saturable with simple Michaelis–Menten kinetics, and the rate of an opposing reaction that deglycosylates RGP1 is simply proportional to the extent of glycosylation. From these data it can be estimated that the steady-state glycosylations by UDP-Glc, -Xyl, and -Gal are approximately in the ratio 10:7:3 (see legend to Fig. 4).
Anti-RGP1 antiserum, raised against the purified smaller member, recognizes the smaller and larger members equally well (Fig. 1B). This was true also of antisera previously raised separately against the smaller and larger membrane-associated RGP1 members (6), and shows that the two members must be structurally very similar, as they appear from the above to be functionally similar. In contrast to the earlier antisera (6), antiserum raised against affinity-purified RGP1 is highly specific for RGP1 (Fig. 1B), and does not crossreact with any chloroplast protein. The antiserum has a very high titer because it detects RGP1 in crude extracts (in which RGP1 comprises only about 0.1% of the total protein) even at 1:100,000 dilution.
Gene Cloning.
By immunoscreening a pea expression library with the anti-RGP1 antiserum, we isolated 32 positive clones from a total of 5 × 105 plaques. These clones all appeared identical as judged by Southern blotting and restriction analysis. In agreement with this, Southern blot analysis of genomic DNA (Fig. 5A) showed that the Rgp1 gene occurs as a single gene in the pea genome. By Northern blot analysis (Fig. 5B) we found apparently only one mRNA of about 1.3 kb. This further suggests that there is only one Rgp1 gene.
Figure 5.
Southern (A) and Northern (B) blots obtained by using Rgp1 cDNA as a probe. (A) Pea genomic DNA (10 μg per lane) was digested by DraI (lane 1), EcoRI (lane 2), EcoRV (lane 3), or HindIII (lane 4) before being subjected to Southern blot analysis. (B) Total RNA was prepared and Northern blot analysis performed (with 10 μg RNA per lane) as described.
The coding region of the 1.3-kb Rgp1 cDNA predicts a polypeptide of 364 amino acids (Fig. 6A) with a molecular mass of 41.5 kDa and a pI of 5.96. The sequences of two tryptic peptides that we obtained from RGP1 match residues 24–37 and 159–173 of the predicted polypeptide, respectively (Fig. 6A). This shows that the cDNA we cloned encodes the RGP1 polypeptide.
Figure 6.
Aminoacyl sequences predicted from base sequence of Rgp1 cDNAs and comparison with other sequences. (A) Top row shows predicted (pea) PsRGP1 sequence; boldface letters denote sequences of two tryptic peptides that we obtained from purified RGP1, which match corresponding sequences predicted from its cDNA. Below, tryptic peptide sequences reported from (corn) ZmRGP1 (“amylogenin”) (15) are shown; these authors designated their peptides with the numbers given next to the upward arrows. The shaded R (position 151 in PsRGP1 sequence) is where Singh et al. (15) determined that a glucosylated arginine occurs in the corresponding sequence for corn peptide T6. (B) Alignment of a 50-amino acid portion of pea (Ps) RGP1, around R-151 (boldface), with portions of EST sequences from Arabidopsis (At, T23020) and rice (Os1, D42010 and Os2, D23283), shaded to show areas of identity. The line labeled Zm shows corresponding portions of tryptic peptides T6 and T4 of Singh et al., which are not represented in the dbest database (the single corn EST corresponds to positions 275–341 of PsRGP1). Gh shows part of the deduced sequence (between position numbers cited at each side) of the UDP-Glc-binding domain of cotton celA (3) in which an R occurs flanked by several residues that match or resemble those in the above sequences.
Several EST sequences (dbest database) from rice and Arabidopsis thaliana and one from maize are homologous to portions of our pea Rgp1 sequence.¶ These related sequences will be distinguished here by the informal designations PsRgp1, ZmRgp1, AtRgp1, and OsRgp1 for pea (P. sativum), corn (Z. mays), A. thaliana, and rice (O. sativa), respectively [formal designation of the pea gene, following recommendations of the Commission on Plant Gene Nomenclature (16), would be Rgp1;Pi.sat].
