Abstract
Objective
In a previous study, we showed that activation of a transfected human erythropoietin receptor (EPOR) in the murine myeloid cell line 32D resulted in the development of morphologic features of granulocytic differentiation and expression of the neutrophil primary granule protein myeloperoxidase. We now studied if EPOR signaling could also mediate secondary granule protein gene expression and investigated the signal transduction requirements for induction of secondary granule gene expression in 32D cells.
Materials and Methods
Wild-type and variant 32D cells expressing normal or chimeric EPORs or receptors for granulocyte colony-stimulating factor (G-CSFRs) were stimulated with EPO or G-CSF and the expression of granulocyte-specific genes was analyzed by Northern blot analysis. To determine the signaling mechanisms required for secondary granule protein gene induction, the activation of STAT pathways following growth factor stimulation was studied by Western blot analysis.
Results
We found that EPO treatment of 32D cells engineered to express EPOR did not result in induction of the secondary granule protein genes encoding lactoferrin and 24p3 lipocalin, the mouse homolog of human N-Gal, or the myeloid transcription factor C/EBPε. Replacement of the intracellular domain of EPOR with the intracellular domain of G-CSFR in a chimeric receptor was associated with EPO-mediated induction of lactoferrin, 24p3 lipocalin, and C/EBPε genes. We found that STAT3 phosphorylation was mediated by the intracellular domain of G-CSFR, but not EPOR. Replacement of one or two of the STAT5 binding sites in the intracytoplasmic domain of the EPOR with STAT3 binding sites resulted in EPO-mediated STAT3 activation and a marked increase in the expression of the 24p3 lipocalin gene. Knockdown of STAT3 protein levels with siRNA caused significant decrease in 24p3 lipocalin gene induction.
Conclusion
These results indicate that EPOR signaling cannot substitute for G-CSFR signaling to stimulate secondary granule protein gene expression in 32D cells. In addition, STAT3 is a critical mediator of 24p3 lipocalin gene expression in these cells.
Granulocyte colony-stimulating factor (G-CSF), through the interaction with its receptor (G-CSFR), is the major hematopoietic growth factor regulating the production of neutrophils. The importance of G-CSF in the regulation of granulopoiesis has been underscored by the observation that mice deficient in the G-CSF or G-CSFR gene, or mice expressing a chimeric G-CSFR/EPOR (erythropoietin receptor), developed severe neutropenia [1–3]. The neutrophils from mice with the chimeric receptor demonstrated reduced chemotaxis and reduced mobilization from the bone marrow to peripheral blood, suggesting that signals mediated by the cytoplasmic domain of EPOR were incapable of completely replacing G-CSFR function. In addition, transgenic mice expressing a truncated murine G-CSFR displayed impaired neutrophilic maturation [4].
These studies and others, including those of different hematopoietic growth factors and receptors, have led to two different theories or models regarding the role of specific growth factors and their receptors in the process of lineage commitment and differentiation: the “instructive” or deterministic model, in which growth factors play a direct role in lineage-specific commitment and differentiation, and the “permissive,” stochastic, or cell-autonomous model, in which growth factors provide the necessary signals for cell proliferation, survival, and maturation in cells already predetermined to differentiate along a given pathway (reviewed in [5–8]).
The binding of G-CSF to its receptor results in tyrosine phosphorylation of bound Janus tyrosine kinases (JAKs), that then activate multiple downstream signaling pathways [9]. The JAK/STAT pathway has been proposed to play a critical role in the control of myeloid proliferation and differentiation [10]. S TAT proteins belong to a family of interactive cytoplasmic transcription factors that, following activation of the appropriate receptor, become tyrosine phosphorylated by JAK family protein tyrosine kinases, undergo dimerization, and translocate to the nucleus to activate gene transcription. Many cytokines and growth factors can activate STAT signaling pathways [11] and at least seven STATs have been identified that are differentially activated by distinct receptors [12]. G-CSF activates STAT3 and to a lesser degree STAT1 and STAT5 [9,13–15]. The relative contribution of these different STATs to G-CSF-dependent neutrophil differentiation has been debated [16–18].
Several transcription factors, such as PU.1 and members of the CCAAT/enhancer-binding protein (C/EBP) family, play key roles in the differentiation of multipotent hematopoietic stem cells to lineage-committed myeloid progenitor cells and their subsequent terminal differentiation. C/EBPα is expressed in early myeloid progenitors and plays a pivotal role in the granulocytic lineage, likely through regulating the promoters of a number of important granulocytic genes, including those encoding the G-CSFR and the primary granule protein myeloperoxidase (MPO) [19–21]. C/EBPε is upregulated at the promyelocyte and myelocyte stages of granulocyte maturation and continues to be expressed thereafter. It plays an important role in mid to late stages of granulocytic differentiation [reviewed in 20].
The function of mature neutrophils is dependent on its granules, which contain characteristic proteins. Two major granules, primary and secondary (specific) granules, are formed at different stages of granulocytic maturation. Primary granules contain several proteolytic enzymes and bactericidal proteins, including cathepsin G, elastase, MPO, and lysozyme. The secondary granules contain a wide variety of different components, including lactoferrin (LF), lysozyme, collagenase, gelatinase, and gelatinase-associated lipocalin (N-Gal). Granule protein gene expression is regulated by a number of transcription factors. Among these factors, PU.1 and C/EBPα are important for the expression of all granule protein genes [22–25], whereas C/EBPε is important for the expression of secondary granule protein genes, such as those encoding LF, neutrophil gelatinase, and neutrophil collagenase [20,26–28].
The growth and differentiation of hematopoietic cells along the erythroid lineage is regulated by the lineage-specific cytokine EPO and its receptor EPOR [29–31]. G-CSFR and EPOR belong to the same superfamily of hematopoietic growth factor receptors and share many common features in their associated signal transduction pathways. In a prior study, we demonstrated that ectopic expression of the EPOR in myeloid 32D cells was associated with EPO-mediated development of myeloid differentiation characteristics including morphological features and expression of the primary granule protein MPO [32]. These results were consistent with those of other studies [33], suggesting that the lineage specificity of a cytokine-associated response can be mediated by activation of noncanonical receptors and/or signaling pathways [reviewed in 5–8,34]. Because the function of mature neutrophils depends on both its primary and secondary granule proteins, we examined if EPOR-mediated signaling could result in increased secondary granule protein gene expression in 32D cells. We found that EPOR-mediated signals could not replace those associated with G-CSFR activation to direct the expression of two different secondary granule protein genes in these cells. In addition, we observed that STAT3 activation, which is mediated by the G-CSFR but not by the EPOR, is essential in 32D cells for cytokine-induced expression of the gene encoding 24p3 lipocalin, the murine ortholog of the human secondary granule protein neutrophil gelatinase-associated lipocalin (N-Gal).
Materials and methods
Cell lines and culture conditions
All cell lines used in this study were derived from murine 32D cells, a factor-dependent myeloid cell line. Cell line 32DEPOR-WT and 32DEPOR-FE express wild-type and C-terminal truncated human EPOR, respectively [32]. Cell lines WT EPOR, ER343-S3, and ER343/401-S3 express, respectively, the wild-type murine EPOR and mutant murine EPORs with modified residues at positions 342–346 and 400–404, changing STAT5 binding sites to STAT3 binding sites [35]. Cell line 32Dwt18 (a gift of Dr. Arati Khanna-Gupta, Yale University, New Haven, CT, USA) was generated in Dr. Daniel Link’s laboratory, Washington University (St. Louis, MO, USA), and expresses a chimeric receptor (extracellular and transmembrane domains of murine EPOR and intracellular domain of human G-CSFR). Because 32D cells are interleukin (IL)-3-dependent myeloid progenitor cells, all of these 32D-derived cell lines were maintained in IMDM medium with 10% heat-inactivated fetal calf serum (FCS), 5% conditioned medium from the WEHI 3B cell line (as a source of IL-3), and 400 μg/mL of G418 for maintaining selection of cell lines containing transfected expression plasmids.
EPO stimulation, preparation of whole-cell lysates, sodium dodecyl sulfate–polyacrylamide gel electrophoresis, and Western blot analysis
Cells were washed with phosphate-buffered saline (PBS) buffer, cultured in IMDM +1% FCS without growth factor, then cultured at 37°C in the presence of added EPO, at the concentrations listed in the text or figure legends, for the indicated periods of time. The culture was terminated by adding an equal volume of Laemmli sample buffer (Bio-Rad Laboratories Inc., Hercules, CA, USA). After boiling for 5 minutes, lysates from 2 × 105 cells were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and electrophoretically transferred to nitrocellulose filters (Millipore Corporation, Burlington, MA, USA). The membranes were blocked in TBST (10 mM Tris-HCl, pH 8, 150 mM NaCl, 0.1% Tween-20) containing 5% milk for 1 hour at room temperature, then incubated with primary antibodies, anti-phospho-STAT3, anti-phospho-STAT5 (Cell Signaling Technology, Beverly, MA, USA), or anti-phospho-JAK2 (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA). The bound antibodies were detected by incubation with horseradish peroxidase–conjugated secondary antibodies and visualized with the ECL detection system (Amersham Biosciences Corp., Piscataway, NJ, USA). Probes were removed from the membranes by incubating with stripping buffer (62.5 mM Tris-HCl, pH 6.7, 2% SDS and 100 mM β-mercaptoethanol) at 50°C for 30 minutes and reprobed with anti-STAT5 (Cell Signaling Technology), anti-STAT3, or anti-JAK2 (Santa Cruz Biotechnology Inc.).
Northern blot analysis
Cells were washed with PBS, transferred to medium containing 2 units/mL of EPO, and harvested at the indicated times. Total RNA was extracted from cells using RNA mini kit (Qiagen Inc., Valencia, CA, USA). RNA (10μg) was fractionated by electrophoresis in 1% agarose gels, transferred to nitrocellulose filters, then probed with mouse MPO cDNA, lactoferrin cDNA, mouse 24p3 lipocalin cDNA, human C/EBPε cDNA, or mouse β-actin cDNA (gifts of Dr. Nancy Berliner), labeled with 32P using Ready-To-Go DNA labeling beads (-dCTP) (Amersham Biosciences). The hybridization was carried out in PerfectHyb Plus hybridization buffer (Sigma-Aldrich, St. Louis, MO, USA) at 68°C for 2 hours. The membranes were then rinsed in washing buffer containing 2× standard saline citrate (SSC) (1× SSC contains 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS for 10 minutes with two changes of buffer at room temperature, followed by washing with a solution of 0.5× SSC +0.1% SDS at 68 °C for 10 minutes. The membranes were exposed using a PhosphorImaging screen and analyzed by PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA).
SiRNA preparation and transfection
All siRNAs were synthesized by Qiagen, Inc. The STAT3 siRNA target sequence is AAGGACATCAGTGGCAAGACC. Ten μg of STAT3 siRNA or control siRNA were used to transfect ER343-S3 and 32Dwt18 cells by electroporation. Cells were grown in medium containing IL-3 and washed twice with PBS buffer. One ×107 cells were suspended in 180 μL of 2× HEPES buffer (0.28 M NaCl, 0.05 M HEPES [(N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid]), 1.5 mM Na2HPO4), mixed with siRNA, transferred to cuvettes, and subjected to electroporation at a voltage of 400 mV and capacitance of 250 uF. After transfection, cells were incubated in IMDM +10% FCS +5% WEHI-conditioned medium for 5 hours, washed with PBS, and transferred to the IMDM medium containing 10%FCS and 2units/mL of EPO. Twenty-four hours after transfection, cells were harvested and STAT3 protein levels were determined by Western blot analyses as described above, using anti-STAT3 antibody and anti-actin antibody (Santa Cruz Biotechnology Inc.). 24p3 lipocalin gene expression was assessed by Northern blot analysis, as described above.
Results
EpoR-mediated signaling in 32D cells results in expression of the gene encoding the primary granule protein MPO but not the expression of two different genes encoding secondary granule proteins
To examine primary and secondary granule protein gene expression in 32D cells, we utilized three different 32D-derived cell lines: 32DEPOR-WT, 32DEPOR-FE, and 32Dwt18. The cell lines 32DEPOR-WT and 32DEPOR-FE express, respectively, the full-length and C-terminal truncated human EPOR [36]. The 32Dwt18 cell line expresses a chimeric receptor that consists of the extracellular and transmembrane domains of the murine EPOR and the intra-cellular domain of the human G-CSFR. This chimeric receptor is activated by EPO, but results in G-CSFR signaling. Therefore, 32Dwt18 cells can differentiate into neutrophils following EPO stimulation [37], and were used as a positive control for neutrophilic differentiation. The cell line 32DEPOR-FE has been reported to be incapable of myeloid differentiation after EPO stimulation due to truncation of its C-terminal domain [32], and was used as a negative control. To determine whether EPOR signaling can mediate the expression of primary and secondary granule protein genes in 32D, the three cell lines were incubated in medium containing 2 units/mL of EPO. Cells were harvested at different times and their RNA isolated for Northern blot analysis (Fig. 1). The mRNA for MPO, a primary granule protein, was significantly increased after EPO stimulation in 32DEPOR-WT and 32Dwt18 cells, but not in 32DEPOR-FE cells (Fig. 1A). This result is consistent with our previous study [32], indicating that signaling mediated by the full-length EPOR can increase primary granule protein gene expression in 32D cells in a manner similar to G-CSFR-mediated signaling.
Figure 1.
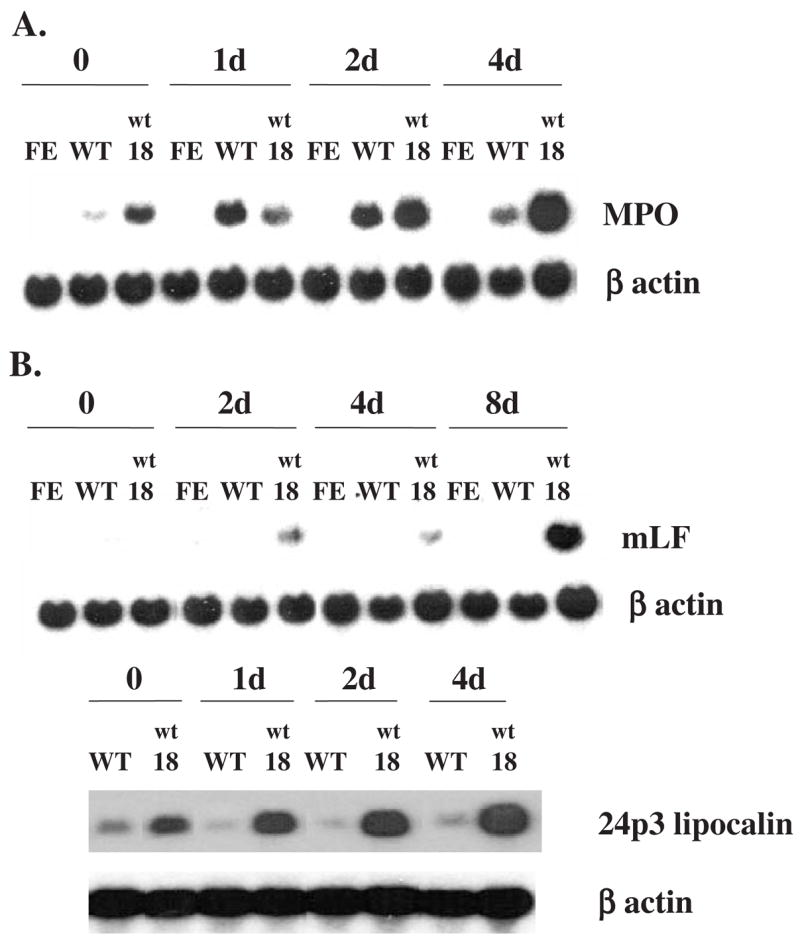
Expression of the mRNAs encoding the primary granule protein myeloperoxidase and secondary granule proteins lactoferrin and 24p3 lipocalin in 32D-derived cell lines after EPO stimulation. The cells were cultured in medium containing 2 units/mL of EPO for the indicated times. Ten ug of total cellular RNA was analyzed by Northern blot hybridization, using as probes 32P-labeled murine cDNAs encoding MPO (A), L F or 24p3 lipocalin (B), and β-actin as a control for the amount of sample. FE: 32DEPOR-FE cells, expressing a C-terminal truncated human EPOR; WT: 32DEPOR-WT cells, expressing wild-type human EPOR; wt18: 32Dwt18 cells, expressing a chimeric EPOR/G-CSFR (see Materials and methods).