For Arabidopsis, a full-length cDNA can be assembled by combining overlapping sequences from ESTs Z38054, T23020, N65528, N65622, and T44917, in that order. The remaining Arabidopsis ESTs appear to be identical with portions of this sequence suggesting only one expressed AtRgp1 gene. This sequence predicts a 357-amino acid long translation product 87% identical to PsRGP1. The ESTs from rice can be separated into two nonidentical sequences, nine of them forming one group (OsRgp1) that is 80–90% identical to the PsRgp1 sequence at the amino acid level. The other two (D23283 and D24192) comprise a sequence (OsRgp2) that is only about 55% identical to either the OsRgp1 or PsRgp1 sequence. These relationships are partly displayed in Fig. 6B, which gives an alignment of a 50-amino acid stretch around PsRGP1 residue R-151.
Intracellular Localization.
Immunogold labeling with anti-PsRGP1 antiserum detects RGP1 in Golgi dictyosomes (Fig. 7). Almost no gold particles occur over the plasma membrane or any other membrane or organelle, including plastids (Fig. 8). Only occasional gold particles occur over the cytoplasmic matrix. They presumably represent cytosolic RGP1 and/or RGP1 that is being synthesized on cytoplasmic ribosomes.
Figure 7.
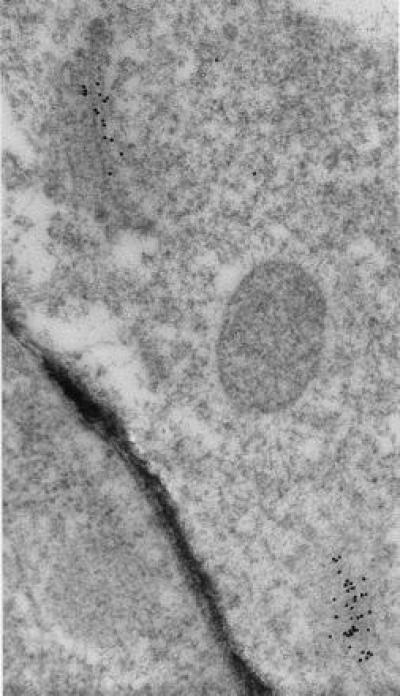
Immunolocalization of RGP1 in a cryofixed pea cell shows heavy labeling of two dictyosomes with almost none over other cell structures or cytoplasm (×60,000). Dictyosome at lower right was sectioned obliquely; its perimeter is shown by the secretory vesicles visible around it. Gold particles occur over only about one-half of the cisternae in each dictyosomal stack.
Figure 8.
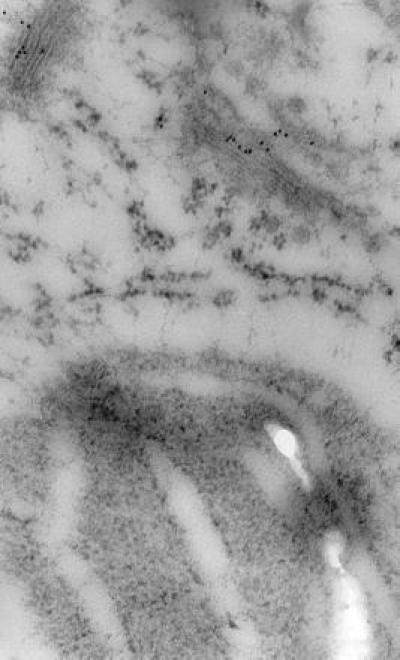
Immunocytochemical localization of RGP1 in part of a cryofixed pea cell showing gold particles over two dictyosomes but none over a plastid containing starch grains nor an endoplasmic reticulum cisterna between the plastid and the nearest dictyosome (×49,000). As in Fig. 7, gold particles occur over about one-half the cisternae in each stack. Swelling of plastid envelope and thylakoids and lack of electron-dense material in these membranes, in endoplasmic reticulum, and in the mitochondrial outer envelope (Fig. 7), resemble published images of other cryofixed plant materials not stained with OsO4 (e.g., ref. 17).
Identical results were obtained using antibodies that had been isolated from the anti-RGP1 antiserum by an affinity purification method using the coli-expressed-cloned Rgp1 gene as noted in Materials and Methods. Preimmune controls showed no Golgi localization of gold particles.