With regard to secondary granule protein gene expression, LF mRNA was increased after EPO stimulation in 32Dwt18 cells expressing the EPOR/G-CSFR chimeric receptor, but not in 32DEPOR-WT or 32DEPOR-FE cells (Fig. 1B), indicating that EPOR-mediated signaling, unlike G-CSFR-mediated signaling, does not result in increased LF gene expression in 32D cells. We also examined the expression in these cells of the gene encoding 24p3 lipocalin, the murine ortholog of human neutrophil gelatinase-associated lipocalin (N-Gal), a secondary granule protein. In humans, N-Gal is synthesized in myelocytes and metamyelocytes and packaged into secondary granules [38]. In mice, 24p3 lipocalin has also been detected in granulocytes prepared from peripheral blood [39]. The results (Fig. 1B) showed that 24p3 lipocalin mRNA was increased after exposure to EPO in 32Dwt18 cells expressing the G-CSFR intracellular signaling domain, but not in 32DEPOR-WT cells expressing the wild-type human EPOR. These results demonstrate that EPOR-mediated signaling cannot replace G-CSFR-mediated signaling to increase the expression in 32D cells of the genes encoding the secondary granule proteins LF and 24p3 lipocalin.
EPOR-mediated signaling failed to increase the expression of the gene encoding CCAAT/enhancer-binding protein epsilon (C/EBPε)
The transcription factor C/EBPα appears to be required for the early stages of myeloid differentiation, whereas C/EBPε appears to be necessary for the later stages of myeloid differentiation, including expression of the secondary granule proteins genes [28]. To understand why EPOR-mediated signaling does not result in increased secondary granule protein gene expression, we examined whether the mRNAs for C/EBPα and C/EBPε are increased in the different 32D-derived cell lines following exposure to EPO (Fig. 2). C/EBPα mRNA levels were slightly increased in 32Dwt18 cells after exposure to EPO, but were unchanged in 32DEPOR-WT and 32DEPOR-FE cells before and after exposure to EPO. Therefore, the level of C/EBPα is unlikely to be the key factor responsible for the difference in secondary granule protein gene expression between 32Dwt18 and the 32DEPOR cells following EPO stimulation. The level of C/EBPα mRNA was consistently much lower in 32Dwt18 cells than in the other 32D-derived cell lines. The significance of this finding is unclear. It may represent genetic drift between different isolates of 32D cells obtained from different laboratories. Different 32D cell isolates may also represent myeloid lineage cells arrested (at baseline) at slightly different stages of maturation. Similar but quantitatively less significant variations of basal levels of other mRNAs were also observed between the different 32D-derived cell lines (Fig. 1 and Fig. 2). Unlike C/EBPα mRNA, C/EBPε mRNA increased in 32Dwt18 cells, but not in 32DEPOR-WT and 32DEPOR-FE cells after exposure to EPO, indicating that EPOR-mediated signaling failed to stimulate C/EBPε gene expression. The lack of increased secondary granule protein gene expression in 32DEPOR cells may be due to the low and unchanged levels in these cells of C/EBPε, which appears to be required for secondary granule protein expression.
Figure 2.
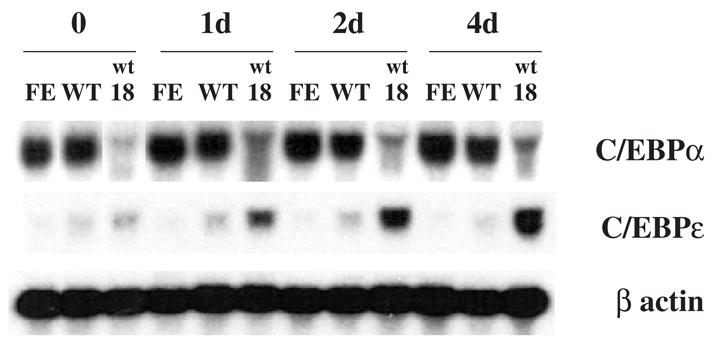
Expression of C/EBPα and C/EBPε mRNA in 32D-derived cell lines after EPO stimulation. Cells were cultured in medium containing 2 units/mL of EPO for the indicated times. Northern blot analysis was carried out as in Figure 1, using as probes 32P-labeled murine cDNAs encoding C/EBPα, C/EBPε, or β-actin. Cell line designations as in Figure 1.
EPOR-mediated signaling is not capable of activating STAT3 in 32D cells
Because EPOR-mediated signaling results in increased expression of the gene encoding the primary granule protein MPO, but not of the genes encoding secondary granule proteins in 32D cells, we used these 32D-derived cell lines as a model system to investigate the signals that are required for secondary granule protein gene expression. Because STAT proteins, particularly STAT3 and STAT5, are critical in myeloid cell differentiation and survival, we examined the patterns of activation of STAT proteins by EPOR- and G-CSFR-mediated signaling in the different 32D-derived cell lines. Figure 3 shows the pattern of tyrosine phosphorylation of STAT proteins in 32DEPOR-WT, 32DEPOR-FE, and 32Dwt18 cells following EPO stimulation. STAT5 was phosphorylated in all three cell lines 5 minutes after EPO stimulation. More significantly, STAT3 was only phosphorylated in 32Dwt18 cells capable of G-CSFR signaling, but not in the two different 32DEPOR cell lines after exposure to EPO, indicating that EPOR-mediated signaling cannot activate STAT3 in 32D cells. Therefore, STAT3 is likely to be required for secondary granule protein gene expression in these cells.
Figure 3.
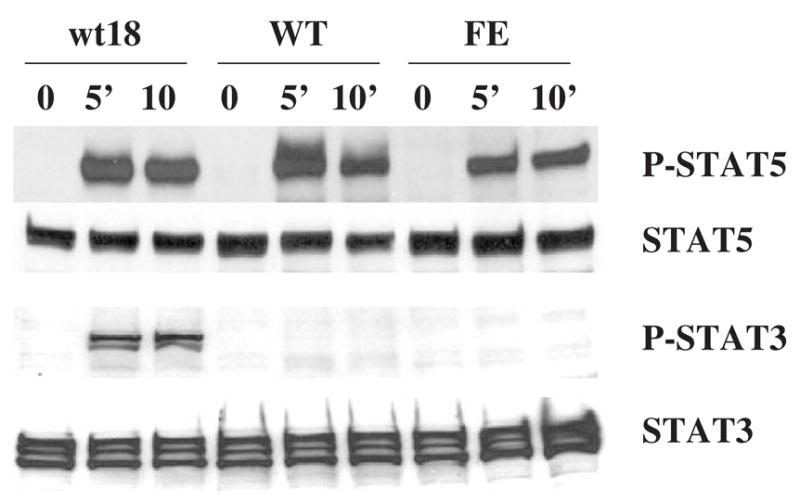
Induced phosphorylation of Stat3 and Stat5 in 32D-derived cell lines after EPO stimulation. Cells were cultured in medium containing 10 units/mL of EPO for the indicated times. Whole-cell lysates were fractionated by SDS-PAGE and analyzed by Western blotting using anti-phospho-Stat3 or anti-phospho-Stat5 antibody. The membranes were then stripped and reprobed with anti-STAT3 or anti-STAT5 antibody. Cell line designations as in Figure 1.
STAT3 phosphorylation mediated by variant EPO receptors containing altered STAT binding sites
To investigate whether STAT3 is a key component mediating increased secondary granule protein gene expression, we utilized 32D cell lines expressing the murine wild-type EPOR (WT EPOR cells) or variant EPOR isoforms that were utilized in a prior study [35] (Fig. 4). The ER343-S3 cell line expresses an altered murine EPOR in which the STAT5 binding site at residues 342–346 was replaced by a STAT3 binding site. The pattern of EPO-dependent JAK/STAT protein tyrosine phosphorylation in ER343-S3 cells was analyzed (Fig. 5). Following EPO stimulation, STAT5 was phosphorylated in WT EPOR cells and, at a lower level, in ER343-S3 cells whose EPOR still contains one of the two STAT5 binding sites. As expected, the alteration of EPOR in ER343-S3 cells caused STAT3 phosphorylation following EPO stimulation, but this did not occur in WT EPOR cells (Fig. 5A). Moreover, the degree of STAT3 phosphorylation in ER343-S3 cells was virtually the same as that in 32Dwt18 cells capable of G-CSFR-mediated signaling after EPO stimulation.
Figure 4.
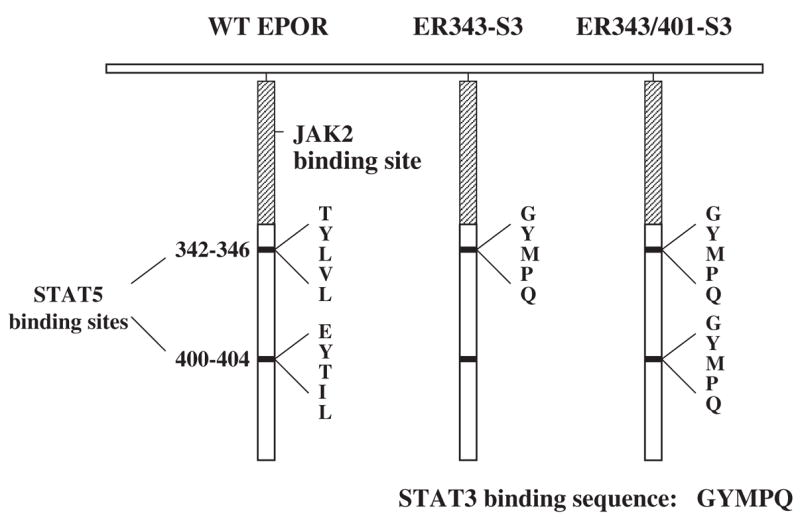
Schematic diagram of the structure of the murine EPOR expressed in cell lines WT EPOR, ER343-S3, and ER343/401-S3. The cytoplasmic region of the murine wild-type (WT) EPOR is diagrammed on the left, showing the position of the JAK2 binding domain (cross-hatched bar) and the regions surrounding the tyrosine residues required for STAT5 binding and activation (Tyr-343 and Tyr-401). ER343-S3 has the sequence GYMPQ (STAT3 binding site) substituted for residues 342–346 of the WT murine EPOR. ER343/401-S3 has the STAT3 binding site substituted at residues 342–346 and 400–404 of the WT murine EPOR. Adapted from Wooten et al. [35].
Figure 5.
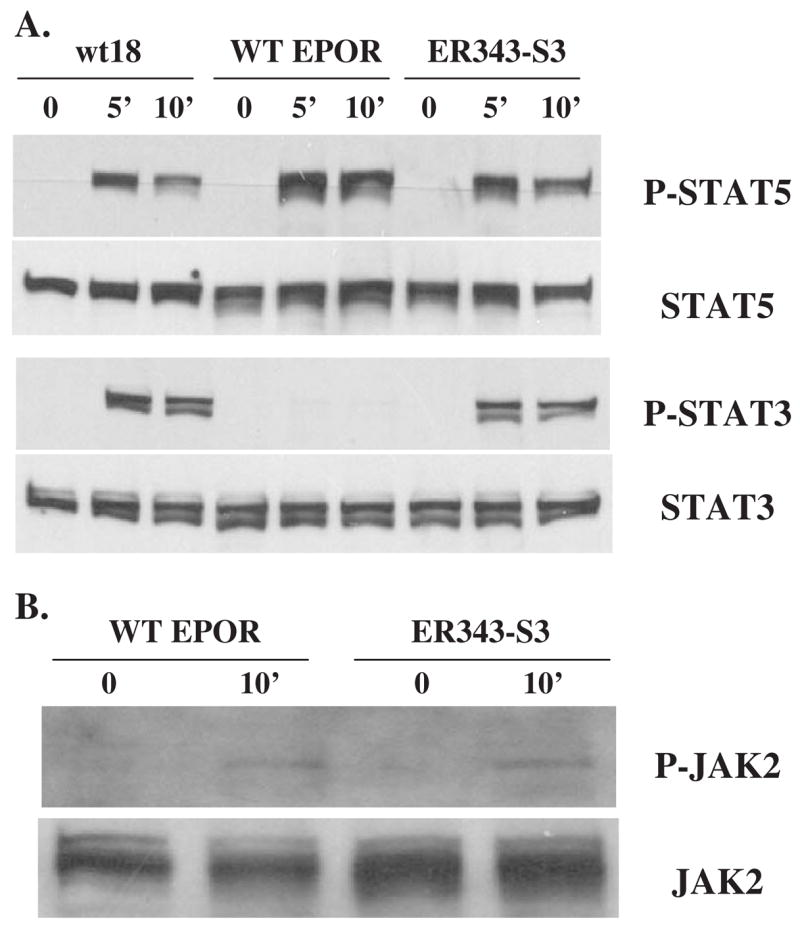
Pattern of Stat3, Stat5, and JAK2 phosphorylation in 32D-derived cells after EPO stimulation. Western blot analysis was carried out as described in Figure 3. Cell line designations as in Figure 1 (wt18) and Figure 4 (WT EPOR and ER343-S3).
STATs are phosphorylated by the JAK2 kinase, which is bound to the EPOR and is activated (autophosphorylated) after ligand binding to the receptor [40,41]. The level of EPO-dependent JAK2 tyrosine phosphorylation was similar in WT EPOR and ER343-S3 cells (Fig. 5B). This result is consistent with previous results [35], indicating that the mutation of the STAT5 binding site does not alter activation of the EPOR and the resulting JAK2 autophosphorylation after exposure to EPO. STAT5 can be activated through non–tyrosine-containing sequences in the membrane proximal region of the EPOR cytoplasmic domain [42].
STAT3 activation mediates cytokine-dependent expression of the gene encoding 24p3 lipocalin
To determine the effect of mutant EPOR-mediated STAT3 activation on secondary granule protein gene expression in 32D cells, WT EPOR and ER343-S3 cells were grown in medium containing 2 units/mL of EPO. Cells were harvested at the indicated times and Northern blots were performed (Fig. 6).
Figure 6.
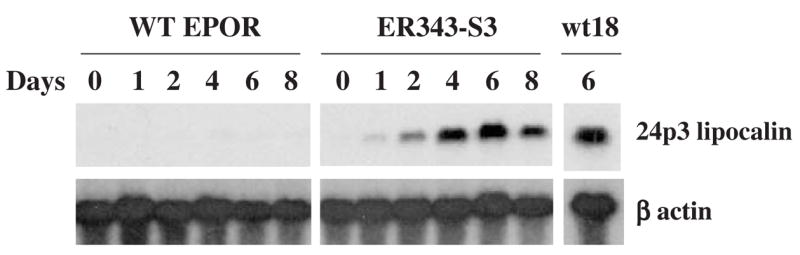
Gene expression of secondary granule protein 24p3 lipocalin in 32D-derived cell lines after EPO stimulation. Northern blot analysis was carried out as described in Figure 1. Cell line designations as in Figure 4.
Expression of 24p3 lipocalin mRNA was markedly increased in ER343-S3 cells after EPO stimulation, whereas no increase of this mRNA occurred in WT EPOR cells (Fig. 6). Moreover, the level of 24p3 lipocalin mRNA in ER343-S3 cells after 6 days of EPO stimulation was approximately the same as that in 32Dwt18 cells, suggesting that STAT3 plays an important role in cytokine-dependent 24p3 lipocalin gene expression.
To further determine if STAT3 signaling is required for the observed increase in 24p3 lipocalin gene expression in 32D cells, we knocked down STAT3 activity by use of STAT3 siRNA. ER343-S3 cells and 32Dwt18 cells were transfected with STAT3 siRNA or control siRNA using electroporation. After transfection, cells were cultured in medium containing IL-3 for 5 hours, then were washed and transferred to medium containing 2 units/mL EPO. After 24 hours, cells were harvested for extraction of protein and total RNA. Western blot analysis of lysates obtained at this time point showed that STAT3 protein levels were reduced by 50% and 70% in 32Dwt18 and ER343-S3 cells, respectively, compared to that in cells transfected with control siRNA (Fig. 7A). The levels of 24p3 lipocalin mRNA following EPO stimulation in cells transfected with STAT3 siRNA were also proportionally reduced, compared to that in the cells transfected with control siRNA (Fig. 7B). Therefore these results indicate that STAT3 signaling is critical for G-CSFR- and variant EPOR-mediated increases in 24p3 lipocalin gene expression.