Immunolabeling consistently indicated that RGP1 occurs in only about one-half of each dictyosome (Figs. 7 and 8), therefore presumably on either the cis or the trans side. Although the preservation of Golgi structure in our immunolabeled micrographs is imperfect, it is at least as good as in previous immunocytochemical studies of plant Golgi in which cis, medial, and trans cisternae have been distinguished and shown to be differentiated biochemically (18–21). As in the works just cited, the trans can be distinguished from the cis cisternae in our micrographs by the trans showing greater electron density and often some curvature, at least of the outermost cisterna. By these criteria it is clear (Figs. 7 and 8) that RGP1 occurs primarily on trans cisternae but possibly to some extent also in medial cisternae, although these are difficult to distinguish categorically by morphology. RGP1 probably occurs also in the trans-Golgi network, although the appearance of the trans-Golgi network is generally vague in this type of preparation. We observed comparable localization in numerous immunolabeled micrographs besides those shown in Figs. 7 and 8.
DISCUSSION
Isolation of RGP1 by affinity chromatography allowed us to raise a potent and highly specific polyclonal anti-RGP1 antiserum, which in turn enabled us to clone and characterize the Rgp1 gene and to demonstrate immunocytochemically a striking localization of RGP1 to trans-Golgi cisternae.
The amino acid sequence inferred for RGP1 (Fig. 6) from the PsRgp1 nucleotide sequence apparently lacks any signal peptide. Nor does it have any membrane-spanning regions, according to hydropathy analysis (not shown). These features suggest that RGP1 is made on cytosolic polysomes and associates postsynthetically with Golgi membranes as a peripheral protein. This is consistent with the occurrence of some of the RGP1 as a soluble protein in pea homogenates, from which we purified this protein in this work, and with our failure to detect any endoplasmic reticulum-associated RGP1 immunocytochemically. However, in these tests we detected much less RGP1 in the cytosol than we had expected on the basis of having found about two-thirds of the total RGP1 in the soluble fraction of homogenates (6). This suggests that much of the soluble RGP1 found in homogenates is released from Golgi membranes during cell fractionation procedures, for example when membranes are pelleted and resuspended (cf., ref. 6).
The immunological crossreactivity, and the finding that the pea genome apparently contains only one gene for this type of polypeptide, indicate that the smaller and larger members of the RGP1 doublet are different forms of the same protein. The smaller member may differ from the larger by being trimmed of a peptide of about 20–30 residues.
Recent reports have described from sweet corn an autoglycosylated (by UDP-Glc) polypeptide of molecular mass considered to be 38 (22) or 42 (15) kDa. This polypeptide, called UDP-glucose:protein glucosyltransferase (22) or amylogenin (15), is thought by these authors to be an intermediate in, or primer for, starch synthesis. The properties of the corn polypeptide, to the extent reported, closely match those we reported for PsRGP1 (6). Sequences of tryptic peptides obtained from the corn polypeptide (15) show an overall 84% identity with corresponding regions of the sequence encoded by the PsRgp1 gene (Fig. 6). Moreover, the N-terminal sequences of two tryptic peptides we obtained from pea RGP1 (Fig. 6) largely correspond with the initial portions of the sequences of tryptic peptides T4 and T8 from the corn protein (15). Part of the amino acid sequence equivalent to the corn EST W21688 sequence is identical to Singh et al.’s (15) peptide T7 plus half of T5, and this EST is 80% identical at the amino acid level to the corresponding part of PsRgp1. These similarities indicate that corn “amylogenin” is homologous to PsRGP1. Therefore, we refer to the corn polypeptide as ZmRGP1. Strong homologies of PsRgp1 also to ESTs from A. thaliana and rice, partially displayed in Fig. 6B, show that the RGP1 gene occurs widely in higher plants.
The sequence of PsRgp1 contains arginine at position 151, which is equivalent to the position within corn tryptic peptide T6 where Singh et al. (15) determined that an arginine in ZmRGP1 becomes glycosylated by UDP-glc (see Fig. 6A). Therefore, we presume that glycosylation of PsRGP1 occurs at R-151.
Interestingly, the portion of the RGP1 sequence around R-151 can be aligned with an 18-amino acid stretch within the UDP-Glc-binding domain of GhCelA (last line in Fig. 6B), the presumed cellulose synthase of cotton (3). Although identity within this stretch is only 39%, conservative substitutions raise the similarity to 67% or higher. Whether this resemblance has significance in the interaction of CelA with UDP-Glc we of course do not yet know. However, we found no comparable match to any part of the sequences of either granule-bound or soluble starch synthases (23, 24), which use primarily ADP-Glc as substrate.
One of the main reasons for assuming that ZmRGP1 functions in starch synthesis was its occurrence in starch-synthesizing endosperm tissue (15, 22). However, as this tissue cellularizes from the liquid endosperm (“milk”) stage, it also synthesizes cell wall polysaccharides. The properties of PsRGP1 suggest that it acts in the synthesis of hemicellulosic polysaccharides, specifically xyloglucan (6), although other evidence (6) indicates that RGP1 is not by itself the xyloglucan backbone-forming enzyme [GS-I (5)]. Besides UDP-Glc, RGP1 can be glycosylated by UDP-Xyl and UDP-Gal, but it does not interact with UDP-Man or GDP-Man. Glc, Xyl, and Gal, but not Man, are constituents of xyloglucan. Furthermore, the maximum extents of RGP1 glycosylation by UDP-Glc, UDP-Xyl, and UDP-Gal are in about the same ratio (see legend to Fig. 4) as the relative proportions of these sugars typically found in xyloglucan (1:0.75:0.25). On the other hand, RGP1 does not interact with ADP-Glc, the principal substrate for starch synthesis in plants (23, 24).
Our observation that RGP1 is localized to Golgi dictyosomes and not plastids further implicates a function in cell wall rather than starch synthesis. Using anti-xyloglucan antisera the trans cisternae of Golgi dictyosomes have been identified as the site of xyloglucan formation (20, 25). Synthesis of pectic polysaccharides, the other important Golgi-synthesized polymers, occurs instead in the cis and medial cisternae (20, 25). Therefore, our finding that RGP1 is localized to trans-Golgi cisternae further strengthens the concept that RGP1 participates in xyloglucan formation. Whereas antibodies against xyloglucan detect this polysaccharide also in Golgi-derived secretory vesicles and in the cell wall (20, 25), the sites respectively of xyloglucan secretory transport and deposition, we do not find RGP1 in either of these latter locations. Its association with the site of xyloglucan synthesis is quite specific.
Therefore, despite the clear homology of PsRGP1 with the ZmRGP1, which Singh et al. (15) called amylogenin, this latter name (which implies a role in starch synthesis) is not appropriate for PsRGP1, and may well not be appropriate for ZmRGP1 either.
Could RGP1 act instead in secretory protein glycosylation? Although xylosylation and galactosylation of proteins occurs in the Golgi (26, 27), Glc seems to be added to plant glycoproteins [as a transient part of high-Man chains (27)] only in the endoplasmic reticulum, where we find no indication that RGP1 occurs (see Fig. 8). Moreover, protein xylosylation and arabinosylation (thus probably also galactosylation) occurs primarily in the cis and medial cisternae (20, 21), rather than in the trans cisternae where RGP1 occurs. Therefore, neither the nucleotide substrate specificity nor the localization of RGP1 seem to agree with a role in protein glycosylation.
Muñoz et al. (28) recently provided evidence for a system for sugar nucleotide transport across pea Golgi membranes that has a substrate specificity (to the extent tested) strikingly similar to that of RGP1. However, the covalent nature of RGP1 glycosylation (6, 15) and the lack of transmembrane domains in RGP1 speak against its simply being a sugar nucleotide transporter, though not necessarily against its being a component of a complex system for coupled sugar nucleotide transport and polysaccharide synthesis.
As suggested earlier (6), RGP1 may act as a primary acceptor at the trans-Golgi cisternal membrane for Glc, Xyl, and Gal residues from UDP sugars after which separate transferases, possibly associated as a complex in the membrane, can transfer these residues to an elongating polymer. We are currently attempting to test this hypothesis.
As a plant polypeptide-based Golgi-specific marker against which an antibody has now been obtained, RGP1 and/or its antibody should be useful in studies that require identification of plant Golgi membranes.
Acknowledgments
We thank Dr. Julie Palmer and Prof. Hans Kende for pea cDNA libraries, Ms. Fran Thomas (Stanford Biological Sciences Department) for assistance with electron microscopy, and Dr. Bob Bensen (Pioneer Hi-Bred International) for help in Southern blotting. This work was supported by a grant to P.M.R. from the U.S. Department of Energy.
ABBREVIATIONS
- EST
expressed sequence tag
- RGP
reversibly glycosylated polypeptide
- GS-I
glucan synthase-I
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U31565).
GenBank EST accession nos. with homology to PsRgp1: Arabidopsis A042694, H37657, H76915, N37306, N65402, N65528, N65622, R30021, R90614, T04300, T20512, T22507, T22943, T23020, T42672, T44394, T44917, T45672, T46245, T46745, Z17897, Z37199, Z38054; rice D15688, D15900, D23283, D24192, D25050, D28284, D39893, D40029, D40099, D40369, D42010; corn W21688.