Figure 7.
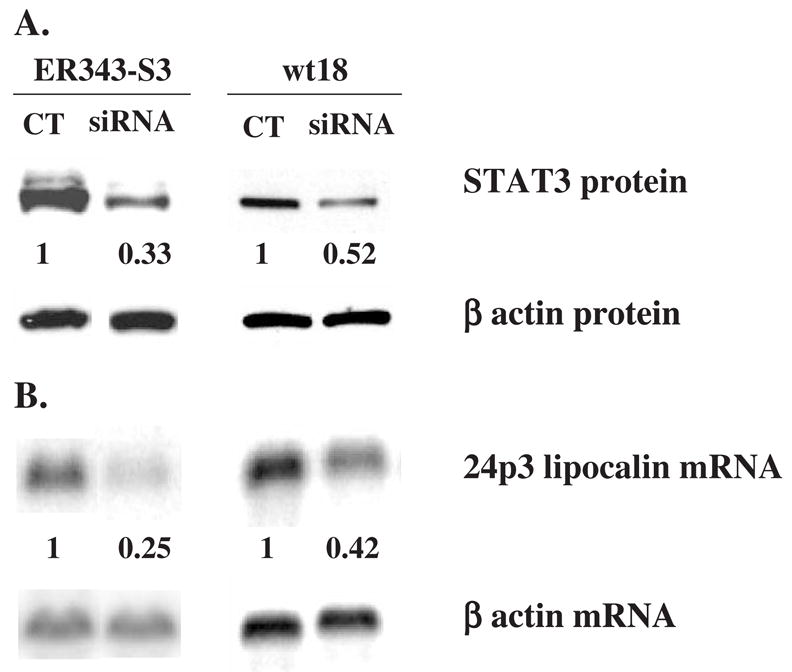
Expression levels of STAT3 protein and 24p3 lipocalin mRNA in EPO-stimulated ER343-S3 and 32Dwt18 cells after transfection with siRNA. Cells were transfected with siRNA by electroporation, cultured in medium containing IL-3 for 5 hours, then washed and transferred to medium containing 2 units/mL of EPO and cultured for an additional 19 hours. Western blot analysis (A) and Northern blot analysis (B) were carried out as described in Figure 3 and Figure 1, respectively. Quantitation of STAT3 protein and 24p3 lipocalin mRNA was performed by PhosphorImager using the ImageQuant software program (Molecular Dynamics) and the average values of the siRNA/control (CT) ratios from at least two separate experiments are shown below the corresponding STAT3 and 24p3 lipocalin images. Cell line designations as in Figure 1 (wt18) and Figure 4 (ER343-S3).
We also examined if 24p3 lipocalin gene expression requires STAT5 activation, in addition to STAT3 activation. The two STAT5 binding sites of EPOR located at residues 342–346 and 400–404 were substituted by two STAT3 binding sites in ER343/401-S3 cells (Fig. 4; Ref. 35). As expected, the level of STAT5 phosphorylation after EPO stimulation was reduced in ER343-S3 and ER343/401-S3 cells compared to that in WT EPOR cells (Fig. 8A). However, there was still a low level of STAT5 phosphorylation even in ER343/401-S3 cells, possibly due to conservation of tyrosine residues 429 and 431 of the EPOR that are capable of binding STAT5 [43–45]. The EPO-dependent phosphorylation level of STAT3 in ER343/401-S3 cells was double that in ER343-S3 cells. The level of EPO-stimulated 24p3 lipocalin gene expression was also increased in ER343/401-S3 cells compared to that in ER343-S3 cells (Fig. 8B). These results suggest that the level of 24p3 lipocalin gene expression correlates with the degree of activation of STAT3, but not with that of STAT5.
Figure 8.
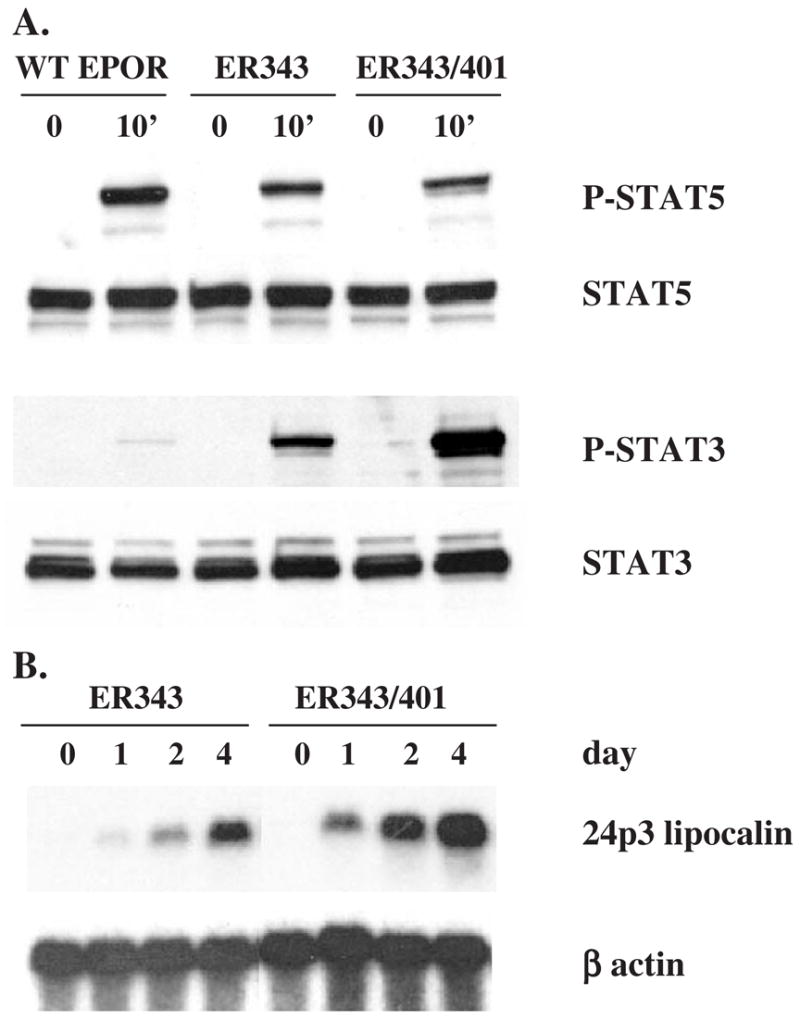
Phosphorylation of STAT3 and STAT5 and expression of 24p3 lipocalin mRNA in 32D-derived cell lines after EPO stimulation. Western blot analysis (A) was carried out as described in Figure 3 and Northern blot analysis (B) was carried out as described in Figure 1. Cell line designations as in Figure 4.
To explore whether STAT3 directly or indirectly activates 24p3 lipocalin gene expression, ER343/401 cells were cultured in medium containing 2 units/mL of EPO in the presence or absence of the protein synthesis inhibitor cycloheximide (CHX), or in the presence of CHX alone. At the indicated times, cells were harvested and Northern blot analysis of their RNA was performed (Fig. 9). After EPO stimulation, 24p3 lipocalin mRNA levels gradually increased in the absence of CHX. The addition of CHX did not inhibit the EPO-stimulated 24p3 lipocalin gene expression, suggesting that STAT3 directly activates 24p3 lipocalin gene expression.
Figure 9.
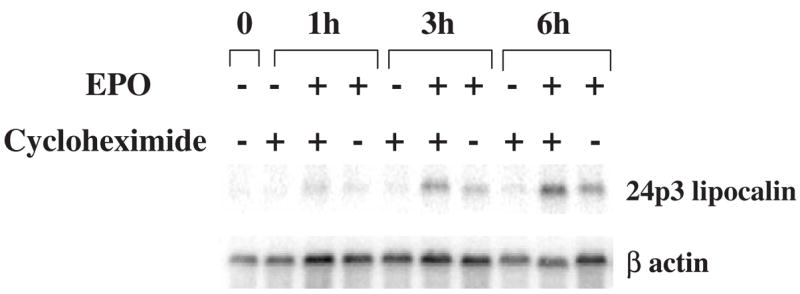
EPO-induced 24p3 lipocalin mRNA expression in ER343/401-S3 cells in the presence or absence of the protein synthesis inhibitor cyclo-heximide (CHX). ER343/401-S3 cells (Fig. 4) were cultured medium containing IL-3, then washed and transferred to medium containing 5 μg/L of CHX, 2 units/mL of EPO, or both EPO + CHX. Cells were harvested at the indicated times, and their total RNA extracted for Northern blot analysis, carried out as described in Figure 1.
Retinoic acid enhances STAT3-mediated 24p3 lipocalin gene expression
Retinoic acid (RA) has been reported to enhance 24p3 lipocalin gene expression in mouse L cells [46]. It is also known that RA plays an important role during myeloid differentiation [47]. Therefore, we examined if RA promotes 24p3 lipocalin gene expression during neutrophilic differentiation of 32D cells. WT EPOR and ER343-S3 cells were incubated in medium containing 2 units/mL of EPO in the presence or absence of 10 μM all-trans retinoic acid (ATRA). Cells were harvested at the indicated times and Northern blot analysis of their RNA was performed (Fig. 10). In WT EPOR cells, 24p3 lipocalin mRNA levels were not increased in the presence of EPO alone, as previously observed in Figure 6, and the addition of ATRA together with EPO also failed to increase 24p3 lipocalin gene expression, indicating that ATRA-mediated signals do not stimulate 24p3 lipocalin gene expression in 32D cells. In ER343-S3 cells, 24p3 lipocalin gene expression was upregulated after exposure to EPO, and the addition of ATRA together with EPO resulted in even higher levels of 24p3 lipocalin mRNA (Fig. 10). When ER343-S3 cells were cultured in medium containing ATRA and IL-3, 24p3 lipocalin gene expression was not increased (data not shown). Taken together, these results indicate that ATRA-mediated signaling enhances the level of 24p3 lipocalin mRNA that is stimulated by STAT3 activation (mediated by the variant EPOR), but it does not play an important role by itself in the activation of 24p3 lipocalin gene expression.
Figure 10.
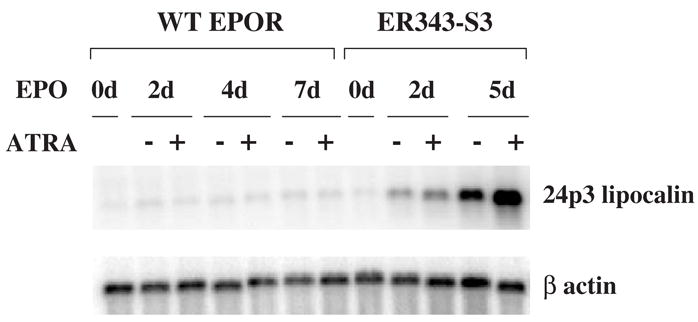
Expression of 24p3 lipocalin mRNA in EPOR WT and ER343-S3 cells after stimulation by EPO in the presence or absence of 10 μM ATRA. EPO stimulation and Northern blot analysis were carried out as described in Figure 9. Cell line designations as in Figure 4.
Discussion
The EPOR and G-CSFR are members of the hematopoietic growth factor receptor superfamily and share a number of common features in their downstream signaling pathways. Jacob et al. [48] demonstrated that bone marrow cells of G-CSFR-deficient mice expressing a retrovirally transduced G-CSFR/EPOR chimeric receptor transgene could differentiate in vitro into morphologically mature, chloroacetate esterase–positive neutrophils expressing the Gr-1 and Mac-1 cell surface markers. However, secondary granule protein gene expression was not studied in these cells. In a previous study [32], we found that ectopic expression of full-length EPOR in myeloid 32D cells led to EPO-dependent granulocytic differentiation, as assessed by morphologic features and expression of the primary granule protein MPO. In the current study, we investigated whether EPO-EPOR signaling can also induce secondary granule protein gene expression that is characteristic of terminal granulocytic differentiation. Our data demonstrated that EPOR-mediated signaling could not replace G-CSFR signaling to stimulate the expression of the secondary granule protein genes lactoferrin and 24p3 lipocalin.
Our findings in 32D cells contrast with those we previously obtained using the promyelocytic EPRO cell line that contains a dominant negative RARα and is capable of myeloid differentiation in the presence of ATRA combined with GM-CSF [49]. In EPRO cells ectopically expressing the EPOR, treatment of the cells with EPO combined with ATRA did result in secondary granule gene expression. Interestingly, in contrast to this finding in EPRO cells, we found that activation of EPOR signaling in 32D cells failed to induce the expression of C/EBP-ε, a myeloid transcription factor that mediates G-CSF-induced granulocytic differentiation [28]. The discrepancy in results is probably due to the fact that these two cell models of myeloid maturation differ in the mechanisms by which myeloid differentiation is induced. As demonstrated by Maun et al. [50], ATRA-mediated differentiation of EPRO cells appears to be STAT3-independent, in contrast to G-CSF-mediated differentiation of G-CSFR-transduced EPRO cells, which is STAT3-dependent and RAR-independent. In our prior study using EPOR-expressing EPRO cells, ATRA plus EPO-mediated myeloid differentiation is likely to have been RAR-dependent but STAT3-independent.
To determine which signaling pathways are involved in induction of secondary granule protein gene expression, we studied the differences in downstream pathways activated by EPOR signaling and G-CSFR signaling in 32D cells. We found that STAT3 was activated by G-CSFR signaling, but not by EPOR signaling, suggesting that STAT3 activation may be required for secondary granule protein gene expression. To test this hypothesis, the two STAT5 binding sites in EPOR were replaced with a STAT3 binding site to produce ER343-S3 cells, allowing the activation of STAT3 upon EPO stimulation. In these cells, granulocytic differentiation and increased expression of the secondary granule protein 24p3 lipocalin were observed following exposure to EPO. This phenomenon was associated with only a minor increase in the expression of C/EBPε (data not shown). It is also noteworthy that lactoferrin mRNA levels also increased slightly in ER343-S3 cells following EPO stimulation (data not shown), but this increase was quantitatively much less significant than that observed with 24p3 lipocalin mRNA. This finding is consistent with the fact that C/EBPε plays an important role in lactoferrin gene expression and its expression did not show a major increase in ER343-S3 cells after EPO stimulation. Although it would appear that 24p3 lipocalin gene expression is STAT3-dependent and C/EBPε-independent, STAT3 binding sites were not identified by nucleotide homology search in the promoter of the gene.
Human N-Gal is a secondary granule bacteriostatic protein that is expressed at the myelocyte/metamylocyte stage of neutrophil maturation [51–53]. The mouse homolog of N-Gal is 24p3 lipocalin, which is expressed in neutrophils as well as in other tissues, such as the uterus, stomach, and lung [38,39]. The factors that regulate N-Gal/24p3 lipocalin gene expression in neutrophils are not known. In this study, we provided the following evidence that G-CSFR signaling stimulates the expression of the 24p3 lipocalin gene through STAT3: 1) 24p3 lipocalin mRNA levels were markedly increased by G-CSFR signaling in 32Dwt18 cells; 2) activation of STAT3 by a modified EPOR in ER343-S3 cells was associated with increased 24p3 lipocalin gene expression; and 3) knockdown of STAT3 protein levels by STAT3 siRNA in ER343-S3 and 32Dwt18 cells was associated with decreased 24p3 lipocalin gene expression. Our results also suggested that STAT3 may directly activate 24p3 lipocalin gene expression, because the protein synthesis inhibitor cycloheximide did not inhibit STAT3-mediated 24p3 lipocalin gene expression.
Retinoic acid plays a role during granulocyte differentiation through interaction with its nuclear receptor, RARα, which in turn modulates the expression of multiple downstream targets by binding to retinoic acid response elements (RAREs) in cellular DNA. The important role of the RA signaling pathway is reflected by the involvement of RARα in the pathogenesis of acute promyelocytic leukemia (APL). It is likely that RARα controls granulopoiesis through several critical regulators of myeloid differentiation, including the C/EBPs, c-myc, and the cyclin-dependent kinase inhibitor p21 [47,54]. The relationship between the RA signaling pathway and G-CSFR-activated pathways during myeloid differentiation may be cooperative, because the combination of retinoic acid and G-CSF can synergistically drive differentiation of RA-resistant t(11;17) promyelocytic leukemia cells toward complete granulocyte differentiation [55–58]. We examined the role of RA in the regulation of expression of the 24p3 lipocalin gene in 32D cells. Our results revealed that RA signaling alone failed to stimulate 24p3 lipocalin gene expression in these cells, but in combination with STAT3 activation (in ER343-S3 cells), it significantly enhanced the level of 24p3 lipocalin mRNA compared to that obtained with STAT3 activation alone (Fig. 10). This finding suggests that RA signaling and G-CSFR signaling may act in concert during granulocyte differentiation.