References
- 1.Ray P M, Eisinger W R, Robinson D G. Ber Dtsch Bot Ges. 1976;89:121–146. [Google Scholar]
- 2.Bolwell G P. Phytochemistry. 1988;27:1235–1253. [Google Scholar]
- 3.Pear J R, Kawagoe Y, Schreckengost W E, Delmer D P, Stalker D M. Proc Natl Acad Sci USA. 1996;93:12637–12642. doi: 10.1073/pnas.93.22.12637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhugga K S, Ray P M. Eur J Biochem. 1994;220:943–953. doi: 10.1111/j.1432-1033.1994.tb18698.x. [DOI] [PubMed] [Google Scholar]
- 5.Ray P M. Biochim Biophys Acta. 1980;629:431–444. doi: 10.1016/0304-4165(80)90149-x. [DOI] [PubMed] [Google Scholar]
- 6.Dhugga K S, Ulvskov P, Gallagher S R, Ray P M. J Biol Chem. 1991;266:21977–21984. [PubMed] [Google Scholar]
- 7.Ray P M. Plant Physiol. 1985;78:466–472. doi: 10.1104/pp.78.3.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson L M, Snyder M, Chang L M S, Davis R W, Campbell J L. Cell. 1985;43:369–377. doi: 10.1016/0092-8674(85)90042-x. [DOI] [PubMed] [Google Scholar]
- 9.Doyle J J, Doyle J L. Focus. 1990;12:13–15. [Google Scholar]
- 10.Athma P, Peterson T. Genetics. 1991;128:163–174. doi: 10.1093/genetics/128.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johal G S, Briggs S P. Science. 1992;258:985–987. doi: 10.1126/science.1359642. [DOI] [PubMed] [Google Scholar]
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Tiwari S C, Polito V S. Protoplasma. 1988;147:5–15. [Google Scholar]
- 14.Attinen H, Kivirikko K I. Biochim Biophys Acta. 1976;429:750–758. doi: 10.1016/0005-2744(76)90322-3. [DOI] [PubMed] [Google Scholar]
- 15.Singh D G, Lomako J, Lomako W M, Whelan W J, Meyer H E, Serwe M, Metzger J W. FEBS Lett. 1995;376:61–64. doi: 10.1016/0014-5793(95)01247-6. [DOI] [PubMed] [Google Scholar]
- 16.Price C A, Reardon E M, Lonsdale D M. Plant Mol Biol. 1996;30:225–227. doi: 10.1007/BF00020109. [DOI] [PubMed] [Google Scholar]
- 17.Lancelle S A, Callahan D A, Hepler P K. Protoplasma. 1986;131:153–165. [Google Scholar]
- 18.Driouich A, Zhang G F, Staehelin L A. Plant Physiol. 1993;101:1363–1373. doi: 10.1104/pp.101.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G F, Driouich A, Staehelin L A. J Cell Sci. 1993;21:819–831. doi: 10.1242/jcs.104.3.819. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G F, Staehelin L A. Plant Physiol. 1992;99:1070–1083. doi: 10.1104/pp.99.3.1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore P J, Swords K M M, Lynch N A, Staehelin L A. J Cell Biol. 1991;112:589–602. doi: 10.1083/jcb.112.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothschild A, Tandecarz J S. Plant Sci. 1994;97:119–127. [Google Scholar]
- 23.Dry I, Smith A, Edwards A, Bhattacharyya M, Dunn P, Martin C. Plant J. 1992;2:193–202. [PubMed] [Google Scholar]
- 24.Marshall J, Sidebottom C, Debet M, Martin C, Smith A M, Edwards A. Plant Cell. 1996;8:1121–1135. doi: 10.1105/tpc.8.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sherrier D J, VandenBosch K A. Plant J. 1994;5:185–195. doi: 10.1104/pp.104.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Driouich A, Levy S, Staehelin L A, Faye L. Plant Physiol Biochem. 1994;32:731–749. [Google Scholar]
- 27.Faye L, Johnson K D, Sturm A, Chrispeels M J. Physiol Plant. 1989;75:309–314. [Google Scholar]
- 28.Muñoz P, Norambuena L, Orellana A. Plant Physiol. 1996;112:1585–1594. doi: 10.1104/pp.112.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]