In summary, we have demonstrated that EPOR-mediated signaling cannot substitute for G-CSFR signaling to stimulate granulocyte secondary granule protein gene expression in 32D cells. In addition, activation of STAT3 is required for induction of 24p3 lipocalin gene expression in these cells. These studies were carried out using established tissue culture cell lines rather than primary myeloid cells. It will be important to confirm these results in future studies using primary myeloid cells, including cells from STAT3-deficient mice.
Acknowledgments
We sincerely thank Drs. Arati Khanna-Gupta and Nancy Berliner for helpful discussions and for providing some of the materials used in these studies. L. Lozavotsky provided skilled technical assistance. This work was support in part by grants K08-DK02566, T32-HL07262, R01-CA77447, and R01-DK19482 from the National Institutes of Health, and grant 0255899Y of the American Heart Association, Texas Affiliate.
References
- 1.Lieschke GJ, Grail D, Hodgson G, et al. Mice lacking granulocyte colony-stimulating factor have chronic neutropenia, granulocyte and macrophage progenitor cell deficiency, and impaired neutrophil mobilization. Blood. 1994;84:1737–1746. [PubMed] [Google Scholar]
- 2.Liu F, Wu HY, Wesselschmidt R, Kornaga T, Link DC. Impaired production and increased apoptosis of neutrophils in granulocyte colony-stimulating factor receptor–deficient mice. Immunity. 1996;5:491–501. doi: 10.1016/s1074-7613(00)80504-x. [DOI] [PubMed] [Google Scholar]
- 3.Semerad CL, Poursine-Laurent J, Liu F, Link DC. A role for G-CSF receptor signaling in the regulation of hematopoietic cell function but not lineage commitment or differentiation. Immunity. 1999;11:153–161. doi: 10.1016/s1074-7613(00)80090-4. [DOI] [PubMed] [Google Scholar]
- 4.Mitsui T, Watanabe S, Taniguchi Y, et al. Impaired neutrophil maturation in truncated murine G-CSF receptor–transgenic mice. Blood. 2003;101:2990–2995. doi: 10.1182/blood.V101.8.2990. [DOI] [PubMed] [Google Scholar]
- 5.D’Andrea AD. Hematopoietic growth factors and the regulation of differentiative decisions. Curr Opin Cell Biol. 1994;6:804–808. doi: 10.1016/0955-0674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 6.Metcalf D. Lineage commitment and maturation in hematopoietic cells: The case for extrinsic regulation. Blood. 1998;92:345–348. [PubMed] [Google Scholar]
- 7.Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92:348–351. [PubMed] [Google Scholar]
- 8.Socolovsky M, Lodish HF, Daley GQ. Control of hematopoietic differentiation: Lack of specificity in signaling by cytokine receptors. Proc Natl Acad Sci U S A. 1998;95:6573–6575. doi: 10.1073/pnas.95.12.6573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avalos BR. Molecular analysis of the granulocyte colony-stimulating factor receptor. Blood. 1996;88:761–777. [PubMed] [Google Scholar]
- 10.Smithgall TE, Briggs SD, Schreiner S, Lerner EC, Cheng H, Wilson MB. Control of myeloid differentiation and survival by Stats. Oncogene. 2000;19:2612–2618. doi: 10.1038/sj.onc.1203477. [DOI] [PubMed] [Google Scholar]
- 11.Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 12.Ihle JN. The Stat family in cytokine signaling. Curr Opin Cell Biol. 2001;13:211–217. doi: 10.1016/s0955-0674(00)00199-x. [DOI] [PubMed] [Google Scholar]
- 13.Tian SS, Lamb P, Seidel HM, Stein RB, Rosen J. Rapid activation of the STAT3 transcription factor by granulocyte colony-stimulating factor. Blood. 1994;84:1760–1764. [PubMed] [Google Scholar]
- 14.Chakraborty A, White SM, Schaefer TS, Ball ED, Dyer KF, Tweardy DJ. Granulocyte colony-stimulating factor activation of Stat3 alpha and Stat3 beta in immature normal and leukemic human myeloid cells. Blood. 1996;88:2442–2449. [PubMed] [Google Scholar]
- 15.Ward AC, Smith L, de Koning JP, van Aesch Y, Touw IP. Multiple signals mediate proliferation, differentiation, and survival from the granulocyte colony-stimulating factor receptor in myeloid 32D cells. J Biol Chem. 1999;274:14956–14962. doi: 10.1074/jbc.274.21.14956. [DOI] [PubMed] [Google Scholar]
- 16.McLemore ML, Grewal S, Liu F, et al. STAT-3 activation is required for normal G-CSF-dependent proliferation and granulocytic differentiation. Immunity. 2001;14:193–204. doi: 10.1016/s1074-7613(01)00101-7. [DOI] [PubMed] [Google Scholar]
- 17.Lee CK, Raz R, Gimeno R, et al. STAT3 is a negative regulator of granulopoiesis but is not required for G-CSF-dependent differentiation. Immunity. 2002;17:63–72. doi: 10.1016/s1074-7613(02)00336-9. [DOI] [PubMed] [Google Scholar]
- 18.Ilaria RL, Jr, Hawley RG, Van Etten RA. Dominant negative mutants implicate STAT5 in myeloid cell proliferation and neutrophil differentiation. Blood. 1999;93:4154–4166. [PubMed] [Google Scholar]
- 19.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha–deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borregaard N, Theilgaard-Monch K, Sorensen OE, Cowland JB. Regulation of human neutrophil granule protein expression. Curr Opin Hematol. 2001;8:23–27. doi: 10.1097/00062752-200101000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21:3295–3313. doi: 10.1038/sj.onc.1205318. [DOI] [PubMed] [Google Scholar]
- 22.Tenen DG, Hromas R, Licht J, Zhang D-E. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:489–519. [PubMed] [Google Scholar]
- 23.McKercher SR, Henkel GW, Maki RA. The transcription factor PU.1 does not regulate lineage commitment but has lineage-specific effects. J Leukoc Biol. 1999;66:727–732. doi: 10.1002/jlb.66.5.727. [DOI] [PubMed] [Google Scholar]
- 24.Flodby P, Barlow C, Kylefjord H, Ahrlund-Richter L, Xanthopoulos KG. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J Biol Chem. 1996;271:24753–24760. doi: 10.1074/jbc.271.40.24753. [DOI] [PubMed] [Google Scholar]
- 25.Wang ND, Finegold MJ, Bradley A, et al. Impaired energy homeostasis in C/EBP alpha knockout mice. Science. 1995;269:1108–1112. doi: 10.1126/science.7652557. [DOI] [PubMed] [Google Scholar]
- 26.Yamanaka R, Barlow C, Lekstrom-Himes J, et al. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon–deficient mice. Proc Natl Acad Sci U S A. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khanna-Gupta A, Zibello T, Sun H, Lekstrom-Himes J, Berliner N. C/EBP epsilon mediates myeloid differentiation and is regulated by the CCAAT displacement protein (CDP/cut) Proc Natl Acad Sci U S A. 2001;98:8000–8005. doi: 10.1073/pnas.141229598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakajima H, Ihle JN. Granulocyte colony-stimulating factor regulates myeloid differentiation through CCAAT/enhancer-binding protein epsilon. Blood. 2001;98:897–905. doi: 10.1182/blood.v98.4.897. [DOI] [PubMed] [Google Scholar]
- 29.Krantz SB. Erythropoietin. Blood. 1991;77:419–434. [PubMed] [Google Scholar]
- 30.D’Andrea AD, Lodish HF, Wong GG. Expression cloning of the murine erythropoietin receptor. Cell. 1989;57:277–285. doi: 10.1016/0092-8674(89)90965-3. [DOI] [PubMed] [Google Scholar]
- 31.Youssoufian H, Longmore G, Neumann D, Yoshimura A, Lodish HF. Structure, function, and activation of the erythropoietin receptor. Blood. 1993;81:2223–2236. [PubMed] [Google Scholar]
- 32.Harris KW, Hu XJ, Schultz S, Arcasoy MO, Forget BG, Clare N. The distal cytoplasmic domain of the erythropoietin receptor induces granulocytic differentiation in 32D cells. Blood. 1998;92:1219–1224. [PubMed] [Google Scholar]
- 33.Stoffel R, Ziegler S, Ghilardi N, Ledermann B, DeSauvage FJ, Skoda RC. Permissive role of thrombopoietin and granulocyte colony-stimulating factor receptors in hematopoietic cell fate decisions in vivo. Proc Natl Acad Sci U S A. 1999;96:698–702. doi: 10.1073/pnas.96.2.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ihle JN. Cytokine receptor signalling. Nature. 1995;377:591–594. doi: 10.1038/377591a0. [DOI] [PubMed] [Google Scholar]
- 35.Wooten DK, Xie X, Bartos D, Busche RA, Longmore GD, Watowich SS. Cytokine signaling through Stat3 activates integrins, promotes adhesion, and induces growth arrest in the myeloid cell line 32D. J Biol Chem. 2000;275:26566–26575. doi: 10.1074/jbc.M003495200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arcasoy MO, Degar BA, Harris KW, Forget BG. Familial erythrocytosis associated with a short deletion in the erythropoietin receptor gene. Blood. 1997;89:4628–4635. [PubMed] [Google Scholar]
- 37.Khanna-Gupta A, Zibello T, Kolla S, Neufeld EJ, Berliner N. CCAAT displacement protein (CDP/cut) recognizes a silencer element within the lactoferrin gene promoter. Blood. 1997;90:2784–2795. [PubMed] [Google Scholar]
- 38.Kjeldsen L, Cowland JB, Borregaard N. Human neutrophil gelatinase-associated lipocalin and homologous proteins in rat and mouse. Biochim Biophys Acta. 2000;1482:272–283. doi: 10.1016/s0167-4838(00)00152-7. [DOI] [PubMed] [Google Scholar]
- 39.Huang HL, Chu ST, Chen YH. Ovarian steroids regulate 24p3 expression in mouse uterus during the natural estrous cycle and the preimplantation period. J Endocrinol. 1999;162:11–19. doi: 10.1677/joe.0.1620011. [DOI] [PubMed] [Google Scholar]
- 40.Watowich SS, Yoshimura A, Longmore GD, Hilton DJ, Yoshimura Y, Lodish HF. Homodimerization and constitutive activation of the erythropoietin receptor. Proc Natl Acad Sci U S A. 1992;89:2140–2144. doi: 10.1073/pnas.89.6.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witthuhn BA, Quelle FW, Silvennoinen O, et al. JAK2 associates with the erythropoietin receptor and is tyrosine phosphorylated and activated following stimulation with erythropoietin. Cell. 1993;74:227–236. doi: 10.1016/0092-8674(93)90414-l. [DOI] [PubMed] [Google Scholar]
- 42.Yoon D, Watowich SS. Hematopoietic cell survival signals are elicited through non–tyrosine-containing sequences in the membrane-proximal region of the erythropoietin receptor (EPOR) by a Stat5-dependent pathway. Exp Hematol. 2003;31:1310–1316. doi: 10.1016/j.exphem.2003.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Damen JE, Wakao H, Miyajima A, et al. Tyrosine 343 in the erythropoietin receptor positively regulates erythropoietin-induced cell proliferation and Stat5 activation. EMBO J. 1995;14:5557–5568. doi: 10.1002/j.1460-2075.1995.tb00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gobert S, Chretien S, Gouilleux F, et al. Identification of tyrosine residues within the intracellular domain of the erythropoietin receptor crucial for STAT5 activation. EMBO J. 1996;15:2434–2441. [PMC free article] [PubMed] [Google Scholar]
- 45.Klingmuller U, Bergelson S, Hsiao JG, Lodish HF. Multiple tyrosine residues in the cytosolic domain of the erythropoietin receptor promote activation of STAT5. Proc Natl Acad Sci U S A. 1996;93:8324–8328. doi: 10.1073/pnas.93.16.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garay-Rojas E, Harper M, Hraba-Renevey S, Kress M. An apparent autocrine mechanism amplifies the dexamethasone- and retinoic acid–induced expression of mouse lipocalin-encoding gene 24p3. Gene. 1996;170:173–180. doi: 10.1016/0378-1119(95)00896-9. [DOI] [PubMed] [Google Scholar]
- 47.Gaines P, Berliner N. Retinoids in myelopoiesis. J Biol Regul Homeost Agents. 2003;17:46–65. [PubMed] [Google Scholar]
- 48.Jacob J, Haug JS, Raptis S, Link DC. Specific signals generated by the cytoplasmic domain of the granulocyte colony-stimulating factor (G-CSF) receptor are not required for G-CSF-dependent granulocytic differentiation. Blood. 1998;92:353–361. [PubMed] [Google Scholar]
- 49.Arcasoy MO, Maun NA, Perez L, Forget BG, Berliner N. Erythropoietin mediates terminal granulocytic differentiation of committed myeloid cells with ectopic erythropoietin receptor expression. Eur J Haematol. 2001;67:77–87. [PubMed] [Google Scholar]
- 50.Maun N, Gaines P, Khanna-Gupta A, et al. G-CSF signaling can differentiate promyelocytes expressing a defective retinoic acid receptor: evidence for divergent pathways regulating neutrophil differentiation. Blood. 2004;103:1693–1701. doi: 10.1182/blood-2002-10-3247. [DOI] [PubMed] [Google Scholar]
- 51.Borregaard N, Sehested M, Nielsen BS, Sengelov H, Kjeldsen L. Biosynthesis of granule proteins in normal human bone marrow cells. Gelatinase is a marker of terminal neutrophil differentiation. Blood. 1995;85:812–817. [PubMed] [Google Scholar]
- 52.Kjeldsen L, Bainton DF, Sengelov H, Borregaard N. Identification of neutrophil gelatinase-associated lipocalin as a novel matrix protein of specific granules in human neutrophils. Blood. 1994;83:799–807. [PubMed] [Google Scholar]
- 53.Goetz DH, Holmes MA, Borregaard N, Bluhm ME, Raymond KN, Strong RK. The neutrophil lipocalin NGAL is a bacteriostatic agent that interferes with siderophore-mediated iron acquisition. Mol Cell. 2002;10:1033–1043. doi: 10.1016/s1097-2765(02)00708-6. [DOI] [PubMed] [Google Scholar]
- 54.Kastner P, Chan S. Function of RARα during the maturation of neutrophils. Oncogene. 2001;20:7178–7185. doi: 10.1038/sj.onc.1204757. [DOI] [PubMed] [Google Scholar]
- 55.Nakamaki T, Sakashita A, Sano M, et al. Granulocyte-colony stimulating factor and retinoic acid cooperatively induce granulocyte differentiation of acute promyelocytic leukemia cells in vitro. Jpn J Cancer Res. 1989;80:1077–1082. doi: 10.1111/j.1349-7006.1989.tb02262.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang F, Zhao HP, Gao XZ, Dai MM, Fan LL. Recombinant human G-CSF and retinoic acid in synergistically inducing granulocyte differentiation of human promyelocytic leukemic cells. Chin Med J (Engl) 1992;105:707–712. [PubMed] [Google Scholar]
- 57.Tsurumi H, Tojo A, Takahashi T, Moriwaki H, Asano S, Muto Y. The combined effects of all-trans retinoic acid and granulocyte colony-stimulating factor as a differentiation induction therapy for acute pro-myelocytic leukemia. Intern Med. 1993;32:648–650. doi: 10.2169/internalmedicine.32.648. [DOI] [PubMed] [Google Scholar]
- 58.Jansen JH, de Ridder MC, Geertsma WM, et al. Complete remission of t(11;17) positive acute promyelocytic leukemia induced by all-trans retinoic acid and granulocyte colony-stimulating factor. Blood. 1999;94:39–45. [PubMed] [Google Scholar]


